Abstract
During activation T cells are thought to change their patterns of gene expression dramatically. To find out whether this is true for T cells activated in animals, the patterns of genes expressed in resting T cells and T cells 8 and 48 hr after activation were examined by using Affymetrix gene arrays. Gene arrays gave accurate comparisons of gene expression in the different cell types because the expression of genes known to vary during activation changed as expected. Of the approximately 6,300 genes assessed by the arrays, about one-third were expressed to appreciable extents in any of the T cells tested. Thus, resting T cells express a surprisingly large diversity of genes. The patterns of gene expression changed considerably within 8 hr of T cell activation but returned to a disposition more like that of resting T cells within 48 hr of exposure to antigen. Not unexpectedly, the activated T cells expressed genes associated with cell division at higher levels than resting T cells. The resting T cells expressed a number of cytokine receptor genes and some genes thought to suppress cell division, suggesting that the state of resting T cells is not a passive failure to respond to extant external stimuli.
Keywords: activated, resting, gene array
A good deal of attention has been paid to gene expression in differentiated T cells of various types. Subtraction techniques, differential display, and gene array analysis all have been used to investigate differences in gene expression between different kinds of T cells (1–6). Most of these experiments have been performed by using established T cell lines, or at least T cells that have been withdrawn from their host and cultured for some time. Relatively few experiments have addressed the question of overall gene expression in T cells in animals. Because of this there are significant holes in our knowledge of these cells. For example, little is known about gene expression in resting T cells. Gene expression in resting T cells is not, however, an uninteresting problem. Although for many years it was thought that resting T cells were in a quiescent stage, existing independently of their surroundings, recent experiments have shown that this is far from true. The survival of small, naïve resting T cells in animals depends on receipt of many life-preserving factors from their environment. For example, the life expectancy of these cells is known to be dependent on low-affinity engagement of their antigen receptors by the MHC protein and probably the peptide that drove their selection in the thymus (7–10). For their survival, small, resting T cells in animals probably also depend on engagement of cytokines such as IL-6 and IL-7 (11–15).
The appearance of antigen or superantigen in animals leads to the rapid activation of specific T cells. These activated cells divide rapidly and then many of the progeny of the dividing cells die within a few days (16, 17). This phenomenon does not occur if the T cells are activated in vitro. Therefore, an understanding of the ways in which activated T cells die in animals depends on activating the cells in vivo, not in vitro.
This paper describes our attempts to deal with these deficiencies in our knowledge. mRNA was purified from resting T cells, or T cells that recently had been activated. Analysis was performed by using Affymetrix gene arrays (Santa Clara, CA). The results showed that resting T cells expressed a surprisingly large diversity of different mRNAs. Within 8 hr of activation in vivo the range of genes expressed by the T cells changed dramatically, although the total number of genes expressed did not change. Two days after activation, the spectrum of genes expressed by the activated cells was much more like that of resting T cells. However, again, the total number of different genes expressed was still about the same, amounting to about one-third of all the genes on the array. Comparison of the genes expressed by resting T cells with T cells 48 hr after activation showed that the latter cells expressed, not surprisingly, many genes associated with cell division. Among the genes expressed at higher levels by resting cells were those coding for a number of cytokine receptors and for several genes thought to inhibit cell division. Hence, the state of resting T cells may depend on active processes that allow the cell to be nurtured by its environment and actively prevent it from entering cell division.
Materials and Methods
Mice and Cells.
T cells were activated in C57BL/10 mice (The Jackson Laboratory) by i.v. injection of 150 μg of the Vβ8.x-specific superantigen, staphylococcal enterotoxin B (SEB; Sigma). This procedure generates T cells bearing Vβ8.x that are fully activated as judged by the fact that, 8 hr later, they all bear CD69 and, 18 hr after injection of SEB, all the Vβ8.x+ T cells begin to divide (ref. 17; unpublished observations). One group of mice was sacrificed 8 hr after injection of SEB. Another group of mice was sacrificed 48 hr after injection of SEB, at the time when the SEB-stimulated T cells had reached their maximum numbers and just before they were to start to die (17). Lymph nodes were harvested from these animals and from control, untreated mice, and T cells prepared by passage through nylon wool columns (18). To purify the T cells more thoroughly, the cell preparations were stained and sorted by using a MoFlo Instrument (Cytomation, Fort Collins, CO). Normal resting T cells were isolated from nonimmunized mice after staining with phycoerythrin (PE)-labeled anti-Cβ, CyChrome-labeled anti-CD4 and anti-CD8, and fluorescein (FL)-labeled anti-IAb and anti-CD69 (PharMingen). Sorting gates were set to collect small Cβ+ cells bearing CD4 or CD8 and to exclude cells bearing IAb or CD69. Sorted cells were analyzed on a FACScan cytofluorograph (Becton Dickinson) to assess their purity.
Activated T cells were isolated from mice previously injected with SEB. Lymph node cells from these animals were passed over nylon wool columns, and these T cell-enriched preparations were sorted to isolate the activated, Vβ8.x-bearing cells after staining with PE-anti Vβ8.x, CyChrome anti-CD4 and anti-CD8, and FL-anti-IAb. Sorting gates were set to include Vβ8.x+ cells bearing CD4 or CD8 and to exclude cells bearing IAb. This procedure thus isolated SEB-stimulated Vβ8.x+ T cells and excluded resting T cells, B cells, and dendritic cells. As above, the effectiveness of the procedure was checked by analysis of the staining profile of the sorted cells. The characteristics of the various sorted cell populations are shown in Table 1. More than 97% of the cells in all three populations bore Cβ (resting T cells) or Vβ8.x. The populations were contaminated with 0.7% or less B cells, dendritic cells, and class II+ macrophages. Residual cells in the SEB-activated populations were resting T cells that bore β other than Vβ8.x (data not shown).
Table 1.
