Abstract
Tangier disease is characterized by low serum high density lipoproteins and a biochemical defect in the cellular efflux of lipids to high density lipoproteins. ABC1, a member of the ATP-binding cassette family, recently has been identified as the defective gene in Tangier disease. We report here the organization of the human ABC1 gene and the identification of a mutation in the ABC1 gene from the original Tangier disease kindred. The organization of the human ABC1 gene is similar to that of the mouse ABC1 gene and other related ABC genes. The ABC1 gene contains 49 exons that range in size from 33 to 249 bp and is over 70 kb in length. Sequence analysis of the ABC1 gene revealed that the proband for Tangier disease was homozygous for a deletion of nucleotides 3283 and 3284 (TC) in exon 22. The deletion results in a frameshift mutation and a premature stop codon starting at nucleotide 3375. The product is predicted to encode a nonfunctional protein of 1,084 aa, which is approximately half the size of the full-length ABC1 protein. The loss of a Mnl1 restriction site, which results from the deletion, was used to establish the genotype of the rest of the kindred. In summary, we report on the genomic organization of the human ABC1 gene and identify a frameshift mutation in the ABC1 gene of the index case of Tangier disease. These results will be useful in the future characterization of the structure and function of the ABC1 gene and the analysis of additional ABC1 mutations in patients with Tangier disease.
Keywords: high density lipoproteins, cholesterol, atherosclerosis
Tangier disease is an autosomal codominant disease characterized by the accumulation of cholesteryl ester in various tissues, most notably the tonsils, liver, spleen, and intestinal mucosa (1, 2). Macrophages, present as foam cells in affected tissue, are the principal cell type that accumulates excess tissue cholesterol (2, 3). The genetic defect in Tangier disease has been proposed to be associated with decreased reverse cholesterol transport (4–8), the pathway by which excess cellular cholesterol is transported back to the liver for excretion. In particular, the efflux of cholesterol to high density lipoprotein (HDL), which is the first step in the reverse cholesterol transport pathway, has been reported to be defective in Tangier disease (4–8).
According to current concepts (4, 9–11), two separate pathways are involved in the efflux of cholesterol from cells to HDL: the aqueous diffusion pathway and the apolipoprotein-mediated pathway. In the aqueous diffusion pathway, cholesterol spontaneously desorbs from the cell membrane and associates with lipoprotein acceptors, such as HDL, in the extracellular space (4, 9). For many cell types this appears to be the principal pathway through which cholesterol efflux occurs (11). Alternatively, in cells such as macrophages (12–14) and fibroblasts (4, 8), cholesterol and phospholipid efflux also can be mediated by lipid-poor or lipid-free apolipoproteins, such as apoA-I, apoA-II, and apoE. Apolipoprotein-mediated efflux is an energy-dependent process that requires the direct interaction of the apolipoprotein with the cell surface, the lipidation of the apolipoprotein, and the subsequent dissociation of the lipid-apolipoprotein complex from the cell (4, 8, 10, 15). Cholesterol loading of cells induces the apolipoprotein-mediated cholesterol efflux pathway (4, 13, 16). Fibroblasts from patients with Tangier disease have been shown to be normal with regard to aqueous diffusion of cholesterol but defective in apolipoprotein-mediated efflux of both cholesterol and phospholipid (4, 8). Lipid efflux from Tangier disease firbroblasts to HDL is also partially reduced (4, 8). In addition to removing cholesterol by aqueous diffusion, a component of lipid efflux by HDL is believed to occur by apolipoproteins that dissociate from HDL and remove lipid from the cell by the apolipoprotein-mediated pathway (4, 8, 10).
Initial studies to elucidate the genetic defect in Tangier disease, using positional cloning, mapped the defective gene to a region on chromosome 9 (17). In a recent report, we (18) and others (48, 49) have identified that mutations in the ABC1 gene are the molecular defect in Tangier disease.
The ABC1 protein is a member of the large ATP-binding cassette family of proteins that transport a wide assortment of molecules, such as ions, sugars, vitamins, lipids, and proteins across the plasma membrane (19–22). Mutations in specific ABC proteins have been described as causes of several human diseases, including cystic fibrosis (23), adrenoleukodystrophy (24), Zellweger disease (25), and macular degeneration (26). The ABC1 gene has been cloned from mice (27) and humans (28) and has been proposed to play a role in macrophage function (27–29). Like other ABC proteins, the ABC1 protein contains two ATP-binding domains and 12 transmembrane domains, which are believed to form a channel-like structure and are essential for the transport function of ABC proteins (20, 23).
In the present study, we determined the genomic organization of human and mouse ABC1 and identified the genetic defect in the original kindred from Tangier Island (1). The two hallmarks of the disease, enlarged lipid laden tonsils and low serum HDL, are based on the initial description of this kindred (1). Subsequent studies of this kindred also have provided key insights into the metabolic and the biochemical abnormalities in Tangier disease. Metabolic kinetic studies established that there is no defect in the biosynthesis of apoA-I in Tangier disease, but rather the low plasma HDL levels are the result of its hypercatabolism (31). More recently, studies of cultured cells from this (4) and other Tangier disease kindreds (5–8) have been pivotal in confirming the importance of the apolipoprotein-mediated pathway in cholesterol and phospholipid cellular efflux in the reverse cholesterol transport pathway. Structural analysis of the ABC1 gene in this study revealed a mutation in the original kindred that results in the production of a nonfunctional truncated ABC1 protein, thus confirming mutations in the ABC1 gene as the cause of Tangier disease.
