Abstract
In haploid Saccharomyces cerevisiae, the mating and invasive growth (IG) pathways use the same mitogen-activated protein kinase kinase kinase kinase (MAPKKKK, Ste20), MAPKKK (Ste11), MAPKK (Ste7), and transcription factor (Ste12) to promote either G1 arrest and fusion or foraging in response to distinct stimuli. This exquisite specificity is the result of pathway-specific receptors, G proteins, scaffold protein, and MAPKs. It is currently not thought that the shared signaling components function under the basal conditions of vegetative growth. We tested this hypothesis by searching for mutations that cause lethality when the STE11 gene is deleted. Strikingly, we found that Ste11, together with Ste20, Ste7, Ste12, and the IG MAPK Kss1, functions in a third pathway that promotes vegetative growth and is essential in an och1 mutant that does not synthesize mannoproteins. We term this pathway the STE vegetative growth (SVG) pathway. The SVG pathway functions, in part, to promote cell wall integrity in parallel with the protein kinase C pathway. During vegetative growth, the SVG pathway is inhibited by the mating MAPK Fus3. By contrast, the SVG pathway is constitutively activated in an och1 mutant, suggesting that it senses intracellular changes arising from the loss of mannoproteins. We predict that general proliferative functions may also exist for other MAPK cascades thought only to perform specialized functions.
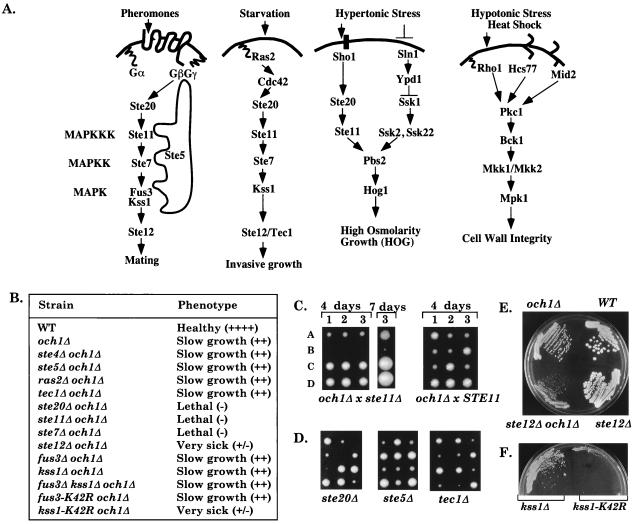
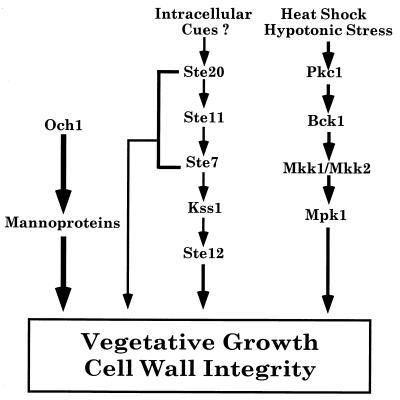
All eukaryotic cells use multiple mitogen-activated protein kinase (MAPK) cascades to respond to external signals to regulate their growth, differentiation, and level of stress (1–3). In Saccharomyces cerevisiae haploid cells, four MAPK cascades respond to different external signals to mediate specialized responses (Fig. 1A) (1, 4, 5). The mating pathway is activated by peptide pheromones and induces cell-cycle arrest and morphological changes required for mating. The invasive growth (IG) pathway is activated by starvation and induces foraging into agar. The high osmolarity glycerol (HOG) pathway increases intracellular glycerol levels in response to hypertonic stress, whereas the protein kinase C (PKC) pathway maintains cell wall integrity and is activated by hypotonic stress and heat shock.
Figure 1.
Summary of genetic interactions with och1Δ. (A) Four MAPK cascades regulate growth and differentiation in haploid S. cerevisiae. (B) Summary of genetic interactions. (C) ste11Δ is synthetically lethal with och1Δ. (Left) Tetrads from DBL60 (STE11/ste11Δ OCH1/och1Δ) dissected onto a YPD plate and germinated at 25°C for 4 days. Ascospores 1A–3A are och1Δ single mutants, 1B–3B are ste11Δ och1Δ double mutants. After 7 days, ascospore 3B generates a microcolony of cells that fail to survive. (Right) Tetrads from DBL52 (OCH1/och1Δ). (D) Tetrads from OCH1/och1Δ diploids heterozygous for the indicated mutation. (E) Streakout of a tetrad from a ste12Δ/STE12 och1Δ/OCH1 diploid on a YPD plate. (F) Streakout (on SC-his) of kss1Δ sls1Δ (BLY443) containing vector control (pRS313) or pKSS1K42R-HIS3.