Characteristics of materials used in gene array experiments
| T cell type | No. of cells sorted | % cells bearing Cβ or Vβ8.x | % cells bearing IAb | Yield RNA, μg | μg RNA/107 cells |
|---|---|---|---|---|---|
| Resting | 13.9 × 107 | 99.6 | 0.4 | 115 | 8.2 |
| 8-hr activated | 2.7 × 107 | 98.6 | 0.7 | 38 | 14.1 |
| 48-hr activated | 6.9 × 107 | 97.4 | 0.5 | 136 | 20.0 |
RNA was isolated from these cells by using rapid total RNA isolation kits (5 Prime → 3 Prime). Between 8.2 and 20 μg of total RNA was isolated per 107 cells from the various populations (Table 1). Poly(A)+ mRNA was purified from each of the preparations by using Oligotex mRNA minikits (Qiagen). The quality of the poly(A)+ mRNA was evaluated on the Affymetrix Gene Arrays as described below.
Preparation of cRNA and Gene Chip Hybridization.
cDNA was synthesized from the poly(A)+ mRNA by using SuperScript Choice kits (GIBCO/BRL) and nucleotide primers that contained a sequence recognized by T7 RNA polymerase. cRNA was prepared in an in vitro transcription reaction by using T7 polymerase (MegaScript T7 kit; Ambion, Austin, TX). The quality of the cRNA prepared from the cells was evaluated by control hybridizations with probes built to match the 5′, middle, and 3′ sequences of β actin, glyceraldehyde phosphate dehydrogenase, and 18S RNA. For all three preparations of cRNA the signals obtained for different regions of the same gene were about the same, i.e., the cRNA contained intact coding sequences. Also, the signals obtained by hybridization to 18S RNA were comparatively low, demonstrating that the poly(A)+ mRNA from which the cRNAs were prepared were relatively pure (data not shown). The cRNAs were not significantly contaminated with the products of B cells, dendritic cells, or macrophages because their cRNAs gave little or no signal with probes for Ig, class II MHC, or macrophage-specific proteins, with the exception of class II IEβ (http://www.kmlab.njc.org).
Results
Normalization of the Measurement of Gene Expression in Resting and Activated T Cells.
Both 8- and 48-hr activated T cells contained about twice as much total RNA, and probably about the same amount more poly(A)+ mRNA, than resting T cells (Table 1). However, activated T cells are much larger than resting T cells; therefore, a doubling in the amount of a particular mRNA does not indicate a doubling in the concentration of that mRNA, or the protein it codes for, in the activated cell. For many proteins, concentration is probably more significant than total number of molecules per cell. Therefore, in the discussions below, we chose not to consider the fact that the bulk amount of RNA was increased between resting and activated cells. Rather, we evaluated the concentration of a given mRNA in a sample relative to the entire pool of poly(A)+ transcripts in that pool. To accomplish this, the gene chip signals were normalized to an overall signal, the average of signals for each cRNA preparation on each chip, before analysis.
Anaylsis of Overall Gene Expression in Resting and Activated T Cells.
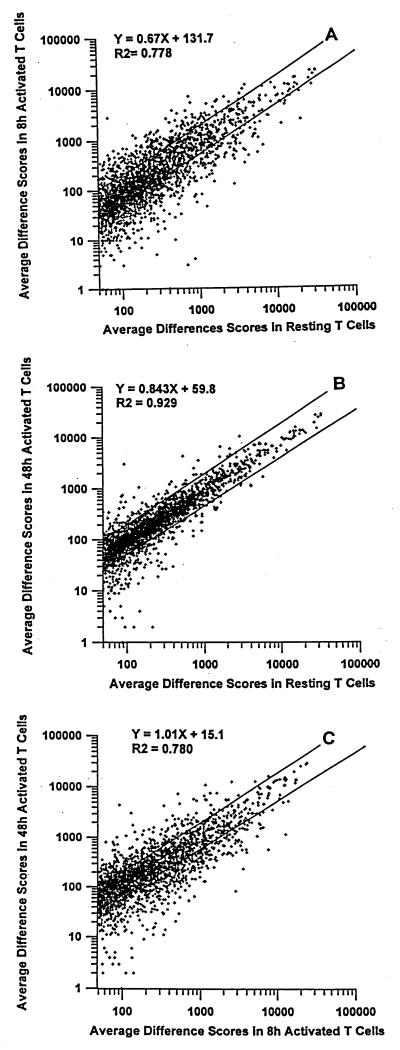
RNA transcript levels for different genes were assessed by using Affymetrix software. The relative abundance of a particular mRNA was expressed as the “average difference.” This is calculated from the difference in fluorescence intensity given by a labeled RNA sample when hybridized to oligos built to match a particular gene sequence vs. when hybridized to oligos mismatched by one base. To get an overall impression of the differences in gene expression between the different types of T cell we plotted the values for average differences obtained for each gene in each type of T cell against their values in the other cells (Fig. 1). Gene expression between resting and 8-hr-activated T cells was quite different as indicated by the scatter in the points on Fig. 1A. Differences in average differences of more than 2-fold for a particular gene between two samples of RNA from different cells are, in general, likely to reflect real differences in gene expression (Affymetrix). Many genes in resting and 8-hr-activated T cells differed by at least this much as indicated in Fig. 1A by the points that lie outside the lines drawn to show 2-fold differences in level. The overall correlation coefficient for Fig. 1A is low, at less than 0.79. Interestingly, the number of genes whose expression decreased upon activation was as large as the number of genes whose expression increased. Hence, transcriptional inhibition in activated T cells was unexpectedly frequent (30).
Figure 1.
Comparison of gene expression in resting and activated T cells. Poly(A) RNA was prepared and converted into fluorescent-labeled cRNA as described in Materials and Methods. The levels of cRNAs derived from different genes were measured by using Affymetrix Gene Arrays and expressed as average differences. The plots compare genes that had average differences greater than 50 in the T cell type against which the comparisons were made. Genes with average differences ≤0 in the index T cell type were omitted from the analyses. These were 57 genes in A, 53 genes in B, and 78 genes in C. The lines drawn on the graphs represent differences in average difference of 2-fold between the two samples considered. (A) Comparison of average differences for gene expression in T cells 8 hr after activation and resting T cells, with 2,758 genes considered. (B) Comparison of average differences for gene expression in T cells 48 hr after activation and resting T cells, with 2,762 genes considered. (C) Comparison of average differences for gene expression in T cells 48 hr after activation and 8 hr after activation, with 2,592 genes considered.
