Abstract
Apolipoprotein E (apoE) is associated with several classes of plasma lipoproteins and mediates uptake of lipoproteins through its ability to interact with specific cell surface receptors. Besides its role in cardiovascular diseases, accumulating evidence has suggested that apoE could play a role in neurodegenerative diseases, such as Alzheimer disease. In vertebrates, apoA-I is the major protein of high-density lipoprotein. ApoA-I may play an important role in regulating the cholesterol content of peripheral tissues through the reverse cholesterol transport pathway. We have isolated cDNA clones that code for apoE and apoA-I from a zebrafish embryo library. Analysis of the deduced amino acid sequences showed the presence of a region enriched in basic amino acids in zebrafish apoE similar to the lipoprotein receptor-binding region of human apoE. We demonstrated by whole-mount in situ hybridization that apoE and apoA-I genes are highly expressed in the yolk syncytial layer, an extraembryonic structure implicated in embryonic and larval nutrition. ApoE transcripts were also observed in the deep cell layer during blastula stage, in numerous ectodermal derivatives after gastrulation, and after 3 days of development in a limited number of cells both in brain and in the eyes. Our data indicate that apoE can be found in a nonmammalian vertebrate and that the duplication events, from which apoE and apoA-I genes arose, occurred before the divergence of the tetrapod and teleost ancestors. Zebrafish can be used as a simple and useful model for studying the role of apolipoproteins in embryonic and larval nutrition and of apoE in brain morphogenesis and regeneration.
Keywords: brain, embryo nutrition, lipoprotein, yolk syncytial layer, zebrafish
Cholesterol and other lipids are transported in the blood by different lipoprotein classes whose basic molecular organization and role in lipid metabolism are similar for all vertebrates (1, 2). Apolipoproteins of the various lipoproteins regulate lipoprotein metabolism and determine the unique roles of these lipoproteins in lipid metabolism. In mammals, a group of eight exchangeable and soluble apolipoproteins (apoC-I, apoC-II, apoC-III, apoC-IV, apoA-I, apoA-II, apoA-IV, and apoE) is characterized by the occurrence of repeated amphipathic helical regions considered to be structural units essential for lipid-binding properties (3, 4). Comparative analysis of sequence data has suggested that the exchangeable mammalian apolipoproteins are all members of a multigene family, and different evolutionary scenarios have been proposed (5–8). These genes appear to have evolved from a common ancestral apolipoprotein gene, apoC-I, through whole-gene duplications, intraexonic amplifications of repeating units, and intragenic deletions. The presence of an apoA-I in bird and trout with a repeat pattern similar to those in human apoA-I (9–11) suggested that all the internal repeats in these sequences arose before the fish–mammal split, which was some 400 million years ago. apoA-I, apoC-III, and apoA-IV genes are closely arranged in a gene cluster in both chicken and mammals (12), and apolipoproteins similar in their biochemical properties to some mammalian apolipoproteins have been identified in lower vertebrates (2, 13–15). However, because no convincing evidence was found for the existence of apoE in nonmammalian vertebrates (14–16), it is generally accepted that this important apolipoprotein is an evolutionary latecomer whose appearance may have coincided with the appearance of mammals.
MATERIALS AND METHODS
Isolation of cDNA Clones from an Embryo Library.
Apolipoprotein cDNA clones have been isolated by whole-mount in situ hybridization screening from a phage λZAPII library constructed from poly(A)+ RNA isolated from 18- to 40-hr-old zebrafish (Danio rerio) embryos (17). Isolated phage plaques were randomly picked. Cloned fragments were automatically excised with helper phage and recircularized to generate subclones in pBluescript SK(−) phagemid vector (18). The clones were selected for further analysis after performing whole-mount in situ hybridization on embryos. The rationale of this procedure was to specifically identify cDNAs derived from tissue-restricted abundance rather than from poorly expressed mRNA. Two different apolipoprotein cDNA clones have been isolated: I-BR102 and E-Bluebag. Because the I-BR102 clone did not include the entire coding region, this clone was used to rescreen the cDNA library according to standard methods (19). Twenty-seven cDNA clones were isolated, and some of them were subsequently selected for further sequencing by the chain–termination method with the Pharmacia T7 sequencing kit. Oligonucleotide primers to internal sequences were used to obtain overlapping bidirectional sequence information.