Surprisingly, subsets of the same kinases are used by the mating, IG, and HOG pathways, even though each pathway has different outputs. The MAPKKKK Ste20 and MAPKKK Ste11 are shared by the mating, IG, and HOG pathways, whereas the MAPKK Ste7 and MAPK Kss1 are shared by the mating and IG pathways (1, 5). Pathway specificity is thought to arise through two major levels of regulation. First, pathway-specific upstream components route the different signals, and pathway-specific MAPKs and transcription factors mediate the different outputs. For example, the pheromone receptors, G protein, Ste5, and Fus3 are specificity factors for the mating pathway, whereas Ras2, Kss1, and the transcription factor Tec1 are specificity factors for the IG pathway (1, 6). Second, the pathways cross-regulate one another to ensure that they are not activated by the wrong signal. For example, Fus3 inhibits misactivation of the IG pathway by mating pheromone (7), whereas Hog1 inhibits misactivation of the mating pathway by hypertonic stress (5, 8).
The analysis of MAPK cascades in yeast and mammalian cells has centered on activities that are induced by a particular external stimulus. Although it is known that the PKC-regulated MAPK cascade has a basal vegetative function that maintains cell wall integrity in the absence of an external stimulus (9), it is currently thought that Ste11 and the downstream kinases in the mating and IG pathways perform specialized functions only in response to their respective stimuli. Here, we show that Ste11 functions under vegetative conditions in a pathway related to the IG pathway that shares functional redundancy with the PKC pathway.
Materials and Methods
Media, Plasmids, and Strains.
Yeast extract/peptone/dextrose (YPD) and synthetic complete (SC) media with 2% dextrose were prepared as described (ref. 10, p. 15). Plasmids made for this study are as follows: (i) pBL1, a 4.2-kb BamHI–XhoI STE11 fragment from pJD11 (D. Jenness, University of Massachusetts Medical Center, Worcester) in BamHI–SalI sites of pRS313 (P. Hieter, University of British Columbia, Vancouver), (ii) pBL2, a 4.2-kb BamHI–XhoI STE11 fragment from pJD11 blunt-ended into the SmaI site of pCH1122 (J. Kranz, Brandeis University, Waltham, MA), (iii) pBL7, an original OCH1-URA3-CEN isolate, (iv) pBL12, a 2.6-kb SalI–OCH1 fragment from pBL7 in pRS315, (v) pBL20, a 4.2-kb BamHI–XhoI STE11 fragment from pBL2 in BamHI–XhoI sites of pBL15 (LEU2-ADE3-CEN). Yeast strains are isogenic to W303a (ADE3: EY698, EY699; ade3: PY1181, PY1183) or BLY39 [MATa ura3–1 leu2–3,112 ade2–1 ade3–1 his3 can1–100 IR+; made from a cross between PY1181 and L5784 (11)]. All deletion derivatives were made with published deletion plasmids. och1Δ was made with pBL-och1Δ∷LEU2 (12), which deletes residues 221–951. Details of all strains and plasmids can be obtained from B.N.L. and E.A.E.
Mutant Hunt and Cloning SLS1/OCH1.
MATa and MATα W303-derived and MATa Sigma-derived ste11Δ ade2 ade3 ura3 strains (BLY33, BLY42, BLY39) harboring pBL2 were mutagenized with ethyl methanesulfonate to a 25–45% survival rate, as described (ref. 10, pp. 277–278), and screened for nonsectoring red colonies on YPD plates, as described (13). sls1–1 (synthetically lethal with ste11Δ) was found among 200,000 BLY33 and BLY42 colonies. sls1–2 was found among 80,000 BLY39 colonies. Four plasmids with inserts overlapping OCH1 were cloned by restoring sectoring to a sls1–1 ste11Δ strain harboring pBL20 (BLY213). URA3 segregated in opposition to sls1 in tetrads derived from crosses between a strain with URA3 integrated into the OCH1 locus (BLY314) and each sls1 mutant (BLY216, BLY122), demonstrating linkage between sls1 and OCH1.
Cell Morphology.
Cells were grown at 30°C to an OD600 of 0.5 in YPD containing 1 M sorbitol. Then they were fixed and stained for DNA with 4′,6-diamidino-2-phenylindole (DAPI), and stained for tubulin with YOL1/34 (Accurate Chemicals), as described (14).
Immunoblots.
Invertase was analyzed as described (15) with 5 μl of total protein extract and rabbit anti-invertase antiserum at a 1:1000 dilution (C. Kaiser, Massachusetts Institute of Technology, Cambridge). Kss1, Fus3, and Ste7 were analyzed as described (16). Ste7 was detected with anti-Ste7 rabbit antiserum (B. Cairns, University of Utah, Salt Lake City, UT; 1:10,000 dilution). Phosphorylated Kss1–HA3 and Fus3 were detected with anti-active MAPK Ab (Promega; 1:1500 dilution). Kss1–HA3 was detected with 12CA5 (Harvard University, Boston; 1:10,000 dilution). Tcm1 was detected with anti-Tcm1 mAb (J. Warner, Albert Einstein College of Medicine, Bronx, NY; 1:10,000 dilution). Immunoblots were developed with the enhanced chemiluminescence system (Amersham Pharmacia).
β-Galactosidase Activity.
Cells were grown to <4 × 106 cells per ml, normalized to equal density, and then grown for 3 hr to 2–4 × 106 cells per ml (OD600 = 0.5–0.6). Extracts were made from 20 ml of cells as described (17).