Materials and Methods
Cell Culture.
Primary human skin fibroblasts were obtained by explant cultures of skin biopsies taken from the upper arm. Patients from which Tangier cell lines were obtained have been described (1). Normal human skin fibroblasts were from the American Type Culture Collection. Fibroblasts were routinely grown in Eagle’s modified minimum essential medium (EMEM) (GIBCO/BRL), supplemented with 10% FCS, 2 mM glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin (EMEM10).
Cholesterol Efflux Assay.
As described (4, 32, 33), fibroblasts grown on 24-well plates were incubated with EMEM10 containing 1 μCi/ml of 1,2-3H-cholesterol (50 Ci/mmol) (DuPont) for 48 hr. After washing the cells, fibroblasts were cholesterol-loaded by a 24-hr incubation with 50 μg/ml of nonlipoprotein cholesterol in EMEM, containing 2 mg/ml of BSA (fraction V) (EMEM/BSA). Efflux medium was prepared by adding the indicated acceptor to EMEM/BSA and then was added to washed cells for a 20-hr incubation period. After the incubation, medium was collected, centrifuged (10,000 × g for 5 min), and counted for radioactivity by liquid scintillation counting. Residual radioactivity in the cell fraction was determined after overnight extraction with isopropanol. Percentage efflux is radioactive counts in the efflux medium divided by the sum of the radioactive counts in the medium plus cell fraction. Results from the blank (EMEM/BSA media only) were subtracted from the radioactive counts obtained in the presence of an acceptor in EMEM/BSA.
Phospholipid Efflux Assay.
As described (4), choline-containing phospholipids were labeled by incubating confluent cholesterol-loaded fibroblasts grown on 24-well plates with EMEM10 medium containing 5 μCi/ml methyl-3Hcholine chloride (84 Ci/mmol, Amersham Pharmacia) for 24 hr. Cells then were washed three times with EMEM/BSA, followed by a 1-hr preincubation in EMEM/BSA, before incubation with the indicated acceptor in EMEM/BSA. After a 20-hr efflux period, medium was collected and centrifuged (10,000 × g; 5 min). Samples of supernatants were extracted with chloroform/methanol (2:1), and radioactivity in the organic phase was quantified by liquid scintillation counting. Radioactivity remaining in the cell fraction was determined after overnight extraction with isopropanol, followed by a 1-hr extraction with hexane/isopropanol (3:2). Percentage efflux was calculated as for cholesterol efflux.
Lipoprotein and Apolipoprotein Isolation and Analysis.
HDL (d = 1.063–1.021 g/ml) was isolated from human plasma by density gradient ultracentrifugation. Apolipoproteins purified from human plasma by column chromatography (34) were determined to be over 99% pure, as assessed by SDS/PAGE and amino-terminal sequence analysis. Plasma lipoprotein analysis was performed on a Hitachi 911 analyzer (Roche Diagnostics), using Roche reagents.
DNA Sequence Analysis.
DNA was extracted from leukocytes from whole blood, using a Promega DNA extraction kit (Promega). Total RNA was extracted from cultured fibroblast with a Qiagen RNA extraction kit (Qiagen, Chatsworth, CA). Reverse transcription–PCR (RT-PCR) was performed by using Superscript One-Step system, as described by the manufacturer (Life Technologies, Gaithersburg, MD). ABC1 cDNA for sequencing was obtained by RT-PCR amplification of four overlapping DNA fragments, using the following primers pairs: (i) 75S (5′-CTA CCC ACC CTA TGA ACA AC-3′) and 2561R (5′-CTC TTC TCA TCA CTT TCC TC-3′), (ii) 1991S (5′-GCT GGT TCA TTA GTA GCC TCA-3′) and 3731R (5′-CCA TCT GAG GTC TCA GCA TCC-3′), (iii) 3651S (5′-TGG CAT CTC AGA GAC GAC CCT-3′) and 4999R (5′-GGT GTT TTG CTT TGC TGA CCC-3′), and (iv) 4741S (5′-GCA ATC AGC TCT TTC CTG AAT-3′) and 6751R (5′-AGC CAC GCC GTA TGA ACA GGA-3′).
Synthetic oligonucleotide primers were synthesized, using phosphoramidite chemistry on a DNA synthesizer (PE Applied Biosytems, Foster City, CA). Sequencing was performed by ABI Prism Big dye cycle sequencing and analyzed on an ABI 310 DNA sequencer.
Identification of exon/intron boundaries was performed by alignment of genomic DNA sequence with the human ABC1 cDNA (28). Genomic DNA sequence was obtained either by direct sequencing of bacterial artificial chromosome (BAC) clones or by PCR amplification of genomic DNA. BAC clones were identified by screening for the ABC1 gene from either Genome Systems human release II library (Genome Systems, St. Louis) or from Research Genetics (Huntsville, AL) library CITB-HSP-D. Primers for genomic sequencing were based on the exon/intron boundaries from the mouse ABC1 gene (GenBank accession no. g495256). Mouse genomic clones were isolated for DNA sequence analysis by screening a phage genomic library from 129SV mouse strain (Stratagene).