Phylogenetic Analysis.
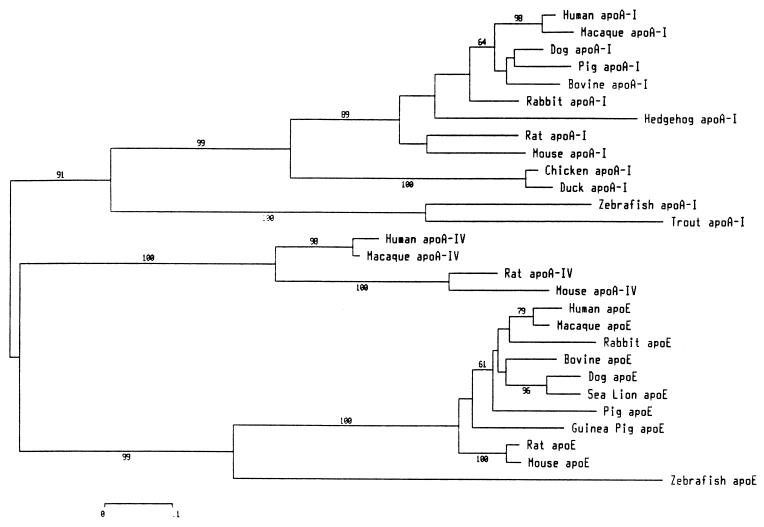
The phylogenetic tree shown (Fig. 2) was constructed with programs from the mega 1.02 package (20). Topology and branch lengths of the phylogenetic tree were estimated using the neighbor-joining (NJ) method (21) based on the number of amino acid substitutions per site (Poisson-correction distance method, complete-deletion option for gap sites). Reliability of the NJ tree topology was evaluated by bootstrap analysis (22) with 5,000 replications. The accession numbers or bibliographic references for the sequences used are: human apoE (spP02649) and apoA-I (spP02647); macaque apoE (spP10517) and apoA-I (spP15568); rabbit apoE (spP18287) and apoA-I (spP09809); bovine apoE (spQ03247) and apoA-I (spP15497); dog apoE (gbC60940) and apoA-I (spP02648); pig apoE (spP18650) and apoA-I (spP18648); guinea pig apoE (spP23529); mouse apoE (spP08226) and apoA-I (spQ00623); rat apoE (spP02650) and apoA-I (spP04639); chicken apoA-I (spP08250); duck apoA-I (gbA61448); hedgehog apoA-I (23); rainbow trout apoA-I1 (11); and sea lion apoE (24). The sequences from zebrafish apoE and A-I reported in this paper were derived from the full-length E-Bluebag and I-7 clones.
Figure 2.
Phylogenetic tree of fish, bird, and mammalian apoA-I, apoA-IV, and apoE protein sequences. The unrooted tree was constructed by the neighbor-joining method based on the number of amino-acid substitutions per site. The alignment is based on the 33-codon block and internal repeats 4–8. The resulting unambiguous alignment contained 126 aligned sites. The numbers on the branches (only those ≥60% are shown) are bootstrap confidence levels.
Whole-Mount in Situ Hybridization.
Embryos were fixed overnight in 4% paraformaldehyde, and the localization of transcripts was performed by whole-mount in situ hybridization as previously described (25). Antisense-digoxygenin-labeled probes were generated using the T7 RNA polymerase after linearization of the vector by digestion with NotI. I-BR102 clone, from nucleotides −20 to +550 of zebrafish apoA-I cDNA, and E-Bluebag clone, from nucleotides −6 to +1321 of zebrafish apoE cDNA, were used as templates to generate the probes. As a control, sense probes were synthesized and did not give any staining. Embryos older than 24 hr were raised in 10% Hanks’ saline plus 0.2 mM 1-phenyl-2-thio-urea to prevent pigment formation. Embryos were mounted in Permount (Fisher Scientific) after dehydration in methanol and clarification in methylsalicylate or, alternatively, mounted in 100% glycerol.