Double- and Triple-Mutant Constructions.
Genes were either deleted in an och1Δ/OCH1 diploid or introduced through crosses. All mutations were verified by Southern blot analysis, except ste mutations, which were verified by mating tests. Twelve or more tetrads were analyzed in each cross. All tetrads were dissected onto YPD plates, both in the absence and in the presence of osmotic support (0.3 M KCl, 1 M sorbitol, or 0.1 M sorbitol) and incubated at 25°C. Growth phenotypes were judged by ascospore germination and streaking of cells on YPD plates, with or without osmotic support. 0.1 M sorbitol was used for bck1, pbs2, and hog1 mutants to bypass problems of germination and recovery of suppressors for bck1 mutants without inhibiting the growth of pbs2 and hog1 mutants (5).
Results
STE11 Is Required for Vegetative Growth in och1 Mutants.
We performed a synthetic lethal screen to identify mutants that require Ste11 for survival. Two mutations were isolated that caused lethality in ste11 null mutants in both W303a and Sigma (IG+) backgrounds. Both mutations caused slow growth and temperature sensitivity at 37°C. Subsequent analysis showed the mutations were recessive and allelic to OCH1 (14), so we will refer to them as och1–1 and och1–2. STE11 was essential for the survival of an och1Δ deletion mutant (see Materials and Methods). och1Δ ste11Δ double-mutant progeny derived from an och1Δ/OCH1 ste11Δ/STE11 heterozygous diploid were inviable, although och1Δ single mutants could grow at permissive temperature (Fig. 1C). Approximately 90% of the och1Δ ste11Δ double mutants either failed to germinate or ceased dividing after one or two cell divisions, whereas ≈10% germinated and formed a microcolony of cells that could not be propagated, even in the presence of osmotic stabilizers that suppressed the och1 temperature-sensitive defect. Thus, STE11 is essential for vegetative growth in an och1 mutant.
STE20, STE7, and STE12 Are Required for Vegetative Growth in an och1 Mutant.
Remarkably, null mutations in the other signaling components that are shared by the mating and IG pathways also interfered with the growth of the och1Δ strain (Fig. 1B, representatives shown in D–E). ste20Δ and ste7Δ mutations caused synthetic lethality, whereas the ste12Δ mutation caused synthetic sickness. In sharp contrast, null mutations in the mating-specific components STE4 and STE5 and the IG-specific components RAS2 and TEC1 had no effect on the growth of the och1Δ strain. These results suggest that Ste20, Ste11, and Ste7 kinases regulate vegetative growth in a pathway that bifurcates at Ste7, with one branch involving activation of the Ste12 transcription factor. Because mating- and IG-specific components were not required for the survival of the och1Δ mutant, Ste20, Ste11, Ste7, and Ste12 must function in a distinct signal-transduction pathway during vegetative growth, which we will refer to as the STE (sterile genes) vegetative growth (SVG) pathway.
och1 Mutants Have Cell-Integrity and Cell-Division Defects.
To determine the function of the SVG pathway, we analyzed the phenotype of och1Δ mutants. OCH1 encodes an α-1,6-mannosyltransferase that initiates outer-chain elongation during N-linked glycosylation in the Golgi (12). Och1 is essential for the synthesis of cell wall mannan, in addition to other mannosylated proteins (12). Phenotypic analysis showed that och1 mutants had reduced cell wall integrity, consistent with the loss of mannan (data not shown). och1 mutants were hypersensitive to agents that interfere with cell wall integrity (i.e., calcofluor white, hygromycin B, and SDS), and more och1 cells appeared as lysed cell ghosts that stained with methylene blue. In addition, growth defects of och1 mutants were suppressed by osmotic stabilizers (1 M sorbitol and 0.3 M KCl) and overexpression of PKC (PKC1).
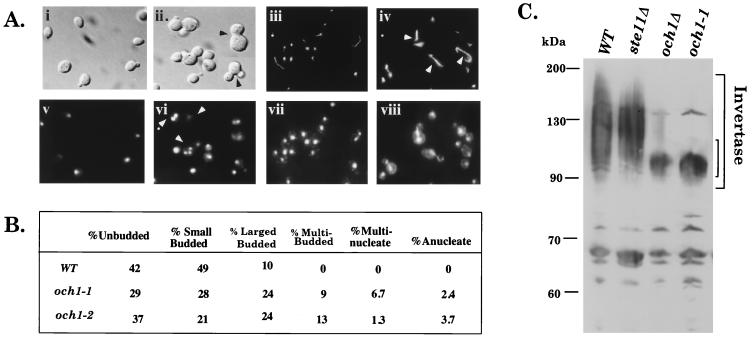
Additional analysis suggested that Och1 is required for other aspects of vegetative growth. och1 cells had cell-division defects, as shown by increased percentages of multibudded cells, large-budded cells, multinucleate and anucleate cells, as well as more elaborate microtubules (particularly in cells that failed to properly segregate their nuclei; Fig. 2 A and B). These defects were not rescued by osmotic stabilizers. och1 haploids also accumulated less glycogen than did wild-type haploids, and och1/och1 diploids sporulated at reduced efficiency (data not shown). Thus, och1 mutants had many defects, any of which could overlap with the SVG pathway.