Results
Characterization of Tangier Disease Kindred.
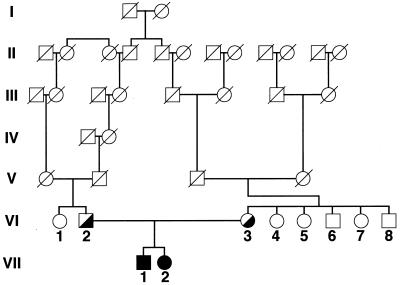
The pedigree of the original Tangier disease kindred (1) shows consanguinity at generations V and VI (Fig. 1). The proband (subject VII-1) has one sibling (subject VII-2), who also is affected with Tangier disease. The parents (subjects VI-2 and VI-3) are obligate heterozygotes for Tangier disease. A recent analysis of the plasma lipoprotein profile of the kindred is shown in Table 1. Characteristic of Tangier disease, the homozygotes (subjects VII-1 and VII-2) had a marked reduction of HDL-C, as well as for apoA-I and apoA-II. The obligate heterozygote parents (subjects VI-2 and VI-3) had approximately a 50% reduction in HDL-C. No samples from subjects VI-7 and VI-8 were available for analysis. Based on lipoprotein analysis and clinical history, there do not appear to be any other homozygotes for Tangier disease in the current living members of the kindred. The heterozygous state of the rest of the family could not be established with certainty based on the plasma lipoprotein analysis (Table 1).
Figure 1.
Pedigree of Tangier disease kindred. Roman numerals indicate the number of the generation. Subjects in generations VI and VII are identified with arabic numbers. Solid symbols indicate homozygotes and half-filled symbols indicate obligate heterozygotes.
Table 1.
Lipoprotein analysis of kindred
| Subject | Total cholesterol | HDL-C | LDL-C, mg/dl | TG | ApoA-I | ApoA-II |
|---|---|---|---|---|---|---|
| VI-1 | 228 | 64 | 125 | 193 | 161 | 46 |
| VI-2 | 127 | 17 | 61 | 158 | 85 | 26 |
| VI-3 | 188 | 23 | 136 | 784 | 118 | 39 |
| VI-4 | 232 | 16 | 136 | 552 | 147 | 40 |
| VI-5 | 200 | 35 | 91 | 371 | 145 | 39 |
| VI-6 | 169 | 27 | 114 | 142 | 99 | 25 |
| VII-1 | 35 | 8 | 28 | 352 | 3 | 2 |
| VII-2 | 86 | 7 | 33 | 471 | 35 | 2 |
Subject number corresponds to number shown in pedigree in Fig. 1. LDL, low density lipoprotein; TG, triglyceride.
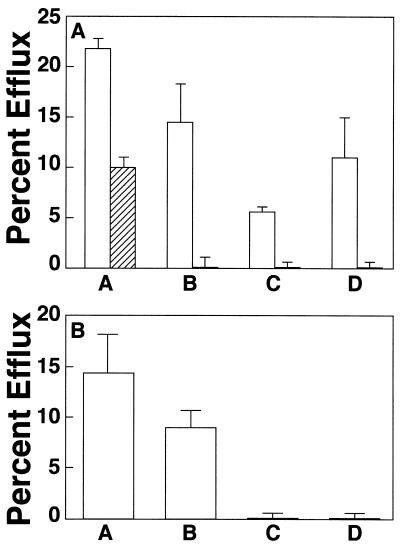
A lipid efflux study was performed on skin fibroblasts from the Tangier disease proband (subject VII-1). As shown in Fig. 2A, fibroblasts from the proband had a lower than normal level of HDL-mediated cholesterol efflux (column A). In contrast, apolipoprotein-mediated cholesterol efflux by either apoA-I (column B) or apoA-II (column C) was nearly absent compared with normal fibroblasts. In addition, phospholipid efflux to apoA-I (column D) also was significantly reduced from the Tangier disease cells. These results are consistent with previous studies (4, 8) that showed a marked reduction of apolipoprotein-mediated lipid efflux from Tangier disease cells, but only a partial reduction in HDL-mediated efflux.
Figure 2.
Lipid efflux study of kindred. (A) Lipid efflux from normal control fibroblast cells (open columns) and proband fibroblast (subject VII-1) (lined columns) was measured, using cholesterol-labeled cells (columns A-C) or phospholipid-labeled cells (column D), with either 100 μg/ml HDL (column A), 10 μg/ml apoA-I (columns B and D), or 10 μg/ml apoA-II (column C) as the acceptor. Results for control cells are the mean percent efflux ± SD from three normal cell lines. Results of the proband are the mean percent efflux ± SD of triplicate determinations. (B) Lipid efflux of kindred. Normal cells (column A), and cells from subjects VI-3 (column B), VII-1 (column C), and VII-2 (column D) were radiolabeled with cholesterol and incubated with 10 μg/ml apoA-I. Effux from control cells is the mean percent efflux ± SD from three normal cell lines. Results for the subjects from the kindred are the mean percent efflux ± SD of triplicate determinations.