RESULTS AND DISCUSSION
Screening of a zebrafish (Danio rerio) cDNA embryo library for genes expressed with a temporal and spatial restriction during embryonic development has led to the isolation of two groups of apolipoprotein sequences. For the first group of cDNA clones, which code for zebrafish apoA-I, sequence information was obtained from nucleotides −26 to +926, excluding the poly(A) tail, and an open reading frame (ORF) of 786 nt was shown coding for 262 amino acids (from nucleotides +1 to +786). A polyadenylylation consensus signal (AATAAA) was found 12 nt upstream from the poly(A) tail. The DNA sequence of the different zebrafish apoA-I clones characterized was found to be heterologous at three positions: nucleotide +684 (T/C), nucleotide +708 (T/C), and nucleotide +846 (G/A). None of the nucleotide differences present in the ORF, which might encode allelic products, caused a change in amino acid. For the second group, one cDNA clone, which codes for zebrafish apoE, has been isolated with sequence information encompassing nucleotides −6 to +1321, excluding the poly(A) tail, and showing an ORF of 843 nt coding for 281 amino acids. Two polyadenylylation consensus signals were found 23 and 164 nt upstream from the poly(A) tail.
An alignment of the deduced amino acid zebrafish apoE and apoA-I sequences with the corresponding human sequences is shown in Fig. 1. Gaps were introduced for maximum alignment of the sequences. Zebrafish apoE and apoA-I sequences showed 27.5% and 25.6% identities to human apoE and apoA-I sequences, respectively. The two zebrafish sequences presented 21.1% of amino acid identity between each other. By analogy to apoE and apoA-I that have been sequenced so far in vertebrates, a putative 18-aa-long signal peptide could be predicted in both zebrafish apolipoproteins. The putative prosegment appeared to be 5 or 6 aa long in zebrafish apoA-I and apoE, respectively. The absence of a prosegment in mammalian apoE could be due to the loss in the mammalian lineage of the conserved site for proteolytic cleavage of the pro-form. The amino acid sequences of mammals’ apoE within this region are extremely poorly conserved, and amino-terminal extensions of this part of the molecule can be observed (24, 26). Both zebrafish apoE and apoA-I mature protein sequences satisfy the common structural features depicted for the exchangeable apolipoproteins (8). The first part includes a block of 33 residues, which consisted of 3 units of 11 amino acids. The second part consists of repeats of 11 or 22 residues predicted to form amphipathic α-helical secondary structures. Zebrafish mature apoE and apoA-I have predicted molecular weights of 29,063 and 27,841, and predicted isoelectric points of 4.95 and 5.14.
Figure 1.
Zebrafish (Ze) apoA-I and apoE predicted amino acid sequences aligned with the corresponding human (Hu) sequences. Human apoA-I and apoE (E3 isoform) sequences are numbered according to the mature proteins. The amino-terminal amino acids of mature zebrafish apoA-I and apoE were postulated to be located at the same position as in human apoA-I. Boundaries of the signal peptide (signal), the propeptide (pro), the unrelated coding regions 1 and 2 (UCR1 and UCR2, respectively), the 33-codon block, and the 11- or 22-residue repeats (4 to 14) of human sequences are indicated above the sequence for human apoA-I and below the sequence for human apoE. Double dots indicate identities and single dots indicate conservative substitutions between human apoA-I and zebrafish apoA-I (Top), between zebrafish apoA-I and apoE (Middle), and between zebrafish apoE and human apoE (Bottom). Residues identical or considered conserved in all apoA-I or apoE sequences currently available in vertebrates (see Materials and Methods) are underlined. The allowed conservative substitutions were defined as follows: A, G; S, T; E, D; R, K, H; Q, N; V, I, L, M; Y, F; W; P; C. Gaps inserted to optimize alignments are indicated by dashes. The lipoprotein receptor-binding region (residues 136–150) of human apoE is double-underlined.