Figure 2.
och1Δ mutants have cell division and glycosylation defects. (A) Morphology of och1 cells. Wild-type (PY1181) panels are i, iii, v, and vii. och1–1 (BLY77) panels are ii, iv, vi, and viii. (i and ii) Nomarski optics. (v and vi) Parallel DAPI staining. (iii and iv) Tubulin detected with YOL1/34. (vii and viii) Parallel DAPI staining. (B) Cell-cycle tally of wild-type and och1 cells. Strains are wild type (PY1181), och1–1 (BLY77), and och1–2 (BLY118). Similar results were found for och1Δ. (C) Immunoblot of invertase from wild type (PY1181), ste11Δ (BLY21), och1Δ (BLY315), and och1–1 (BLY77).
The SVG Pathway Regulates FKS2, a Cell Wall Gene.
Microscopic examination of the och1Δ ste11Δ double-mutant progeny that germinated did not provide clues as to the possible function of the SVG pathway because they resembled the och1Δ single mutant. In addition, Ste11 was not required for the transcription of either OCH1 or its homolog, HOC1 (18), during vegetative growth (data not shown), nor was it required for N-linked outer-chain elongation as assessed by the mobility of secreted invertase on SDS gels (Fig. 2C; the sharper and faster mobility of invertase from och1 mutants is because of the absence of outer-chain mannose).
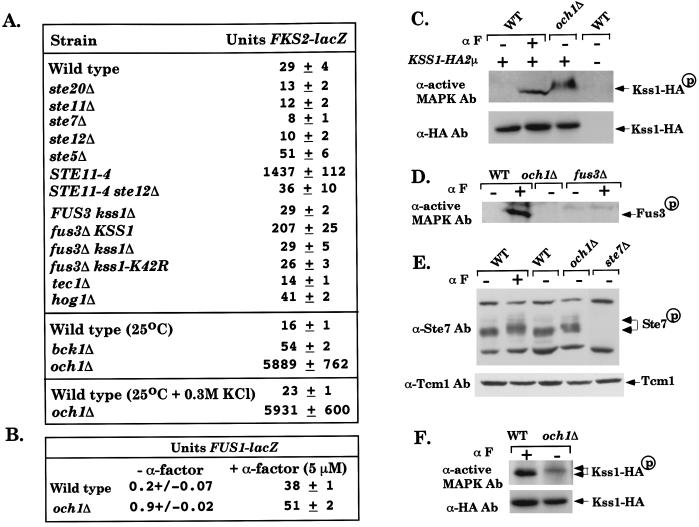
We tested whether the SVG pathway regulates cell wall integrity through transcriptional activation of genes required for the synthesis of either glucan or chitin, which form the cell wall with mannan (19). This possibility was suggested by the observation that osmotic support rescued the poor growth of an och1Δ ste12Δ double mutant (data not shown). Of several genes tested, FKS2 was found to be positively regulated by Ste11 during vegetative growth (Fig. 3A). FKS2 encodes a subunit of (1,3)-β-glucan synthase that functions redundantly with the more abundant Fks1 (1,3)-β-glucan synthase (20). FKS2 expression was reduced ≈2- to 3-fold in ste20Δ, ste11Δ, ste7Δ, and ste12Δ mutants, with no reduction in a ste5Δ mutant (Fig. 3A). Moreover, FKS2 expression increased 44-fold in a STE11–4 strain expressing a constitutively activated form of Ste11 (21), and this increase required STE12 (Fig. 3A). Thus, the SVG pathway operates during vegetative growth in wild-type cells and may play a role in regulating cell wall integrity. The SVG pathway regulates additional targets besides the FKS2 gene, because a fks2Δ mutation did not impair the growth of an och1Δ mutant (data not shown).
Figure 3.
The SVG pathway regulates FKS2 and is activated in an och1 mutant. (A) STE20, STE11, STE7, STE12, and TEC1 are required for FKS2 expression. Strains: PY1181 (WT), BLY409 (ste20Δ), BLY21 (ste11Δ), FP56 (ste7Δ), BLY380 (ste12Δ), BLY263 (ste5Δ), EY1298 (STE11–4 far1Δ), BLY426 (STE11–4 ste12Δ far1Δ), BLY405 (kss1Δ), BLY407 (fus3Δ), EY966 (fus3Δ kss1Δ), BLY535 (fus3Δ kss1Δ + pKSS1K42R-HIS3), BLY369 (tec1Δ), BLY457 (hog1Δ), C699-59 (bck1Δ), and BLY 341 (och1Δ). All strains contained FKS2-lacZ on a 2μ plasmid (pDM5) (33) and were grown in SC − uracil at 30°C or at 25°C, with or without 0.3 M KCl where indicated. β-Galactosidase activity (Miller units) was assayed in triplicate, and the average ± SD of three experiments is shown. (B) FUS1-lacZ expression is not induced in the och1Δ mutant. PY1181 (WT) and BLY470 (och1Δ) containing FUS1-lacZ on a CEN plasmid, induced for 120 min with α factor. (C) Kss1-HA3 is constitutively phosphorylated in the och1Δ mutant. Kss1-HA3 is on a 2μ plasmid (7). α F indicates α factor. The altered mobility of Kss1-HA3-P from the och1Δ strain is because of gel conditions (see F). (D) Fus3 is not phosphorylated in the och1Δ mutant. (E) Ste7 is hyperphosphorylated in an och1Δ mutant. Arrows indicate the portion of Ste7 that is shifted upward in lanes 2 and 4. (F) Mobility of phosphorylated Kss1-HA3 in OCH1 and och1Δ strains. Samples in C, D, and F are 50% saturated ammonium sulfate cuts of 2 mg of whole cell extract. Samples in E are 50 μg of whole cell extract.