The results of a cholesterol efflux study using apoA-I as acceptor are shown for the affected sibling and one of the parents. Compared with normal cells (Fig. 2B, column A), cells from the parent, (column B: subject VI-3) had only a partial reduction of apolipoprotein efflux. In contrast, the proband (column C; subject VII-1) and the affected sibling (column D; subject VII-2) both had almost a complete absence of apolipoprotein-mediated efflux from cells. These results are consistent with the assigned genotypes for Tangier disease shown in the pedigree (Fig. 1).
Organization of the ABC1 Gene.
The organization of the human ABC1 gene was investigated by alignment of the genomic sequence with the previously published cDNA sequence for ABC1 (28). Genomic DNA sequence was obtained either by direct sequencing of bacterial artificial chromosome (BAC) clones spanning the ABC1 gene or by sequence analysis of PCR-amplified fragments of genomic DNA. A total of 49 splice junctions were observed in the human ABC1 gene (Table 2). The location of the splice sites and the overall structure were very similar for the human and mouse ABC1 genes. The sequences of all identified donor and acceptor splice sites are consistent with consensus sequences for splice junctions and contain the invariant AG (5′ splice) and GT (3′ splice) sites. The exons, ranging in size from 33 to 249 bp, are relatively small compared with other genes but similar to those in other ABC genes (19–22). The coding region resides in exons 3–49. Based on the insert sizes of the BACs used for the analysis, the ABC1 gene is estimated to span over 70 kb.
Table 2.
Intron-exon boundaries of the ABC1 gene
| Exon num. | Exon size, bp | 5′ splice site | 3′ splice site | Position, bp | Exon num. | Exon size, bp | 5′ splice site | 3′ splice site | Position, bp5 |
|---|---|---|---|---|---|---|---|---|---|
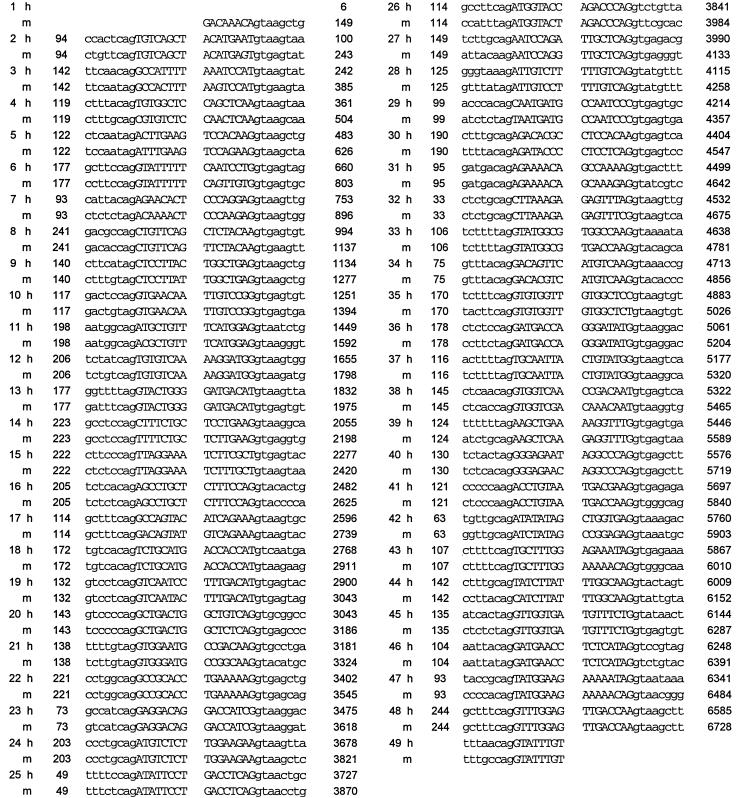 |
h, human; m, mouse.
DNA Sequence Analysis of ABC1 Gene in Tangier Disease.
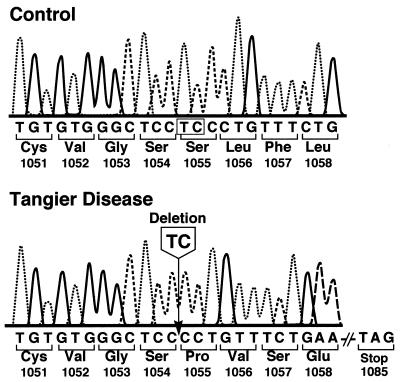
Sequence analysis was performed on DNA from the proband for the ABC1 gene. Four overlapping PCR fragments were produced by reverse transcription–PCR, as described in Materials and Methods, and both strands were sequenced. A 2-bp TC deletion at nucleotides 3283 and 3284 was identified in the cDNA from the proband (Fig. 3). The deletion causes a frameshift, starting at amino acid 1055 and results in a premature stop codon after amino acid 1084. The truncated protein encoded by the mutated ABC1 gene would be predicted to be nonfunctional, because it contains only approximately half of the amino acids as the intact protein. The truncation would occur between the predicted sixth and seventh transmembrane domains in the linker region of the ABC1 protein (Fig. 4). No other mutations were discovered in the ABC1 cDNA sequence of the proband.