It is now established that in human apoE the basic amino acid residues between residues 136 and 150, clustered into a surface patch on one long α-helix (27), are necessary for low-density lipoprotein receptor binding (26, 28). This receptor-binding region is strongly conserved in zebrafish apoE (residues 117–131) and contains seven positively charged amino acids instead of eight in human apoE (Fig. 1). Computer-based analysis of zebrafish apoE indicates that this region has a very low hydrophobicity together with a large hydrophobic moment (data not shown). The apoE alleles commonly found in humans and associated with a number of clinical states are distinguished by cysteine–arginine interchanges in the amino acid sequence at residues 112 (Cys → Arg) and 158 (Arg → Cys). The presence of a basic residue occupying in most mammalian species (26), but also in zebrafish apoE (Lys-93), the equivalent of amino acid position 112 of human apoE, indicates that apoE4 (Arg-112, Arg-158) instead of apoE3 (Cys-112, Arg-158) is most likely the ancestral allele in human. In human apoE2 (Cys-112, Cys-158), the defective binding to the low-density lipoprotein receptor occurs by disruption of a single salt bridge between Asp-154 and Arg-158 and formation of a new one between Arg-150 and Asp-154 (29). In zebrafish apoE, two charged amino acid residues (Lys-131, Glu-135) are present at positions equivalent to amino acids 150 and 154 of human apoE, but a basic residue is absent from the equivalent position 158 (Thr-139). It would be very interesting to compare the conformation of the two receptor-binding regions together with the ability of both proteins to compete for the same lipoprotein receptor.
The evolutionary relationships between zebrafish apoE and apoA-I and other available sequences from long exchangeable apolipoproteins are presented in Fig. 2. The phylogenetic tree was constructed using parts of the amino acid sequences that can be unambiguously aligned (i.e., the common 33-codon block and internal repeats 4–8; see Fig. 1). In contradiction to previous phylogenetic analysis predicting the absence of apoE in teleost fish (30), the clustering pattern of Fig. 2 provides convincing evidence that both apoE and apoA-I could be regrouped with the corresponding apolipoproteins of higher vertebrates. In addition to the presence of an low-density lipoprotein receptor-binding domain in zebrafish apoE, some amino acid residues, markers of apoA-I or apoE, are conserved in zebrafish apoA-I and apoE (Fig. 1). In addition, a small apolipoprotein (LAL1) found in lamprey blood plasma (31) seems to be more closely related to zebrafish apoE than to zebrafish apoA-I. The extent of amino acid identities with zebrafish apoE and apoA-I were 26.1% and 16.9%, respectively (65 amino acid positions compared, including the 33-common-codon block, the first 11 amino acids of repeat 4, and the entire repeat 5). The presence of both apoE and apoA-I in a teleost fish supports the concept that the duplication events responsible for the presence of an apolipoprotein multigene family (7) occurred at the basis of vertebrate evolution. Different exchangeable apolipoproteins with lipid-binding properties and diverse, specific functional domains permit the appearance of a complex transport system for the transfer of exogenous or endogenous lipids toward different tissues in the context of the establishment of a closed circulatory system.
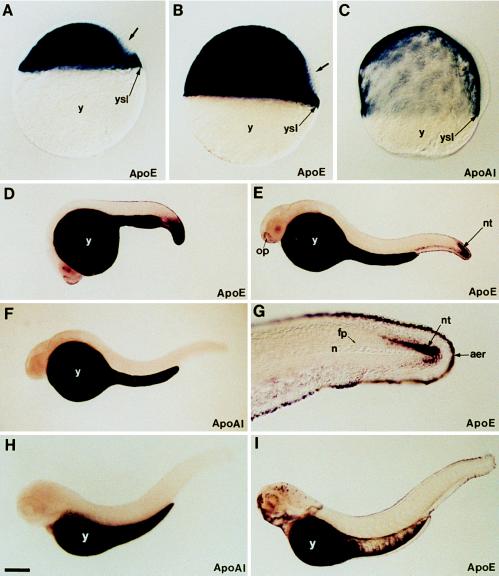
The expression pattern of apoE and apoA-I genes determined by whole-mount in situ hybridization revealed a very high level of transcripts in the yolk syncytial layer (YSL) during embryonic and early larval development (Fig. 3). ApoA-I transcripts were observed from the gastrula stage (see ref. 32 for details about stages of embryonic development of the zebrafish) to the end of embryogenesis exclusively in the YSL (Fig. 3 C, F, and H). Similarly to apoA-I gene, but starting at the blastula stage, the expression of apoE gene was very strong in the YSL from this very early developmental stage until larval development (Fig. 3 A, B, D, E, and I). The YSL is an extraembryonic structure that is formed during the blastula stage and is responsible for the degradation and transfer to the embryo and early larva of the yolk reserves contained in the yolk cell. It is most likely that apoA-I and apoE gene expression in the YSL is associated with the synthesis and secretion of lipoproteins. Fish YSL is a lipoprotein-secreting tissue (33, 34) that plays an essential role in the transport of yolk nutrients to the developing embryo. Apolipoproteins may play a similar role in embryonic nutrition of birds and mammals as suggested by apolipoprotein gene expression and lipoprotein secretion by the yolk sac (35–38).