Kss1 Is the SVG Pathway MAPK.
We next tested whether one or both of the MAPKs that regulate mating and IG function in the SVG pathway. MAPK dependency was tested by comparing the effects of FUS3/KSS1 null mutations with those of catalytically inactive kinases. Growth of the och1Δ mutant was not impaired by null mutations in FUS3/KSS1 or by a catalytically inactive form of Fus3 (encoded by fus3-K42R). In contrast, catalytically inactive forms of Kss1 (encoded by kss1-K42R, kss1-T183M, and kss1-Y185F) severely inhibited growth, as found for the ste12Δ mutation (Fig. 1 B and F, and data not shown). Thus, Kss1 functions in at least one branch of the SVG pathway, similar to the IG pathway.
FKS2 expression was elevated 7-fold in a fus3Δ mutant, indicating that Fus3 inhibits the SVG pathway during vegetative growth (Fig. 3A). By contrast, Kss1 positively regulated FKS2 expression, as shown by greatly reduced FKS2 expression in kss1Δ fus3Δ and kss1-K42R fus3Δ strains, compared with the KSS1fus3Δ strain. Inactive and unactivatable forms of Kss1 (encoded by kss1-K42R and kss1-Y185F) did not inhibit FKS2 expression below basal levels, suggesting that Kss1 regulates FKS2 through both positive and negative functions, as reported for IG genes (7, 22). We therefore tested whether basal expression of FKS2 requires the Tec1 transcription factor that dimerizes with Ste12 and is regulated by Kss1 in the IG pathway (7, 22, 23). FKS2 expression was reduced in a tec1Δ mutant to the same degree as in the ste12Δ mutant, suggesting that Kss1 regulates the FKS2 gene through effects on Ste12/Tec1.
The SVG Pathway Functionally Overlaps with the PKC Pathway.
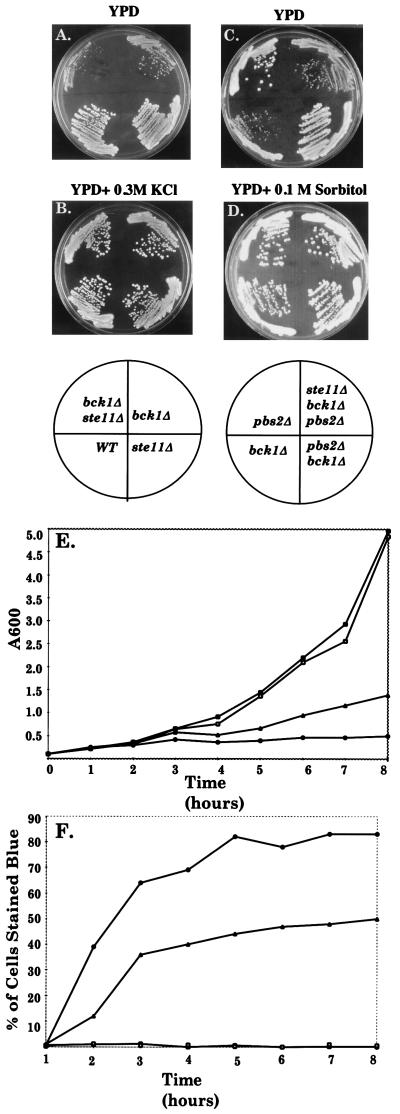
We next determined whether the SVG pathway is functionally related to the PKC pathway that is essential for cell wall integrity (9). Null mutations in either BCK1 (MAPKKK) or MPK1 (MAPK) of the PKC pathway (Fig. 1A) were lethal in an och1Δ mutant, and overexpression of PKC1 suppressed the poor growth of an och1–1 mutant (data not shown). In addition, simultaneous inactivation of the PKC and SVG pathways caused synthetic growth defects. A bck1Δ ste11Δ strain grew more slowly than a bck1Δ strain (Fig. 4 A and E), and its poor growth was suppressed by osmotic support (Fig. 4B). Moreover, a greater number of the bck1Δ ste11Δ cells lysed compared with the bck1Δ cells (Fig. 4F). Thus, SVG and PKC pathways provide additive functions that promote cell wall integrity in parallel with Och1. However, FKS2 expression was not reduced in a bck1Δ mutant, but rather was elevated 3-fold (Fig. 3A), arguing that the two pathways do not function identically.