Figure 3.
DNA sequence analysis of ABC1 gene in proband. DNA sequences of control subject and proband (subject VII-1) are shown for part of exon 22 of the ABC1 gene. Arrow indicates the site of the TC deletion.
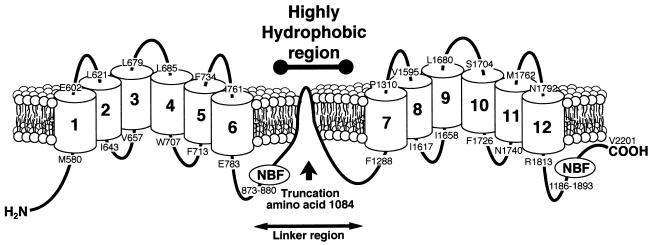
Figure 4.
Diagram of the secondary structure of the ABC1 transporter. Arrow indicates the site of the TC deletion and the approximate location of the carboxyl terminus of the truncated protein. NBF indicates the positions of the nucleotide-binding folds.
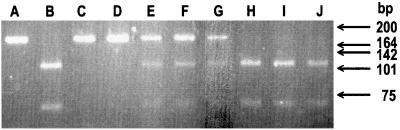
The 2-bp deletion shown in Fig. 3 results in a loss of a Mnl1 restriction site. The genotypes of the rest of the kindred, in regard to the mutation discovered in the proband, were evaluated by restriction fragment polymorphism (Fig. 5). A 172-bp fragment was amplified by PCR from genomic DNA of the indicated members of the kindred, digested with Mnl1, and analyzed by electrophoresis. The position of the 172-bp fragment amplified by PCR from control DNA and not digested with Mnl1 is shown in Fig. 5, lane A. Mnl1 cleavage of the control DNA fragment in Fig. 5, lane A resulted in two bands of approximately 111 and 61 bp (Fig. 5, lane B), as expected. The PCR-amplified band from the proband (subject VII-1, Fig. 5, lane C) was not cut by Mnl1, as predicted by the DNA sequence analysis (Fig. 3), Complete absence of the 111- and 61-bp fragments in Fig. 5, lane C indicates that the proband is homozygous for the TC deletion shown in Fig. 5. The affected sibling (subject VII-2, Fig. 5, lane D) had a pattern similar to that of the proband, thus showing that she is also homozygous for the same mutation. The obligate heterozygote parents (Fig. 5, lanes E and F) had DNA fragment patterns expected for a heterozygote. They had one mutant allele, which was resistant to the Mnl1 digestion and migrated as a 172-bp band. The other apparently normal allele yielded bands of approximately 111 and 61 bp after digestion with Mnl1. The DNA fragment pattern in Fig. 5, lane G from subject VI-6 is also consistent with heterozygosity for the mutation. The lipoprotein analysis of subject VI-6 is similar to those of the obligate heterozygote parents (Table 1). The other members of the kindred (Fig. 5, lanes H-J) had normal patterns of DNA fragments and apparently do not have the mutation. The presence of the 2-bp deletion was confirmed in all subjects identified to be heterozygotes or homozygotes by DNA sequence analysis.
Figure 5.
Restriction digest polymorphism analysis of Tangier disease kindred. A 172-bp fragment was amplified from DNA extracted from peripheral blood leukocytes by PCR, using a sense primer starting at nucleotide 3181 (5′GGCCGCACCATTATTCTCTCT3′) and a reverse primer starting at nucleotide 3353 (5′GATTCCACATCCTTTCTTGACC3′). PCR fragments were digested with Mnl1 and analyzed on 3% Nusieve/0.5% Me Seakem gel. Letters above each lane correspond to the following samples: undigested control DNA (A), Mnl1-digested DNA from control (B), subject VII-1 (C), subject VII-2 (D), subject VI-3 (E), subject VI-2 (F), subject VI-6 (G), subject VI-4 (H), subject VI-5 (I), and subject VI-1 (J).
Discussion
In this report, we describe the organization of the human ABC1 gene and identify an ABC1 mutation in the index case for Tangier disease. The organization of the ABC1 gene is similar to that of other ABC genes (19–22) and contained 49 exons that spanned greater than 70 kbp in length (Table 2). Because of the frequency of mutations in splice site junctions (35), particularly in a gene like ABC1 with a relatively large number of splice sites, the description of the ABC1 genomic organization will be useful in the future identification of ABC1 mutations in Tangier disease patients. The mutation described in the proband of the kindred is a 2-bp deletion that results in a frameshift (Fig. 3) and a truncated protein that is predicted to be nonfunctional, because it would be only approximately half the size of the intact protein. The genotype analysis of the rest of the kindred is consistent with this mutation being the cause of Tangier disease (Fig. 5). The identification of an ABC1 mutation in the original Tangier disease kindred thus establishes that defects in the ABC1 gene are a cause for Tangier disease. Additional studies, however, will be necessary to determine whether there are other genetic defects that can lead to the Tangier disease phenotype.