Comparison of overall gene expression between resting T cells and T cells 48 hr after activation with SEB in mice showed fewer differences. Fewer genes differed in average differences by more than 2-fold, and the correlation coefficient was higher, at greater than 0.92. Again, some genes were expressed at higher levels in 48-hr-activated cells than in resting T cells and vice versa (Fig. 1B).
Not surprisingly, given the data in Fig. 1 A and B, T cell gene expression also was quite different when data from cells 8 hr after activation and 48 hr after activation were compared (Fig. 1C). The significance of differences in gene expression in two samples of mRNA was calculated by Affymetrix software by using a combination of actual values of the average differences for that gene in the two samples and the value of the subtraction of the average differences. The parameter thus derived is called the sort score.
To get an overview of the differences in gene expression between resting and activated T cells, we counted the numbers of genes in each comparison that had sort scores greater than 2 in such comparisons and an average difference value in the higher-expressing tissue greater than 100. This cutoff gave a conservative estimate of the numbers of genes that actually changed their expression levels between resting and activated T cells. The results are shown in Table 2. Also shown in Table 2 is the number of RNAs in each sample that had average difference scores of greater than 100.
Table 2.
Activation changes the spectrum but not the diversity of genes expressed in T cells
| T cell type
|
|||
|---|---|---|---|
| Resting | 8-hr activated | 48-hr activated | |
| No. genes with average | 2,057 | 1,852 | 2,056 |
| differences >100 | |||
| No. genes increased* | 143 | 36 | |
| No. genes decreased* | 139 | 15 | |
Genes noted had average differences in the higher-expressing T cells >2 and sort scores ≥2 and are compared with their level in resting T cells.
These data confirm the impression given by Fig. 1. All three types of T cells expressed about the same number of RNAs with average difference scores of greater than 100. RNA expression between resting and 8-hr-activated cells was quite different, with about 280 of the 6,319 genes evaluated as being significantly differently expressed, some expressed more highly in activated cells, and some expressed at greater levels in resting cells. There was much less difference between 48-hr-activated and resting T cells with only 51 genes differing, at this level of sensitivity, in their level of expression. Thus, shortly after exposure to antigen in animals, target T cells dramatically change their mRNA composition. At later times this pattern returns to a composition that is much closer to, but not the same as, that of resting T cells.
Comparison of Individual Gene Expression in Resting and Activated T Cells.
Individual genes were evaluated to find out whether changes in gene expression were consistent with expectations. Signals for housekeeping genes such as HPRT and β-tubulin were unaffected by activation. The levels of CD3, CD4, and CD8 proteins in T cells are known to be unchanged by activation, and the expression of their genes likewise was unaffected. On the other hand, expression of the α-chains for both the IL-6 receptor and IL-7 receptors was reduced at the protein level by activation, and the levels of expression of the genes for these proteins, as detected by the Gene Arrays, also was reduced in activated samples. Finally, surface expression of the α-chain of the IL-2 receptor was transiently increased and that of CD62L was transiently decreased during activation, a result that mirrored the RNA expression data (http://www.kmlab.njc.org; and data not shown).
Occasionally there were discrepancies between the protein and gene expression data. For example, the Gene Array data indicated that expression of the gene for the IL-2 receptor β-chain increased after activation. Levels of this protein on the surface of T cells did not increase, however, until 48 hr after activation. This discrepancy may be because protein synthesis and expression on the cell surface will always, of course, be delayed by comparison with mRNA induction. Thus, for the IL-2 receptor β-chain, 8 hr of activation may have been early enough to observe mRNA induction but too early to observe increases in surface protein. Similarly, mRNA for CD62L fell precipitously by 8 hr after T cell activation. However, cell surface levels of the protein were only halved at this time, again demonstrating a significant temporal delay in levels of protein consequent to changes in mRNA level (http://www.kmlab.nationaljewish.org; and data not shown).
The results for genes that changed in expression level more than 2-fold and that had average differences of greater than 100 in the higher-expressing tissue, between resting and 48-h-activated T cells, are shown in Tables 3 and 4. Expressed sequence tags (ESTs) were omitted from this list because, in our experience, data from ESTs do not necessarily represent values for the gene to which they are thought to be similar.