Figure 4.
Genetic interactions between bck1, ste11, and pbs2 mutations. (A) ste11Δ inhibits the growth of a bck1Δ strain. (B) Rescue of a ste11Δ bck1Δ double mutant by 0.3 M KCl. (C) pbs2Δ suppresses a bck1Δ strain. (D) bck1Δ and bck1Δ pbs2Δ ste11Δ strains are rescued by 0.1 M sorbitol. A–D, Ascospore progeny derived from DBLY71 (BCK1/bck1Δ STE11/ste11Δ PBS2/pbs2Δ). Plates were incubated at 25°C. (E) ste11Δ reduces the growth rate of a bck1Δ strain. (F) ste11Δ increases lysis of a bck1Δ strain. (E and F) Strains PY1181 (STE11), BLY21 (ste11Δ), BLY480 (bck1Δ), and BLY478 (ste11Δ bck1Δ) were grown at 25°C in YPD with 1 M sorbitol medium to logarithmic phase, washed once with YPD medium, resuspended in YPD, and assayed for OD600 and methylene blue uptake (0.01% methylene blue in 0.2% sodium citrate and 0.1 M sorbitol). A total of 300 cells were counted for each time point. Strains are wild type (□), ste11Δ (■), bck1Δ (▴), and ste11Δ bck1Δ (●).
By contrast, the SVG pathway is functionally distinct from the HOG pathway, which also utilizes Ste20 and Ste11 (Fig. 1A). A pbs2Δ mutation conferred wild-type growth to the bck1Δ mutant instead of a growth defect (Fig. 4C; hog1Δ gave identical results), and this growth enhancement required Ste11 (compare pbs2Δ bck1Δ with pbs2Δ bck1Δ ste11Δ). In addition, FKS2 expression was not reduced in a hog1Δ mutant (Fig. 3A).
The SVG Pathway Is Constitutively Induced in the och1 Mutant.
We tested whether the SVG pathway is induced in the och1Δ mutant that requires it for survival. Remarkably, Kss1 was constitutively phosphorylated in the och1Δ mutant, as shown with an Ab that recognizes phosphorylated MAPKs (Fig. 3C, note that very low levels of phosphorylated Kss1 could occasionally be detected in the untreated wild-type cells). By contrast, the mating MAPK Fus3 was not phosphorylated in the och1Δ mutant (Fig. 3D) and was inactive, based on the absence of FUS1 gene induction (Fig. 3B). Kss1 phosphorylation correlated with hyperphosphorylation of Ste7 (Fig. 3E), a substrate known to be feedback-phosphorylated by active Fus3 and Kss1 (24), and a 200-fold induction of FKS2-lacZ (Fig. 3A). Moreover, FKS2-lacZ was equivalently induced in osmotic support conditions that suppressed the och1Δ growth defect (Fig. 3A). Collectively, these results suggest that the SVG pathway is activated as a result of the loss of mannoproteins.
Discussion
Our results lead us to conclude that Ste20, Ste11, Ste7, Kss1, and Ste12 promote normal vegetative growth in a distinct signal-transduction pathway we term SVG for STE vegetative growth (Fig. 5). The existence of this pathway is consistent with biochemical analysis showing that both Ste7 and Kss1 have basal kinase activity during vegetative growth (7, 24–26). Basal vegetative functions for these shared kinases provide an additional explanation for the requirement for specificity factors such as Ste5, which may segregate the kinases away from these general constitutive functions.
Figure 5.
Model for the control of proliferation by the Ste11 vegetative pathway.
The SVG pathway is functionally distinct from the mating and IG pathways that also utilize Ste20, Ste11, Ste7, Kss1, and Ste12, based on the absence of a requirement for the pathway-specific genes RAS2, TEC1, STE4, STE5, and FUS3 for the survival of the och1 mutant. The SVG pathway is also distinct from the HOG pathway, which utilizes Ste20 and Ste11 (5, 27), underscoring its functional specificity. Nevertheless, it seems likely that the SVG pathway is a manifestation of the IG pathway that regulates genes controlled by Ste12/Tec1 heterodimers (23). However, Ste12 must regulate additional genes independently of Tec1 to promote vegetative growth, because Tec1 is not required for vegetative growth in an och1Δ mutant.
During vegetative growth, the SVG pathway is inhibited by the same proteins that cross-regulate the mating and IG pathways. Based on FKS2 expression, Fus3 inhibits the pathway quite strongly, whereas Ste5 modestly inhibits the pathway. Ste5 may inhibit the pathway through sequestration of Ste11 and Ste7 (28). However, possible stimulatory effects from release of Ste11 and Ste7 may be masked by simultaneous release of Fus3, which inhibits the pathway. Hog1 also modestly inhibits FKS2 expression as shown for the mating pathway (5, 8). The ability of a hog1 mutation to suppress the growth defect of the bck1 null may be partly because of an increase in the activity of the SVG pathway.