Although mutations in the ABC1 gene now have been identified as a cause for Tangier disease, the exact physiologic role of the ABC1 protein and how a defect in the protein can lead to a loss of apolipoprotein-mediated efflux is still not known. Prior studies of ABC proteins, however, suggest several possibilities. The ABC1 protein along with ABC2, ABC3, and ABCR are members of the ABCA subfamily (36–39) that, unlike other ABC proteins, are not present in prokaryotes or in yeast, which suggests a specialized role for these proteins in multicellular organisms (20, 36, 40, 41). All members of the ABCA subfamily also contain a relatively large hydrophobic region between the sixth and seventh transmembrane domains (Fig. 4). The exact orientation of the hydrophobic region in relation to the plasma membrane is not known, but it has been suggested that it may facilitate the transport of lipid across the membrane bilayer (36, 38, 39). The ligands for transport by the ABC1 subfamily are not known except for ABCR, which transports retinoids that are conjugated to phosphatidylethanolamine in the retinal rod photoreceptor cells (39). In addition to the ABCR, several other ABC proteins also transport lipids (42–44). For example, disruption of the mdr2 gene in mice leads to defective secretion of phospholipid and cholesterol into bile (45).
If ABC1 transports lipid, a defect in ABC1 function could directly lead to a disruption of lipid efflux from cells. In one simple model, apoA-I and other apolipoproteins could interact directly with the ABC1 protein. ApoA-I then could undergo lipidation with free cholesterol and/or phospholipid transported by the ABC1 protein, and the lipidaton of apoA-I would lead to the formation and dissociation of nascent HDL from the cell. A functional defect in ABC1 therefore would lead to low plasma HDL, because of the rapid catabolism of lipid poor apoA-I (4, 8, 31).
Several other aspects of ABC1 expression are consistent with a direct role for ABC1 in lipid efflux from cells. Macrophages abundantly express the ABC1 protein (27, 28, 30) and are the predominant cell type that is affected by Tangier disease (2). Unlike many other cells, macrophages can efflux lipid by the apolipoprotein-mediated pathway (13, 16, 46). Macrophages may be particularly prone to lipid accumulation in the absence of the apolipoprotein-mediated pathway, because of their scavenger role in the clearance of cellular debris, such as lipid-rich membranes and oxidized lipids. The induction of ABC1 expression in macrophages after cholesterol loading (28) is also consistent with a requirement for cells to be cholesterol-loaded before they are able to undergo apolipoprotein-mediated efflux (4, 8).
Previous studies also have identified several other potential roles for ABC1 that may relate indirectly to lipid efflux. Both ABC1 and Ced7, a similar protein expressed in Caenorhabditis elegans, have been proposed to be important in the phagocytosis of apoptotic cells (29, 30). The mechanism by which this occurs is not known but has been proposed to involve a function of ABC1 in the transport of adhesion proteins to the cell surface (29, 30). Alternatively, the translocation of lipids by ABC1 may modify the plasma membrane of macrophages and facilitate the process of phagocytosis. Like other ABC proteins, ABC1 also has been proposed to play a part in the transport and secretion of leaderless proteins, such as IL-1β (47). Leaderless proteins lack a signal peptide and are secreted from the cell through a pathway that bypasses the classical endoplasmic reticulum-Golgi pathway. The ABC1 protein therefore might mediate lipid efflux from cells indirectly by its role in the transport of a cell surface protein that is essential for lipid efflux.
Now that the genetic defect of Tangier disease has been identified, it will be useful to carefully examine the causes for the phenotypic heterogeneity of the disease in regard to the development of premature cardiovascular disease. Minimal atherosclerosis is present in the original kindred with Tangier disease (1), but premature cardiovascular disease has been identified in another Tangier disease kindred that also has a ABC1 gene mutation (17). Additional studies will be required to elucidate the genetic, as well as the environmental factors, that may modulate the phenotypic expression of Tangier disease.
In the first description (1) of Tangier disease in 1961, one of the authors (D.S.F) wrote the following: “One of the remarkable opportunities that patients with rare genetically determined diseases offer is an occasional view of normal processes obtainable in no other way. Possibly Tangier disease will provide us with just such a view into the now-clouded aspects of fat transport and metabolism.” It took nearly 40 years to unravel the biochemical and genetic defect in Tangier disease but now this prediction has come true. The identification of ABC1 mutations as the defect in Tangier disease has provided a key insight into the normal process of reverse cholesterol transport. Future studies aimed at understanding the function of ABC1 in lipid efflux represent a promising new direction for atherosclerosis research.
Acknowledgments
The technical contribution of Carole Lafargue, Edouard Turlotte, Delphine Lagnezux, and Mary L. Van Tine is acknowledged. We also thank the members of the family described in this paper, who have generously participated in this and many other studies of Tangier disease.