Table 3.
RNAs increased in 48-hr-activated vs. resting T cells
| Accession no. | Description | Average differences
|
||
|---|---|---|---|---|
| Resting | 8-hr activated | 48-hr activated | ||
| Extracellular matrix and cell adhesion | ||||
| X16834 | Carbohydrate-binding protein, Mac-2 | 110 | 71 | 271 |
| U08020 | Alpha-1 Type 1 collagen | 60 | 176 | 175 |
| U25652 | Alpha-1 Type XII collagen | 41 | 44 | 127 |
| L24430 | Osteocalcin precursor, gla protein | 103 | 16 | 244 |
| D00622 | Heparin-binding protein 44 | 86 | 41 | 221 |
| Cell surface receptors/transporters/proteins | ||||
| X71788 | Blr-1, receptor for chemokine BLC | 34 | 86 | 123 |
| X85214 | Ox40 | 238 | 1,481 | 637 |
| X05719 | CTLA-4 | 831 | 1,477 | 3,706 |
| X98113 | CD4-like cell surface glycoprotein | 4 | 184 | 235 |
| X04653 | Ly-6E.1 | 781 | 5,881 | 1,951 |
| M99377 | Alpha-2 adrenergic receptor | 43 | 18 | 125 |
| M63436 | GABA-A receptor alpha-1 subunit | 20 | 43 | 119 |
| L01776 | Neuronal calcium channel | 42 | 41 | 132 |
| U65593 | K+ channel beta 4 subunit | 39 | 54 | 147 |
| Cell structure/vesicle movement/secretion | ||||
| W29468 | Myosin light chain 2 | 29 | 178 | 147 |
| X97650 | Myosin 1 | 103 | 43 | 293 |
| W13586 | Atrial/fetal myosin light chain | 35 | 2 | 186 |
| X54511 | Mbh1, gelsolin actin-binding protein | 190 | 93 | 448 |
| M26251 | Vimentin | 2,172 | 537 | 4,810 |
| D12646 | kif4, kinesin-like protein | 47 | 37 | 135 |
| Y09632 | Kinesin-like protein 174 | 33 | 30 | 190 |
| M16455 | Calpactin-1 light chain (p11) | 1,586 | 336 | 4,362 |
| D10024 | Calpactin-1 heavy chain | 143 | 110 | 1,086 |
| U66865 | Vacuolar protein-sorting hom. (VPS45) | 35 | 58 | 126 |
| M62418 | Clathrin-associated protein 19 (AP19) | 143 | 188 | 333 |
| L33726 | Fascin | 17 | 19 | 117 |
| W29418 | Fast skeletal troponin C | 36 | 32 | 152 |
| Signal transduction | ||||
| U03856 | CD45-associated protein (cd45-ap) | 1,294 | 1,304 | 2,640 |
| U28168 | Familial adenomatous polyposis | 287 | 285 | 677 |
| D00208 | S100A4 + Ca2+-binding protein, pEL98 | 311 | 61 | 802 |
| AA120244 | S100 Ca2+-binding protein, A13 | 553 | 142 | 1,292 |
| X66449 | Calcyclin | −33 | −122 | 338 |
| M19380 | Calmodulin (Cam III) | 324 | 389 | 929 |
| X65138 | Eph-related receptor tyr kinase | −23 | 50 | 122 |
| U38196 | Mpp-1 | 82 | 61 | 218 |
| Chromatin and nuclear structure | ||||
| X12944 | HMG-17 chromosomal protein | 2,015 | 2,553 | 4,436 |
| X58069 | Histone H3.2-F, H2a.1-F H2b-F | 222 | 319 | 725 |
| X16705 | Iamin B | 89 | 75 | 558 |
| Z46757 | High mobility group 2 protein | 1,606 | 1,106 | 6,829 |
| Cell division | ||||
| X62154 | P1 protein (P1.m) | 45 | 282 | 147 |
| D13545 | Primase large subunit | −7 | 61 | 113 |
| J04620 | Primase small subunit | 78 | 290 | 308 |
| D17384 | DNA polymerase α subunit | 1 | 42 | 105 |
| D12513 | DNA topoisomerase IIα | 50 | 55 | 355 |
| U19604 | DNA ligase I | 23 | 75 | 212 |
| D86726 | mMIS5 | 426 | 1,113 | 1,116 |
| D13473 | RecA-like protein MmRad51 | −3 | 24 | 141 |
| L26320 | Flap endonuclease-1 (FEN-1) | 19 | 187 | 141 |
| Z26580 | Cyclin A | 96 | 80 | 561 |
| X64713 | Cyclin B | 10 | 17 | 123 |
| X66032 | Cyclin B2 | 37 | 19 | 653 |
| X75888 | Cyclin E | 105 | 142 | 215 |
| U63337 | Cyclin-dependent kinase-2α | 41 | 107 | 185 |
| D26091 | mCDC47 | 572 | 1,387 | 1,488 |
| U58633 | p34 CDC2 + B68 | 19 | −5 | 555 |
| U20497 | Cdk4 and Cdk6 inhibitor p19 | 177 | −30 | 491 |
| D21099 | Stk-1 | 10 | −12 | 206 |
| u50378 | Ku70 | 59 | 281 | 165 |
| K02927 | Ribonucleotide reductase M1 | 157 | 323 | 541 |
| X15666 | Ribonucleotide reductase M2 | 74 | 27 | 263 |
| W08120 | Thioredoxin | 2,142 | 7,378 | 6,254 |
| X77731 | Deoxycytidine kinase | 35 | 28 | 178 |
| M68489 | Cytosolic thymidine kinase | −79 | −61 | 347 |
| M13019 | Thymidylate synthase | −38 | 132 | 640 |
| M63445 | Methylenetetrahydrofolate DeH | 65 | 234 | 139 |
| L08266 | Fanconi’s anemia complementation | 23 | 49 | 104 |
| X14805 | Cytosine-5-methyltransferase | 116 | 146 | 314 |
| Transcription | ||||
| X61385 | T cell transcription factor, TSF1 | 526 | 730 | 1,234 |
| Z54283 | Oct-binding factor 1 | 42 | 46 | 138 |
| M16449 | Myb | 338 | 254 | 845 |
| X70472 | B-Myb | −8 | 44 | 158 |
| X72310 | DRTF-polypeptide-1 | 69 | 425 | 224 |
| M83380 | RelB | 86 | 152 | 187 |
| U19 799 | IkB-β | 53 | 357 | 125 |
| M36146 | Zfp-35 | 33 | 14 | 110 |
| X72697 | Xmr meiosis-regulated protein | 179 | 258 | 464 |
| U32394 | Mad3 | 59 | 20 | 134 |
| U46187 | KRAB + Zinc finger protein | 1 | 13 | 104 |
| U41741 | USF | 51 | 94 | 135 |
| Y07836 | Basic helix–loop–helix protein | 74 | 291 | 225 |
| M13018 | Cysteine-rich intestinal protein (CRIP) | 90 | −28 | 226 |
| D26090 | Hox-3.1, Hox 3.2-Hox-3.1 intergenic region | 353 | 1,107 | 1,250 |