Figure 3.
Expression pattern of apoE and apoA-I genes during embryonic and early larval development of zebrafish. (A and B) ApoE expression at 30% epiboly and shield stages, respectively. (C) ApoA-I expression at 70% epiboly. Both genes are expressed in the yolk syncytial layer (ysl) at these early stages while the yolk cell (y) does not contain any transcripts. The dorsal domain, which does not express apoE, is indicated by an arrow. (D) At 20 hr of development, apoE transcripts are located in the yolk syncytial layer and in the caudal region. (E) At 24 hr, apoE transcripts are detected in the yolk syncytial layer that covers the yolk cell in cells surrounding the olfactory placodes (op), in the ventral caudal mesoderm, and in the caudal part of the neural tube (nt). (G) High magnification of the tail region of a 24-hr-old embryo showing apoE transcripts in the apical ectodermal ridge (aer) of the fin and in caudal neural tube. Notocord (n) and floor plate (fp) are unlabeled. At 24 hr (F) and 3 days (H) of development, the expression of apoA-I gene is restricted to the yolk syncytial layer, while a new domain of apoE gene expression appears at 3 days in the head region, in the facial ectoderm, and in some cells of the retina and brain (I). Embryos were mounted in Permount (A–C) or in 100% glycerol and viewed with differential interference contrast optics (A–C and G) or with a dissecting microscope (D–F, H, and I). [Bar = 100 μm (A–C); 200 μm (D–F, H, and I); 50 μm (G).]
In addition to their presence in the YSL, apoE transcripts were also detected at the blastula stage in some embryonic cells (Fig. 3A). The presence of apoE mRNA expression at this early stage, revealed by in situ hybridization, was confirmed by reverse transcriptase–PCR assay (data not shown). ApoE transcripts were observed in the deep cell layer (the cells that will form the embryo proper) but not in enveloping layer cells (Fig. 3A). In the deep cell layer, the staining is homogeneous except in the dorsal part of the blastula. ApoE transcripts were not found in dorsal cells, with the exception of the more dorsal marginal cells. At gastrulation stage, cells of the dorsal unlabeled domain give rise to the embryonic shield (the zebrafish homologous structure of the dorsal blastopore lip of amphibians) (Fig. 3B). During gastrulation and neurulation stages, apoE transcripts progressively disappeared from the embryonic cells. At 20 hr after fertilization, apoE transcripts were detected in the posterior part of the tail, in the caudal-most part of neural tube, in caudal paraxial mesoderm, and in the ventral mesenchyme of the tail (Fig. 3D). ApoE expression was also observed in some cells of the epidermis, anteriorly surrounding the olfactory placodes, and in the more axial epidermal cells that line the whole embryo and that give rise to the apical ectodermal ridge (AER) of dorsal caudal and anal fins (Fig. 3D). At 24 hr, the expression in the tail region appeared restricted to the most posterior cells of the developing spinal cord, to cells of the presumptive AER, and to a few cells of the most caudal paraxial mesoderm (Fig. 3 E and G). Expression in the caudal neurectoderm and the mesoderm disappeared at the end of tail elongation but was still observed in the AER of uneven fins and in cells surrounding the anus (Fig. 3I). ApoE transcripts were found in the AER of the pectoral fins, which start to form after 36 hr of development. Anteriorly, expression becomes more complex in the epiderm. A very strong expression was observed in cells surrounding the mouth opening and sensorial organs of the face, i.e., olfactory vesicules and anterior lateral line organs (Fig. 4 A and C). Expression was also observed in epidermal cells covering the heart, in ectodermal cells of the gill arch, and in epiblastic cells covering the mouth cavity (Fig. 4 A–C).