Abbreviations
- HDL
high density lipoprotein
- EMEM
Eagle’s modified minimum essential medium
- apoA
apolipoprotein A
References
- 1.Fredrickson D S, Attrocchi P H, Avioli L V, Goodman D S, Goodman H C. Ann Intern Med. 1961;55:1016–1031. [Google Scholar]
- 2.Assmann G, von Eckardstein A, Brewer H B., Jr . In: The Metabolic and Molecular Basis of Inherited Disease. Scriver C R, Beaudet A L, Sly W S, Valle D, editors. New York: McGraw–Hill; 1995. pp. 2053–2072. [Google Scholar]
- 3.Ferrans V J, Fredrickson D S. Am J Pathol. 1975;78:101–158. [PMC free article] [PubMed] [Google Scholar]
- 4.Remaley A T, Schumacher U K, Stonik J A, Farsi B D, Nazih H, Brewer H B., Jr Arterioscler Thromb Vasc Biol. 1997;17:1813–1821. doi: 10.1161/01.atv.17.9.1813. [DOI] [PubMed] [Google Scholar]
- 5.Schmitz G, Bruning T, Williamson E, Nowicka G. Eur Heart J. 1990;11, Suppl. E:197–211. doi: 10.1093/eurheartj/11.suppl_e.197. [DOI] [PubMed] [Google Scholar]
- 6.Rogler G, Truembach B, Klima B, Lackner K J, Schmitz G. Arterioscler Thromb. 1995;15:683–690. doi: 10.1161/01.atv.15.5.683. [DOI] [PubMed] [Google Scholar]
- 7.Walter M, Gerdes U, Seedorf U, Assmann G. Biochem Biophys Res Commun. 1994;205:850–856. doi: 10.1006/bbrc.1994.2742. [DOI] [PubMed] [Google Scholar]
- 8.Francis G A, Knopp R H, Oram J F. J Clin Invest. 1995;96:78–87. doi: 10.1172/JCI118082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rothblat G H, De la Llera-Moya M, Atger V, Kellner-Weibel G, Williams D L, Phillips M C. J Lipid Res. 1999;40:781–796. [PubMed] [Google Scholar]
- 10.Oram J F, Yokoyama S. J Lipid Res. 1996;37:2473–2491. [PubMed] [Google Scholar]
- 11.Johnson W J, Mahlberg F H, Rothblat G H, Phillips M C. Biochim Biophys Acta. 1991;1085:273–298. doi: 10.1016/0005-2760(91)90132-2. [DOI] [PubMed] [Google Scholar]
- 12.Vega G L, Grundy S M. J Intern Med. 1989;226:5–15. doi: 10.1111/j.1365-2796.1989.tb01347.x. [DOI] [PubMed] [Google Scholar]
- 13.Sakar S W, Williams D L, Stoudt G W, Phillips M C, Rothblat G H. Biochim Biophys Acta. 1999;1438:85–98. doi: 10.1016/s1388-1981(99)00041-4. [DOI] [PubMed] [Google Scholar]
- 14.Hara H, Yokoyama S. J Biol Chem. 1991;266:3080–3086. [PubMed] [Google Scholar]
- 15.Mendez A J. J Lipid Res. 1997;38:1807–1821. [PubMed] [Google Scholar]
- 16.Smith J D, Miyata M, Ginsberg M, Grigaux C, Shmookler E, Plump A S. J Biol Chem. 1996;271:30647–30655. doi: 10.1074/jbc.271.48.30647. [DOI] [PubMed] [Google Scholar]
- 17.Rust S, Walter M, Funke H, von Eckardstein A, Cullen P, Kroes H Y, Hordijk R, Geisel J, Kastelein J, Molhuizen H O, et al. Nat Genet. 1998;20:96–98. doi: 10.1038/1770. [DOI] [PubMed] [Google Scholar]
- 18.Rust S, Rosier M, Funke H, Real J, Amoura Z, Piette J C, Deleuze J F, Brewer H B, Jr, Duverger N, Denefle P, Assmann G. Nat Genet. 1999;22:352–355. doi: 10.1038/11921. [DOI] [PubMed] [Google Scholar]
- 19.Dean M, Allikmets R. Curr Opin Genet Dev. 1995;5:779–785. doi: 10.1016/0959-437x(95)80011-s. [DOI] [PubMed] [Google Scholar]
- 20.Decottignies A, Goffeau A. Nat Genet. 1997;15:137–145. doi: 10.1038/ng0297-137. [DOI] [PubMed] [Google Scholar]
- 21.Allikmets R, Gerrard B, Hutchinson A, Dean M. Hum Mol Genet. 1996;5:1649–1655. doi: 10.1093/hmg/5.10.1649. [DOI] [PubMed] [Google Scholar]
- 22.Schwiebert E M. Am J Physiol. 1999;276:C1–C8. doi: 10.1152/ajpcell.1999.276.1.C1. [DOI] [PubMed] [Google Scholar]
- 23.Riordan J R, Rommens J M, Kerem B, Alon N, Rozmahel R, Grzelczak Z, Zielenski J, Lok S, Plavsic N, Chou J L. Science. 1989;245:1066–1073. doi: 10.1126/science.2475911. [DOI] [PubMed] [Google Scholar]
- 24.Mosser J, Douar A M, Sarde C O, Kioschis P, Feil R, Moser H, Poustka A M, Mandel J L, Aubourg P. Nature (London) 1993;361:726–730. doi: 10.1038/361726a0. [DOI] [PubMed] [Google Scholar]