| M75953 | Homeobox+, PMUR10F | 28 | 28 | 137 |
| M34857 | Hox-2.5 | 17 | 5 | 125 |
| U52951 | Putative transcrip., reg., mEnx-1 | 59 | 349 | 348 |
| Protein synthesis/degradation | ||||
| U39302 | 26S proteosome sub. 4 ATPase | 233 | 783 | 484 |
| W11011 | Ubiquitin-like protein | −147 | 172 | 121 |
| U48830 | Subtilisin-like convertase-7 | 96 | 44 | 218 |
| Glycolysis/ATP production | ||||
| M32599 | Glyceraldehyde-3-phos. DeH | 2,571 | 3,730 | 5,162 |
| X53333 | Triose phosphate isomerase | 375 | 2,723 | 1,399 |
| AA028501 | Cytochrome c oxidase VIII-H | 14 | −16 | 105 |
| Redox control/control of oxidation damage | ||||
| X82067 | Thioredoxin-dep. peroxide red’ase | 315 | 684 | 657 |
| D49956 | 8-oxo-dGTPase | 88 | 252 | 177 |
| X61147 | Iron-responsive element-binding protein | 52 | 113 | 132 |
| M68896 | Androgen-regulated protein, arMEP24 | 44 | 14 | 119 |
| U48420 | Theta class glutathione transferase type 2 | −34 | −20 | 113 |
| Cell life and death | ||||
| L16462 | A1, Bcl2-like protein | 583 | 510 | 1,257 |
| U54803 | Caspase 3 | 85 | 86 | 719 |
| L37296 | BAD | 12 | 54 | 115 |
| X95591 | C1D | 65 | 33 | 136 |
| X73985 | Calretinin | 122 | 141 | 331 |
| Secreted products | ||||
| X86374 | TAG7, TNF-like cytokine | 147 | 35 | 335 |
| X04072 | Granzyme B | 12 | 3,561 | 238 |
| M13226 | Granzyme A | 88 | 336 | 763 |
| X53257 | Neurotrophin 3 | 23 | −2 | 171 |
| AA124831 | Eosinophil second. gran. protein (mEAR-2) | 80 | 80 | 198 |
| X04573 | Preproelastase | 67 | 60 | 140 |
| X04574 | Preprotrypsin | 61 | 34 | 213 |
| D00466 | Apoplipoprotein E | 78 | 98 | 160 |
| J02644 | Type 1 epidermal keratin | 88 | 83 | 186 |
| Miscellaneous | ||||
| M23236 | Proline-rich protein (MP-2) | 55 | 135 | 148 |
| L21027 | A10 | 209 | 1,307 | 463 |
| U69488 | Viral envelop-like protein (G7e) | 80 | 48 | 415 |
| M34897 | Ecotropic viral integration site 2 ORF | 39 | −103 | 121 |
| M21332 | RNA-binding protein | 1 | 196 | 212 |
| M26270 | Stearoyl-CoA desaturase (SCD2) | 94 | 311 | 437 |
| U42385 | FGF-inducible gene 16 (FIN16) | 34 | 87 | 195 |
| D21099 | Putative ser/thre kinase, Stk1 | 10 | −12 | 206 |
| U10484 | Lymphoid membrane protein, Jaw1 | 46 | 220 | 938 |
| W13002 | β-galactosidase-binding protein | 526 | 91 | 4,460 |
| X82786 | Ki-67 (MIB1) | 36 | 21 | 895 |
| L42293 | Acyl CoA:cholesterol acetyltrans’ase | 72 | 95 | 152 |
| U13837 | Vacuolar ATPase subunit A | 38 | 80 | 101 |
| U232332 | p13MTPC1 | −7 | 12 | 134 |
| X58523 | MIPP | 45 | 28 | 105 |
| X06917 | Aspartate aminotransferase | 250 | 700 | 547 |
| J03857 | B 29 | 56 | 23 | 114 |
| L09192 | Pyruvate carboxylase homo. protein | 1 | 70 | 139 |
Table 4.
RNAs decreased in 48-hr-activated vs. resting T cells
| Accession no. | Description | Average differences
|
||
|---|---|---|---|---|
| Resting | 8-hr activated | 48-hr activated | ||
| Extracellular matrix and cell adhesion | ||||
| U12236 | α M290 integrin, binds β7 integrin | 530 | 53 | 253 |
| X64550 | RHAMM, hyaluronan receptor | 111 | 66 | 40 |
| X84037 | E selectin ligand-1 | 119 | 107 | 54 |
| X58251 | Pro-alpha-2(1) collagen | 174 | 11 | 52 |
| D00613 | Matrix Gla protein | 108 | 38 | 23 |
| Cell surface receptors/transporters/proteins | ||||
| M27960 | IL-4 receptor | 1,634 | 519 | 559 |
| M29697 | IL-7 receptor α-chain | 897 | 83 | 228 |
| X53802 | IL-6 receptor | 549 | 70 | 271 |
| U69599 | IFNγ receptor second chain, ifngr2 | 383 | 203 | 104 |
| D63679 | Decay accelerating factor | 255 | 28 | 91 |
| U36757 | Thrombin receptor | 120 | 96 | 56 |
| X62701 | Urokinase-type plasminogen acti R | 111 | 180 | 17 |
| X99581 | Leucocyte 7 transmembrane R | 1,411 | 557 | 396 |
| X15643 | β-2-Adrenergic receptor | 139 | 33 | 42 |
| X62600 | α-1-Acid glycoprotein, AGP/EB | 318 | 157 | 27 |
| X61433 | Na/K ATPase β-subunit | 649 | 184 | 241 |
| D78572 | LIG-1 | 145 | 77 | 60 |
| D83206 | p24 | 164 | 70 | 70 |
| U49720 | Blue cone pigment | 246 | 90 | 39 |
| X81582 | IGF binding protein 4 | 1,077 | 1,029 | −54 |
| U35836 | Tumor-ass. glycoprotein E4, Tage4 | 111 | 14 | −25 |
| X85992 | Semaphorin C | 134 | 40 | 55 |
| Cell structure/vesicle movement/secretion | ||||
| AA123361 | Rab6/rab5-ass. protein, rab6 | 2,940 | 961 | 1,205 |
| M13444 | α-Tubulin isotype M-α-4 | 1,315 | 1,448 | 629 |
| Signal transduction | ||||
| X02452 | Ki-ras | 368 | 120 | 142 |
| M63630 | GTP-binding protein, IRG-47 | 292 | 462 | 112 |
| U19119 | G protein-like LRG-47 | 221 | 284 | 66 |
| U15636 | U2, T cell GTP-binding protein | 915 | 605 | 155 |
| X51829 | MyD116 | 2,998 | 924 | 1,286 |
| Y08361 | RIL | 175 | 7 | −12 |
| U38252 | Proline-rich RING finger protein | 926 | 631 | 330 |
| X63039 | RSP-1 | 257 | 138 | 114 |
| U58497 | Mnb protein kinase, Dyrk | 183 | 59 | 80 |
| U58885 | SH3-containing protein SH3P8 | 131 | 125 | 26 |
| U58882 | SH3 domain-containing protein, Lasp-1 | 133 | −45 | 28 |
| U58512 | Rho-associated protein kinase | 108 | 44 | 45 |