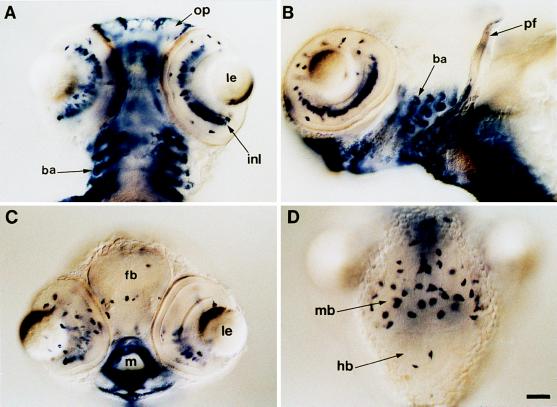
Figure 4.
apoE gene expression in the head region at early larval stage of zebrafish. Ventral (A) and ventro-lateral (B) views of an early larva (4 days of development), focusing on the head region. ApoE transcripts accumulate in cells surrounding the olfactory placodes (op), in branchial arches (ba), and in cells of the inner nuclear layer (inl) of the retina. pf, pectoral fin. (C) Transverse optical cross-section of the head of an early larva at the eye level. Expression of the apoE gene is observed in the retina, in some cells of the forebrain (fb), and in cells surrounding the mouth opening (m). le, lens. (D) Dorsal view of the head region showing the expression of apoE gene in a limited number of cells located both in the hindbrain (hb) and in the tectal part of the midbrain (mb). Early larvae were mounted in Permount and viewed with differential interference contrast optics. (Bar = 50 μm.)
After 3 days of development and in addition to the expression in these ectodermal derivatives, apoE gene starts to be transcribed in a limited number of cells both in brain and in the eyes (Figs. 3I and 4). In brain, most of these apoE-expressing cells are located in the tectal region of the mesencephalon, where neurons of the retina project their axons, but some are also observed in telencephalon or rhombencephalon (Fig. 4 C and D). apoE gene expression was also observed in cells of the inner nuclear layer of the retina (Fig. 4 A and B). The number of these brain cells containing apoE transcripts increased during early larval stages. Unlike other mammalian apolipoproteins, which are expressed primarily in the intestine and liver, apoE mRNA is present in most mammalian adult tissues, with the brain containing the second highest abundance of apoE mRNA (28). Although not fully understood, apoE appears to play a role in both normal and pathological brain function. Within the central nervous system of mammals, apoE is synthesized and secreted mainly by astrocytes both in brain and in the eyes, but not by neurons (39–42). ApoE may play a role in local transport of lipids, and its synthesis is implicated in neuronal growth and in repair after injury of both peripheral and central neurons (28, 41, 43, 44). Considerable knowledge concerning the embryonic development of the central nervous system and the neuroanatomy of the adult zebrafish has accumulated in recent years (45, 46). It remains to be determined whether apoE gene expression is limited to a subpopulation of neuronal or nonneuronal cells during zebrafish brain morphogenesis. This animal could be used as a model organism to identify the mechanisms whereby apoE is involved in normal and abnormal pathophysiology within the nervous system.
In summary, the present manuscript shows that apoE can be found in a nonmammalian vertebrate. The presence of both apoA-I- and apoE-expressed genes in zebrafish indicates, in contradiction to previous predictions, that the duplication events from which apoE and apoA-I genes arose occurred before the divergence of the tetrapod and teleost ancestors. Deduced amino acid sequences of zebrafish apoE and apoA-I show 27.5% and 25.6% identities to human apoE and apoA-I sequences, respectively. Some amino acid residues, markers of apoA-I or apoE, are conserved in zebrafish apoA-I and apoE. Analysis of the deduced amino acid sequences showed the presence of a region enriched in basic amino acids in zebrafish apoE similar to the lipoprotein receptor-binding region of human apoE. The temporal and timing of expression of apoA-I in the zebrafish embryo is different from that of apoE. Although very high levels of apoA-I transcripts were found in the YSL, those of apoE were located in the YSL but also in the deep cell layer during blastula stage, in numerous ectodermal derivatives after gastrulation, and after 3 days of development in a limited number of cells both in brain and in the eyes. Finally, zebrafish can be used as a simple and useful model for studying the role of apolipoproteins in embryonic and larval nutrition and of apoE in brain morphogenesis and regeneration.
Acknowledgments
We thank E. Lubzens for critically reading the manuscript. This work was supported by funds from the Centre National de la Recherche Scientifique, the Institut National de la Santé et de la Recherche Médicale, the Université Paris-Sud, and the Centre Hospitalier Universitaire Régional.