Several lines of evidence argue that the SVG pathway functions, in part, to maintain cell wall integrity in parallel with the PKC pathway. First, Ste11 functions in parallel with Och1, which regulates the synthesis of mannan, an important structural component of the cell wall. och1 mutants have reduced cell wall integrity, and osmotic support suppresses the poor growth of an och1 ste12 double mutant. Second, Ste11 regulates vegetative growth additively with Bck1, the MAPKKK in the PKC pathway. The poor growth of a bck1 ste11 double mutant is suppressed by osmotic stabilizers, and bck1 ste11 double mutants lyse more than bck1 single mutants (Fig. 4). Third, the SVG pathway positively regulates the expression of FKS2, a glucan synthase gene involved in cell wall synthesis (20). The SVG and PKC pathways may regulate vegetative growth through related mechanisms. First, FKS2 expression is also up-regulated by Bck1 during heat shock through a distinct promoter element (29). Second, a hyperactive form of Ste7 (Ste7P368) can weakly suppress a bck1 mutant (30). Third, kinases from both pathways interact by two-hybrid analysis with the same morphogenesis proteins, Spa2 and Sph1 (31, 32).
The SVG pathway may have other functions besides cell wall integrity, because Och1 is essential for the N-linked glycosylation of other mannoproteins besides cell wall mannan. For example, och1 defects in nuclear segregation and microtubule structure may not be indirect consequences of an impaired cell wall (Fig. 2 A and B). Several observations suggest that the SVG pathway has additional targets besides the control of cell wall integrity (Fig. 5). First, the inviability of an och1 ste11 double mutant is not remediated by osmotic support, although an och1 ste12 double mutant is suppressed. Second, although the pathway regulates Tec1, a tec1 mutation does not impair the growth of an och1 mutant. Third, two-hybrid interactions between Ste11 and Ste7 and the homologous regulators of polarized morphogenesis Spa2 and Sph1 (31, 32) suggest that Ste11 and Ste7 may regulate morphogenesis.
The constitutive activation of the SVG pathway in the och1 null (Fig. 3) raises the interesting possibility that the SVG pathway is activated by intracellular signals that arise from structural defects in the cell wall that are the result of the loss of mannoproteins. This possibility is further supported by the observation that FKS2 expression is also elevated during vegetative growth in both fks1 and bck1 mutants that have cell wall defects (29) (Fig. 3). We note that, while the amount of phosphorylated Kss1 appears to be lower in och1 cells than in wild-type cells treated with α factor, a greater percentage of Kss1 appears to be the slower migrating species of a doublet (Fig. 3F). This observation suggests that, on average, a greater percentage of Kss1 has both TEY sites phosphorylated and there may be higher specific activity in the och1 mutant than in the presence of α factor. Such a scenario would allow the SVG pathway to effect greater change during vegetative growth as a consequence of minor stimulatory events. Further analysis will determine whether the Ste11 vegetative pathway is regulated by physical changes in the cell wall.
To date, the analysis of MAPK cascades in yeast and other organisms has focused on functions and activities that are induced by extracellular signals and routed by specialized regulatory proteins (1, 3). Our analysis suggests that, in S. cerevisiae, MAPK cascades also regulate proliferation in addition to specialized functions, through basal levels of activity that may be modulated by intracellular cues to maintain homeostasis. These findings may be applicable to more complex systems and may be important in defining the functions of MAPK cascades in less genetically tractable systems.
Acknowledgments
We thank J. Broach, K. Cunningham, M. Cyert, B. Errede, G. Fink, D. Levin, Y. Jigami, D. Pellman, H. Saito, P. Silver, and C. Douglas (Merck Research Labs) for generously providing plasmids and strains; J. Horecka for advice on growth of och1 strains; C. Kaisar for anti-invertase Ab; F. Posas for help with PCR knock outs; and D. Koepp and C. Tai for advice on synthetic lethal screens. We also thank Elion Laboratory members for helpful discussions; and E. Harlow, D. Morisato, D. Pellman, and H. Saito for comments on the manuscript. This work was supported by Leukemia Society of America Fellowship 5583-98 (to B.N.L.) and National Institutes of Health Grant GM4962 (to E.A.E.)
Abbreviations
- MAPK
mitogen-activated protein kinase
- IG
invasive growth
- HOG
high osmolarity glycerol
- PKC
protein kinase C
- YPD
yeast extract/peptone/dextrose
- DAPI
4′,6-diamidino-2-phenylindole
- SVG
STE vegetative growth
References
- 1.Gustin M C, Albertyn J, Alexander M R, Davenport K. Microbiol Mol Biol Rev. 1998;62:1264–1300. doi: 10.1128/mmbr.62.4.1264-1300.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ip I T, Davis R J. Curr Opin Cell Biol. 1998;10:205–219. doi: 10.1016/s0955-0674(98)80143-9. [DOI] [PubMed] [Google Scholar]
- 3.Widmann C, Gibson S, Jarpe M B, Johnson G L. Physiol Rev. 1999;79:1–35. doi: 10.1152/physrev.1999.79.1.143. [DOI] [PubMed] [Google Scholar]
- 4.Rajavel M, Philip B, Buehrer B M, Errede B, Levin D E. Mol Cell Biol. 1999;19:3969–3976. doi: 10.1128/mcb.19.6.3969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O’Rourke S M, Herskowitz I. Genes Dev. 1998;12:2874–2886. doi: 10.1101/gad.12.18.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elion E A. Science. 1998;281:1625–1626. doi: 10.1126/science.281.5383.1625. [DOI] [PubMed] [Google Scholar]
- 7.Madhani H D, Styles C A, Fink G R. Cell. 1997;91:673–684. doi: 10.1016/s0092-8674(00)80454-7. [DOI] [PubMed] [Google Scholar]
- 8.Hall J P, Cherkasova V, Elion E A, Gustin M C, Winter E. Mol Cell Biol. 1996;16:6715–6723. doi: 10.1128/mcb.16.12.6715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Levin D E, Bartlett-Heubusch E. J Cell Biol. 1992;116:1221–1229. doi: 10.1083/jcb.116.5.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guthrie, C. & Fink, G. R., eds. (1991) Methods Enzymol. 194.