| U56909 | Tousled-like kinase | 123 | 85 | 49 |
| L01695 | Calmodumin-dep. p.diesterase, PDE1B | 254 | 73 | 54 |
| M96163 | Serum-inducible kinase, SNK | 130 | 33 | 39 |
| U18310 | SEK1 | 123 | 70 | 34 |
| Chromatin and nuclear structure | ||||
| X70887 | Protein like transition protein 2, TP2 | 752 | 1,597 | 349 |
| U62673 | Histones H2a(A)-613, H2a(B)-613, H2b-613 | 103 | 87 | 31 |
| J03482 | Histone H1 | 273 | 55 | 130 |
| U40796 | DNA repair enzyme, ERCC5 | 224 | 34 | 80 |
| Cell cycle | ||||
| U44426 | D52, cell cycle inhibitor | 161 | 180 | 80 |
| Z14986 | S-Admethionine decarbox’ase | 1,314 | 728 | 601 |
| Transcription | ||||
| U25096 | Kruppel-like factor LKLF | 5,727 | 583 | 1,754 |
| U70662 | Kruppel-like factor EZF, Zie | 708 | 529 | 155 |
| U36340 | BKLF | 389 | 101 | 184 |
| U06924 | STAT1 | 1,468 | 873 | 437 |
| J03236 | junB | 11,687 | 2,121 | 5,821 |
| J04115 | c-Jun | 234 | 25 | −25 |
| X14897 | FosB | 514 | 235 | 151 |
| X98096 | Transcription factor BFCOL1 | 1,195 | 286 | 567 |
| X62940 | TSC-22 | 237 | 50 | 68 |
| M58564 | TIS11 | 552 | 311 | 241 |
| U73329 | Dix7, Distal less homeobox gene | 158 | 60 | 60 |
| M82974 | Hen1 | 127 | 59 | 37 |
| U28071 | Hoxc-5 | 105 | 20 | 36 |
| U20282 | Stromelysin PDGF-resp. elem binding ? | 277 | 121 | 94 |
| X61753 | Heat shock transcription factor 1 | 140 | 350 | 14 |
| U13878 | Neural-restrictive silencer factor | 174 | 60 | 85 |
| M34476 | Retenoic acid receptor gamma-A | 198 | 61 | 82 |
| Protein production and degradation | ||||
| W13646 | Polyubiquitin, TI-225 | 3,991 | 12,610 | 1,170 |
| Z19579 | slah-1A | 314 | 81 | 52 |
| L40406 | Heat shock protein 105 kD β | 1,343 | 2,599 | 451 |
| U63323 | Translation init. factor, Eif4g2 | 1,532 | 1,187 | 726 |
| U70674 | B2 element and 18S RNA seq. | 122 | 7 | 2 |
| U16162 | Prolyl-4-hydroxylase alpha-1 | 143 | 204 | 65 |
| U35646 | Aminopeptidase | 171 | 35 | 75 |
| Cell life and death | ||||
| U43678 | Ataxia telangiectasia gene | 106 | 76 | 21 |
| Intermediary metabolism | ||||
| X85983 | Camitine acetyl transferase | 138 | 67 | 64 |
| U53142 | Constitutive nitric oxide synthase | 113 | −10 | −36 |
| X51905 | Lactate dehydrogenase-B | 141 | 55 | 57 |
| U00978 | Type 1 inosine monophosphate DeH | 261 | 596 | 130 |
| Secreted products | ||||
| L38580 | Galanin | 106 | 55 | 14 |
| M11943 | Wnt-1 | 2,334 | 145 | 107 |
| X96618 | Stromal cell protein-inducing RAG | 431 | 184 | 181 |
| Miscellaneous | ||||
| M64292 | TIS21 | 1,623 | 317 | 305 |
| V00727 | Replication-defective murine sarcoma virus | 3,956 | 643 | 1,209 |
| U34072 | Steroid DeH, Ke 6, Ke 6a, Ke 6b | 343 | 24 | 110 |
| U43085 | Glucocorticoid-atten. resp. gene 39, GARG-39 | 197 | 30 | 54 |
| D30785 | Neuropsin, serine protease | 575 | 146 | 276 |
| x96639 | EXT1, analog of human multiple exostosis | 173 | 46 | 33 |
| M29011 | Immunoglobulin α-chain switch region | 123 | −36 | 27 |
| D87744 | Atrophin 1 (DRPLA) | 157 | 48 | 73 |
| U42386 | FIN14, FGF-inducible gene | 1,259 | 564 | 565 |
| X59379 | Nexin II, amyloid beta precursor | 102 | 16 | 13 |
| X67140 | SR calcium ATPase | 227 | 175 | 110 |
| M21532 | PCD-5 | 188 | 65 | 79 |
| X57199 | Lysosomal acid phosphatase | 429 | 206 | 82 |
| Z46720 | Perinuclear-bind. protein, PICK-1 | 159 | −26 | 18 |
| M10021 | Cytochrome P1-450 | 113 | 22 | −8 |
Many genes contributing to cell division were expressed at higher levels in activated vs. resting T cells, as expected. These included the genes for DNA polymerase and primase, for the cyclins, and for many of the enzymes involved in synthesis of DNA precursors. Eight-hour-activated T cells contained higher levels of mRNA for cyclins D and G1, presaging their entry into mitosis about 16 hr later. Forty-eight-hour-activated T cells contained elevated levels of mRNA for cyclins A1, B, and E. Also increased in activated cells were mRNAs for Myb and Myb-B, DNA-binding proteins involved in the stimulation of cell division (refs. 19 and 20; Table 3).
Conversely, some genes that are expressed preferentially in nondividing cells and/or whose products are thought to prevent cell division were expressed at higher levels in resting than activated T cells. Included in these were the genes for the retinoid acid receptor RAR, Dyrk, a protein involved in terminal differentiation and cessation of proliferation, and the proliferation inhibitory transcription factors D52 and TSC-22 (refs. 6 and 21–23; Table 4).
There were large changes in expression of other transcription factors. For example, as reported previously, resting T cells expressed the gene for the Kruppel-like transcription factor, LKLF, at higher levels than activated T cells (ref. 24; Table 4) Less expected was the fact that this also applied to other Krueppel-related transcription factors, EZF and BKLF. Also noteworthy was the increased expression of RelB (19, 25) and IkB-β and several Hox genes in activated cells.