ABBREVIATIONS
- apo
apolipoprotein
- YSL
yolk syncytial layer
Footnotes
References
- 1.Chapman M J. Methods Enzymol. 1986;128:70–147. doi: 10.1016/0076-6879(86)28063-5. [DOI] [PubMed] [Google Scholar]
- 2.Babin P J, Vernier J M. J Lipid Res. 1989;30:467–489. [PubMed] [Google Scholar]
- 3.Segrest J P, Garber D W, Brouillette C G, Harvey S C, Anantharamaiah G M. Adv Protein Chem. 1994;45:303–369. doi: 10.1016/s0065-3233(08)60643-9. [DOI] [PubMed] [Google Scholar]
- 4.Allan C M, Walker D, Segrest J P, Taylor J M. Genomics. 1995;28:291–300. doi: 10.1006/geno.1995.1144. [DOI] [PubMed] [Google Scholar]
- 5.Barker W C, Dayhoff M O. Comp Biochem Physiol. 1977;57B:309–315. doi: 10.1016/0305-0491(77)90060-8. [DOI] [PubMed] [Google Scholar]
- 6.Boguski M S, Birkenmeier E H, Elshoubagy N A, Taylor J M, Gordon J I. J Biol Chem. 1986;261:6398–6407. [PubMed] [Google Scholar]
- 7.Luo C-H, Li W-H, Moore M N, Chan L. J Mol Biol. 1986;187:325–340. doi: 10.1016/0022-2836(86)90436-5. [DOI] [PubMed] [Google Scholar]
- 8.Li W-H, Tanimura M, Luo C-H, Datta S, Chan L. J Lipid Res. 1988;29:245–271. [PubMed] [Google Scholar]
- 9.Byrnes L, Luo C-C, Li W-H, Yang C-Y, Chan L. Biochem Biophys Res Commun. 1987;148:485–492. doi: 10.1016/0006-291x(87)91137-5. [DOI] [PubMed] [Google Scholar]
- 10.Gu Z-W, Xie Y-H, Yang M, Sparrow J T, Wang K, Li Y, Li W-H, Gotto A M, Yang C-Y. J Protein Chem. 1993;12:585–591. doi: 10.1007/BF01025123. [DOI] [PubMed] [Google Scholar]
- 11.Delcuve G P, Sun J M, Davie J R. J Lipid Res. 1992;33:251–262. [PubMed] [Google Scholar]
- 12.Lamon-Fava S, Sastry R, Ferrari S, Rajavashisth T B, Lusis A J, Karathanasis S K. J Lipid Res. 1992;33:831–842. [PubMed] [Google Scholar]
- 13.Babin P J. Biochem J. 1987;246:425–429. doi: 10.1042/bj2460425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barakat H A, St. Clair R W. J Lipid Res. 1985;26:1252–1268. [PubMed] [Google Scholar]
- 15.Hermier D, Sellier N, Rousselot-Pailley D, Forgez P. Eur J Biochem. 1995;234:586–591. doi: 10.1111/j.1432-1033.1995.586_b.x. [DOI] [PubMed] [Google Scholar]
- 16.Mehta K D, Brown M S, Bilheimer D W, Goldstein J L. J Biol Chem. 1991;16:10415–10419. [PubMed] [Google Scholar]
- 17.Fjose A, Izpisua-Belmonte J-C, Fromental-Ramain C, Duboule D. Development. 1994;120:71–81. doi: 10.1242/dev.120.1.71. [DOI] [PubMed] [Google Scholar]
- 18.Short J M, Fernandez J A, Sorge J A, Huse W D. Nucleic Acids Res. 1988;16:7583–7600. doi: 10.1093/nar/16.15.7583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 20.Kumar S, Tamura K, Nei M. Comput Appl Biosci. 1994;10:189–191. doi: 10.1093/bioinformatics/10.2.189. [DOI] [PubMed] [Google Scholar]
- 21.Saitou N, Nei M. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 22.Felsenstein J. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 23.Sparrow D A, Laplaud P M, Saboureau M, Zhou G, Dolphin P J, Gotto A M, Sparrow J T. J Lipid Res. 1995;36:485–495. [PubMed] [Google Scholar]
- 24.Davis R W, Pierotti V R, Lauer S J, Hubl S T, McLean J W, Witztum J L, Young S G. J Lipid Res. 1991;32:1013–1023. [PubMed] [Google Scholar]