- 11.Roberts R L, Fink G R. Genes Dev. 1994;8:2974–2985. doi: 10.1101/gad.8.24.2974. [DOI] [PubMed] [Google Scholar]
- 12.Nakayama K, Nagasu T, Shimma Y, Jigami Y. EMBO J. 1992;11:2511–2519. doi: 10.1002/j.1460-2075.1992.tb05316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bender A, Pringle J R. Mol Cell Biol. 1992;11:1295–1300. doi: 10.1128/mcb.11.3.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elion E A, Trueheart J, Fink G R. J Cell Biol. 1995;130:1283–1296. doi: 10.1083/jcb.130.6.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaiser C A, Botstein D. Mol Cell Biol. 1986;6:2382–2391. doi: 10.1128/mcb.6.7.2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elion E A, Satterberg B, Kranz J E. Mol Biol Cell. 1993;4:495–510. doi: 10.1091/mbc.4.5.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Farley F W, Satterberg B, Goldsmith E J, Elion A E. Genetics. 1999;151:1425–1444. doi: 10.1093/genetics/151.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Neiman A M, Mhaiskar V, Manus V, Galibert F, Dean N. Genetics. 1997;145:637–645. doi: 10.1093/genetics/145.3.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cid V J, Duran A, Rey D F, Synder M P, Nombela C, Sanchez M. Microbiol Rev. 1995;59:345–386. doi: 10.1128/mr.59.3.345-386.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mazur P, Morin N, Baginsky W, el-Sherbeini M, Clemas J A, Nielsen J B, Foor F. Mol Cell Biol. 1995;15:5671–5681. doi: 10.1128/mcb.15.10.5671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stevenson B J, Rhodes N, Errede B, Sprague G F., Jr Genes Dev. 1992;6:1293–1304. doi: 10.1101/gad.6.7.1293. [DOI] [PubMed] [Google Scholar]
- 22.Cook J G, Bardwell L, Thorner J. Nature (London) 1998;390:85–88. doi: 10.1038/36355. [DOI] [PubMed] [Google Scholar]
- 23.Madhani H D, Fink G R. Science. 1997;275:1314–1317. doi: 10.1126/science.275.5304.1314. [DOI] [PubMed] [Google Scholar]
- 24.Zhou Z, Gartner A, Cade R, Ammerrer G, Errede B. Mol Cell Biol. 1993;13:2069–2080. doi: 10.1128/mcb.13.4.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bardwell L, Cook J G, Voora D, Baggott D M, Martinez A R, Thorner J. Genes Dev. 1998;12:2887–2898. doi: 10.1101/gad.12.18.2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ma D, Cook J G, Thorner J. Mol Biol Cell. 1995;6:889–909. doi: 10.1091/mbc.6.7.889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Posas F, Saito H. Science. 1997;276:1702–1705. doi: 10.1126/science.276.5319.1702. [DOI] [PubMed] [Google Scholar]
- 28.Choi K Y, Satterberg B, Lyons D M, Elion E A. Cell. 1994;78:499–512. doi: 10.1016/0092-8674(94)90427-8. [DOI] [PubMed] [Google Scholar]
- 29.Zhao C, Jung U S, Garrett-Engele P, Roe T, Cyert M S, Levin D E. Mol Cell Biol. 1998;18:1013–1022. doi: 10.1128/mcb.18.2.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yashar B, Irie K, Printen J A, Stevenson B J, Sprague G F, Jr, Matsumoto K, Errede B. Mol Cell Biol. 1995;15:6545–6553. doi: 10.1128/mcb.15.12.6545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roemer T, Vallier L, Sheu Y J, Synder M. J Cell Sci. 1998;4:479–494. doi: 10.1242/jcs.111.4.479. [DOI] [PubMed] [Google Scholar]
- 32.Sheu Y J, Santos B, Fortin N, Costigan C, Synder M. Mol Cell Biol. 1998;18:4053–4069. doi: 10.1128/mcb.18.7.4053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matheos D P, Kingsbury T J, Ahsan U S, Cunningham K W. Genes Dev. 1997;11:3445–3458. doi: 10.1101/gad.11.24.3445. [DOI] [PMC free article] [PubMed] [Google Scholar]