Many papers have shown that members of the Fos/Jun family are very important inducers of gene expression in activated T cells (26). Others have shown that expression of these genes is changed during T cell development, increased upon T cell activation, and decreased in anergic cells (27–29). The analysis in this paper and in a previous report (30) showed a dramatic lowering in levels of mRNA for proteins of the Fos/Jun family and for one of the kinases upstream of activation of these proteins, SEK1. These results suggest that the function of this branch of the MAP kinase signaling pathway may be significantly curtailed in activated T cells.
mRNAs coding for some of the secreted products of T cells, such as the granzymes, increased as the cells were activated. Surprisingly, however, it appeared that resting T cells also make transcripts for some secreted proteins. Overall, the decrease in expression of one of the integrins and a ligand for E-selectin and the increase in expression of enzymes such as trypsin and elastase, which could be involved in tissue penetration, gave the impression that, as is known to occur, the activated T cells were preparing themselves for greater mobility than their resting precursors.
Finally, there were several examples in which expression of a gene in resting T cells appeared to be replaced by expression of a related gene in activated T cells. For example, activated T cells contained more mRNA for Alpha 2 type I collagen and for the transcription factor BFCOL1, which activates this gene (31), and less mRNA for Alpha 1 types I and XII collagen than resting T cells. Likewise, expression of genes related to the basement membrane-binding proteins osteocalcin and matrix Gla protein seesawed in the two types of T cells, as did genes for some of the Hox proteins.
Discussion
Recently, immunologists’ view of the resting T cell has undergone a revolution. Our data and other recent reports have demonstrated that the resting T cell is constantly receiving signals from its environment in the animal. These signals help to keep the cell alive and to guide the cell to various locations (7–14, 32). Given this new appreciation of the activity of resting T cells it perhaps is not surprising to find that mRNAs for many different proteins are expressed at detectable levels in these cells, a result that has been suggested before (30).
Among the factors that are detected by resting T cells, and that help to keep them alive, are IL-4 and IL-7 (12–14). IL-4 also can stimulate the proliferation of activated T cells. However, it is not a good proliferative factor for resting cells. Perhaps the failure of resting T cells to divide in response to IL-4 is because resting cells express proteins such as Dyrk, D52, TSC-22, and LKLF, which may be inhibitors of cell division (21–24). If so, cell division by resting T cells may be, in a sense, actively inhibited by factors within the cell.
Activation caused an approximate doubling in the amount of total RNA per cell, and it is interesting to notice that expression of many of the RNAs in T cells increased concordantly when the cells were activated. This was very striking 48 hr after activation, a time when analysis showed that, in spite of the increase in RNA per cell, once the amounts of RNA per cell had been normalized, the levels of expression of many different genes in resting and activated T cells were the same. Such a result suggests that a global mechanism of transcription regulation was induced by T cell activation, increasing expression of many genes similarly. Such a global mechanism could involve components of the transcription apparatus, p53, or other proteins that concordantly regulate many populations of genes and/or mRNA stability (33, 34).
It is not surprising that many genes change in their level of expression after T cells had been activated for only 8 hr in animals. Perhaps this rapid response is, in part, due to the nature of the antigen we used. Superantigen does not need to be processed by antigen-presenting cells before it can interact with T cell receptors, and, therefore, T cell responses to such material probably will be a few hours faster than that of T cells to conventional protein antigens.
We were surprised to find that, although many genes increase in expression immediately after T cell activation, approximately the same number decrease. Thus, the variety of mRNAs in resting and activated T cells, as previously suggested (30), is about the same in magnitude. Again, such a result may reflect the recently appreciated high receptivity of resting T cells.
We did not expect to find that the mRNA content of T cells 48 hr after exposure to superantigen in animals would be more similar to that of resting T cells than to that of recently (8-hr) activated cells. Forty-eight hours after injection of superantigen, target T cells are still dividing vigorously, although they will stop dividing very shortly thereafter (data not shown). At 48 hr, the activated T cells are also about to die by apoptosis (16, 17), an event that we thought might involve induction of quite a few new genes. We have shown that exposure to reactive oxygen species is the major cause of the death of these cells (35). Nevertheless, we did not find increases in expression of any of the genes on the microarrays that code for proteins that might cause increases in reactive oxygen species concentrations.
There is the question of whether the kinetics and nature of the changes in gene expression that we report here, during T cell responses to superantigen, reflect the kinds of change that take place during T cell responses to conventional peptide antigens and/or infectious organisms. The kinetics of T cell responses to peptides and proteins administered in the absence of adjuvants are very similar to those of T cells responding to superantigens. Therefore, we believe that such responses will prove to be very similar to those described in this paper. Under nonlaboratory conditions, however, T cells usually encounter superantigens and conventional antigens in the presence of infections. Infectious organisms and laboratory adjuvants induce components of innate immunity. Such components directly or indirectly affect T cell responses such that the responses are larger in magnitude and the responding T cells are more long-lived (17, 36). Infectious agents therefore change gene expression in activated T cells, and the pattern of gene expression in activated T cells described in this paper thus probably is not identical to that which will be found in T cells activated in the presence of bacterial or viral products.
Acknowledgments
We thank Drs. Clive Slaughter and Steven Madden, University of Texas Southwestern Medical Center, Dallas, for their help in preparation of the cRNA samples, for conducting the Affymetrix gene array hybridizations, and for their patient advice afterward. We thank Drs. Louis Staudt, Gary Johnson, and James Hagman for reading the manuscript and for their very helpful suggestions. We also thank Bill Townend at the National Jewish Medical and Research Center for his help with cell sorting. This work was supported by Public Health Service Grants AI-17134, AI-18785, and AI-22295.