- 25.Thisse C, Thisse B, Halpern M E, Postlethwait J H. Dev Biol. 1994;164:420–429. doi: 10.1006/dbio.1994.1212. [DOI] [PubMed] [Google Scholar]
- 26.Weisgraber K H. Adv Prot Chem. 1994;45:249–302. doi: 10.1016/s0065-3233(08)60642-7. [DOI] [PubMed] [Google Scholar]
- 27.Wilson C, Wardell M R, Weisgraber K H, Mahley R W, Agard D A. Science. 1991;252:1817–1822. doi: 10.1126/science.2063194. [DOI] [PubMed] [Google Scholar]
- 28.Mahley R W. Science. 1988;240:622–630. doi: 10.1126/science.3283935. [DOI] [PubMed] [Google Scholar]
- 29.Dong L-M, Parkin S, Trakhanov S D, Rupp B, Simmons T, Arnold K S, Newhouse Y M, Innerarity T L, Weisgraber K H. Nat Struct Biol. 1996;3:718–722. doi: 10.1038/nsb0896-718. [DOI] [PubMed] [Google Scholar]
- 30.Powell R, Higgins D G, Wolff J, Byrnes L, Stack M, Gannon F. Gene. 1991;104:155–161. doi: 10.1016/0378-1119(91)90245-7. [DOI] [PubMed] [Google Scholar]
- 31.Pontes M, Xu X, Graham D, Riley M, Doolittle R F. Biochemistry. 1987;26:1611–1617. doi: 10.1021/bi00380a019. [DOI] [PubMed] [Google Scholar]
- 32.Kimmel C B, Ballard W W, Kimmel S R, Ullmann B, Schilling T F. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- 33.Vernier J M, Sire M F. Biol Cell. 1977;29:45–54. [Google Scholar]
- 34.Walzer C, Schönenberger N. Cell Tissue Res. 1979;196:75–93. doi: 10.1007/BF00236349. [DOI] [PubMed] [Google Scholar]
- 35.Shi W-K, Heath J K. J Embryol Exp Morphol. 1984;81:143–152. [PubMed] [Google Scholar]
- 36.Noble R C, Cocchi M. Prog Lipid Res. 1990;29:107–140. doi: 10.1016/0163-7827(90)90014-c. [DOI] [PubMed] [Google Scholar]
- 37.Plonné D, Winkler L, Franke H, Dargel R. Biochim Biophys Acta. 1992;1127:174–185. doi: 10.1016/0005-2760(92)90275-z. [DOI] [PubMed] [Google Scholar]
- 38.Farese R V, Cases S, Ruland S L, Kayden H J, Wong J S, Young S G, Hamilton R L. J Lipid Res. 1996;37:347–360. [PubMed] [Google Scholar]
- 39.Boyles J K, Pitas R E, Wilson E, Mahley R W, Taylor J M. J Clin Invest. 1985;76:1501–1513. doi: 10.1172/JCI112130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pitas R E, Boyles J K, Lee S H, Foss D, Mahley R W. Biochim Biophys Acta. 1987;917:148–161. doi: 10.1016/0005-2760(87)90295-5. [DOI] [PubMed] [Google Scholar]
- 41.Poirier J. Trends Neurosci. 1994;17:525–530. doi: 10.1016/0166-2236(94)90156-2. [DOI] [PubMed] [Google Scholar]
- 42.Amaratunga A, Abraham C R, Edwards R B, Sandell J H, Schreiber B M, Fine R E. J Biol Chem. 1996;271:5628–5632. doi: 10.1074/jbc.271.10.5628. [DOI] [PubMed] [Google Scholar]
- 43.Strittmatter W J, Roses A D. Proc Natl Acad Sci USA. 1995;92:4725–4727. doi: 10.1073/pnas.92.11.4725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weisgraber K H, Mahley R W. FASEB J. 1996;10:1485–1494. doi: 10.1096/fasebj.10.13.8940294. [DOI] [PubMed] [Google Scholar]
- 45.Eisen J S. Cell. 1996;87:969–977. doi: 10.1016/s0092-8674(00)81792-4. [DOI] [PubMed] [Google Scholar]
- 46.Wullimann M F, Rupp B, Reichert H. Neuroanatomy of the Zebrafish Brain: A Topological Atlas. Basel: Birkhäuser; 1996. [Google Scholar]