- 25.Gartner J, Moser H, Valle D. Nat Genet. 1992;1:16–23. doi: 10.1038/ng0492-16. [DOI] [PubMed] [Google Scholar]
- 26.Allikmets R, Shroyer N F, Singh N, Seddon J M, Lewis R A, Bernstein P S, Peiffer A, Zabriskie N A, Li Y, Hutchinson A, et al. Science. 1997;277:1805–1807. doi: 10.1126/science.277.5333.1805. [DOI] [PubMed] [Google Scholar]
- 27.Luciani M F, Denizot F, Savary S, Mattei M G, Chimini G. Genomics. 1994;21:150–159. doi: 10.1006/geno.1994.1237. [DOI] [PubMed] [Google Scholar]
- 28.Langmann T, Klucken J, Reil M, Liebisch G, Luciani M-F, Chimini G, Kaminski W E, Schmitz G. Biochem Biophys Res Commun. 1999;257:29–33. doi: 10.1006/bbrc.1999.0406. [DOI] [PubMed] [Google Scholar]
- 29.Luciani M-F, Chimini G. EMBO J. 1996;15:226–233. [PMC free article] [PubMed] [Google Scholar]
- 30.Wu Y-C, Horvitz H R. Cell. 1998;93:951–960. doi: 10.1016/s0092-8674(00)81201-5. [DOI] [PubMed] [Google Scholar]
- 31.Schaefer E J, Anderson D W, Zech L A, Lindgren F T, Bronzert T B, Rubalcaba E A, Brewer H B., Jr J Lipid Res. 1981;22:217–228. [PubMed] [Google Scholar]
- 32.Oram J F. Methods Enzymol. 1986;129:645–659. doi: 10.1016/0076-6879(86)29096-5. [DOI] [PubMed] [Google Scholar]
- 33.Rothblat G H, Bamberger M, Phillips M C. Methods Enzymol. 1986;129:628–644. doi: 10.1016/0076-6879(86)29095-3. [DOI] [PubMed] [Google Scholar]
- 34.Brewer H B, Jr, Ronan R, Meng M, Bishop C. Methods Enzymol. 1986;129:223–246. doi: 10.1016/0076-6879(86)28070-2. [DOI] [PubMed] [Google Scholar]
- 35.Rogan P K, Faux B M, Schneider T D. Hum Mutat. 1998;12:153–171. doi: 10.1002/(SICI)1098-1004(1998)12:3<153::AID-HUMU3>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 36.Illing M, Molday L L, Molday R S. J Biol Chem. 1997;272:10303–10310. doi: 10.1074/jbc.272.15.10303. [DOI] [PubMed] [Google Scholar]
- 37.Croop J M. Methods Enzymol. 1998;292:101–115. doi: 10.1016/s0076-6879(98)92010-9. [DOI] [PubMed] [Google Scholar]
- 38.Allikmets R, Wasserman W W, Hutchinson A, Smallwood P, Nathans J, Rogan P K, Schneider T D, Dean M. Gene. 1998;215:111–122. doi: 10.1016/s0378-1119(98)00269-8. [DOI] [PubMed] [Google Scholar]
- 39.Sun H, Molday R S, Nathans J. J Biol Chem. 1999;274:8269–8281. doi: 10.1074/jbc.274.12.8269. [DOI] [PubMed] [Google Scholar]
- 40.Deeley R G, Cole S P C. Cancer Biol. 1997;8:193–204. doi: 10.1006/scbi.1997.0070. [DOI] [PubMed] [Google Scholar]
- 41.van Veen H W, Konings W N. Cancer Biol. 1997;8:183–191. doi: 10.1006/scbi.1997.0064. [DOI] [PubMed] [Google Scholar]
- 42.Ruetz S, Brault M, Dalton W, Gros P. Biochemistry. 1997;36:8180–8188. doi: 10.1021/bi970564o. [DOI] [PubMed] [Google Scholar]
- 43.Raggers R J, van Helvoort A, Evers R, van Meer G. J Cell Sci. 1999;112:415–422. doi: 10.1242/jcs.112.3.415. [DOI] [PubMed] [Google Scholar]
- 44.Zhou Y, Gottesman M M, Pastan I. Mol Cell Biol. 1999;19:1450–1459. doi: 10.1128/mcb.19.2.1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shneider B L. J Pediatr. 1999;28:1124–1131. [Google Scholar]
- 46.Hara H, Komaba A, Yokoyama S. Lipids. 1992;27:302–304. doi: 10.1007/BF02536480. [DOI] [PubMed] [Google Scholar]
- 47.Andrei C, Dazzi C, Lotti L, Torrisi M R, Chimini G, Rubartelli A. Mol Biol Cell. 1999;10:1463–1475. doi: 10.1091/mbc.10.5.1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brooks-Wilson A, Marcil M, Clee S M, Zhang, Lin-Hua, Roomp K, van Dam M, Yu L, Brewer C, Collins J A, Molhuizen H O F, et al. Nat Genet. 1999;22:336–345. doi: 10.1038/11905. [DOI] [PubMed] [Google Scholar]
- 49.Bodzioch M, Orso E, Klucken J, Langmann T, Bottcher A, Diederich W, Drobnik W, Barlage S, Buchler C, Porsch-Ozcurumez M, et al. Nat Genet. 1999;22:347–351. doi: 10.1038/11914. [DOI] [PubMed] [Google Scholar]