Abbreviation
- SEB
staphylococcal enterotoxin B
References
- 1.Zipfel P F, Irving S G, Kelly K, Siebenlist U. Mol Cell Biol. 1989;9:1041–1048. doi: 10.1128/mcb.9.3.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zheng W, Flavell R A. Cell. 1997;89:587–596. [Google Scholar]
- 3.Choi J W, Lee S Y, Choi Y. Cell Immunol. 1996;168:78–84. doi: 10.1006/cimm.1996.0051. [DOI] [PubMed] [Google Scholar]
- 4.Liu A Y, Torchia B S, Migeon B R, Siliciano R F. Genomics. 1997;39:171–184. doi: 10.1006/geno.1996.4463. [DOI] [PubMed] [Google Scholar]
- 5.Renner C, Pfitzenmeier J P, Gerlach K, Held G, Ohnesorge S, Sahin U, Bauer S, Pfreundschuh M. J Immunol. 1997;159:1276–1283. [PubMed] [Google Scholar]
- 6.Ishaq M, Zhang Y M, Natarajan V. J Biol Chem. 1998;273:21210–21216. doi: 10.1074/jbc.273.33.21210. [DOI] [PubMed] [Google Scholar]
- 7.Kirberg J, Berns A, von Boehmer H. J Exp Med. 1997;186:1269–1275. doi: 10.1084/jem.186.8.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rooke R, Waltzinger C, Benoist C, Mathis D. Immunity. 1997;7:123–134. doi: 10.1016/s1074-7613(00)80515-4. [DOI] [PubMed] [Google Scholar]
- 9.Tanchot C, Lemonnier F A, Perarnau B, Freitas A A, Rocha B. Science. 1997;276:2057–2062. doi: 10.1126/science.276.5321.2057. [DOI] [PubMed] [Google Scholar]
- 10.Bender J, Mitchell T, Kappler J, Marrack P. J Exp Med. 1999;190:367–374. doi: 10.1084/jem.190.3.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Teague T K, Marrack P, Kappler J, Vella A T. J Immunol. 1997;158:5791–5799. [PubMed] [Google Scholar]
- 12.Boise L H, Minn A J, June C H, Lindsten T, Thompson C B. Proc Natl Acad Sci USA. 1995;92:5491–5496. doi: 10.1073/pnas.92.12.5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Akbar A N, Borthwick N J, Wickremasinghe R G, Panayoitidis P, Pilling P, Bonfill M, Krajewski S, Reed J C, Salmon M. Eur J Immunol. 1996;26:294–299. doi: 10.1002/eji.1830260204. [DOI] [PubMed] [Google Scholar]
- 14.Vella A T, Dow S, Potter T A, Kappler J, Marrack P. Proc Natl Acad Sci USA. 1998;95:3810–3815. doi: 10.1073/pnas.95.7.3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boursalian T E, Bottomly K. J Immunol. 1999;162:3795–3801. [PubMed] [Google Scholar]
- 16.Kawabe Y, Ochi A. Nature (London) 1991;349:245–248. doi: 10.1038/349245a0. [DOI] [PubMed] [Google Scholar]
- 17.Vella A T, McCormack J E, Linsley P S, Kappler J W, Marrack P. Immunity. 1995;2:261–270. doi: 10.1016/1074-7613(95)90050-0. [DOI] [PubMed] [Google Scholar]
- 18.Julius M H, Simpson E, Herzenberg L. Eur J Immunol. 1973;3:645–649. doi: 10.1002/eji.1830031011. [DOI] [PubMed] [Google Scholar]
- 19.Kelly K, Siebenlist U. J Biol Chem. 1988;263:4828–4831. [PubMed] [Google Scholar]
- 20.Torelli G, Selleri L, Donelli A, Ferrari S, Emilia G, Venturelli D, Moretti L, Torelli U. Mol Cell Biol. 1985;5:2874–2877. doi: 10.1128/mcb.5.10.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Becker W, Joost H G. Prog Nucleic Acid Res Mol Biol. 1999;62:1–17. doi: 10.1016/s0079-6603(08)60503-6. [DOI] [PubMed] [Google Scholar]
- 22.Byrne J A, Nourse C R, Basset P, Gunning P. Oncogene. 1998;16:873–881. doi: 10.1038/sj.onc.1201604. [DOI] [PubMed] [Google Scholar]
- 23.Kawamata H, Nakashiro K, Uchida D, Hino S, Omotehara F, Yoshida H, Sato M. Brit J Cancer. 1998;77:71–80. doi: 10.1038/bjc.1998.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuo C T, Veselits M L, Leiden J M. Science. 1997;277:1986–1990. doi: 10.1126/science.277.5334.1986. [DOI] [PubMed] [Google Scholar]
- 25.Kahn-Perles B, Lipcey C, Lecine P, Olive D, Imbert J. J Biol Chem. 1997;272:21774–21783. doi: 10.1074/jbc.272.35.21774. [DOI] [PubMed] [Google Scholar]
- 26.Jain J, Valge-Archer V E, Rao A. J Immunol. 1992;148:1240–1250. [PubMed] [Google Scholar]
- 27.Chen F, Chen D, Rothenberg E. Int Immunol. 1999;11:677–688. doi: 10.1093/intimm/11.5.677. [DOI] [PubMed] [Google Scholar]
- 28.Shin H M, Han T M. Mol Immunol. 1999;36:197–203. doi: 10.1016/s0161-5890(99)00030-9. [DOI] [PubMed] [Google Scholar]
- 29.Sundstedt A, Dohlsten M. J Immunol. 1998;161:5930–5936. [PubMed] [Google Scholar]
- 30.Alizadeh A, Eisen M, Botstein D, Brown P O, Staudt L M. J Clin Immunol. 1998;18:373–379. doi: 10.1023/a:1023293621057. [DOI] [PubMed] [Google Scholar]
- 31.Hasegawa T, Takeuchi A, Miyaishi O, Isobe K, de Crombrugghe B. J Biol Chem. 1997;272:4915–4923. doi: 10.1074/jbc.272.8.4915. [DOI] [PubMed] [Google Scholar]
- 32.Jung T M, Gallatin W M, Weissman I L, Dailey M O. J Immunol. 1988;141:4110–4117. [PubMed] [Google Scholar]
- 33.Sauer F, Tijan R. Curr Opin Genet Dev. 1997;7:176–181. doi: 10.1016/s0959-437x(97)80126-8. [DOI] [PubMed] [Google Scholar]
- 34.el-Deiry W S. Semin Cancer Biol. 1998;8:3445–3457. doi: 10.1006/scbi.1998.0097. [DOI] [PubMed] [Google Scholar]
- 35.Hildeman D A, Mitchell T C, Teague T K, Henson P, Day B J, Kappler J, Marrack P. Immunity. 1999;10:735–744. doi: 10.1016/s1074-7613(00)80072-2. [DOI] [PubMed] [Google Scholar]
- 36.Mitchell T, Kappler J, Marrack P. J Immunol. 1999;162:4527–4535. [PubMed] [Google Scholar]