Abstract
Inheritance of an inactivated form of the VHL tumor suppressor gene predisposes patients to develop von Hippel–Lindau disease, and somatic VHL inactivation is an early genetic event leading to the development of sporadic renal cell carcinoma. The VHL gene was disrupted by targeted homologous recombination in murine embryonic stem cells, and a mouse line containing an inactivated VHL allele was generated. While heterozygous VHL (+/−) mice appeared phenotypically normal, VHL −/− mice died in utero at 10.5 to 12.5 days of gestation (E10.5 to E12.5). Homozygous VHL −/− embryos appeared to develop normally until E9.5 to E10.5, when placental dysgenesis developed. Embryonic vasculogenesis of the placenta failed to occur in VHL −/− mice, and hemorrhagic lesions developed in the placenta. Subsequent hemorrhage in VHL −/− embryos caused necrosis and death. These results indicate that VHL expression is critical for normal extraembryonic vascular development.
Germ-line mutations in the VHL tumor suppressor gene predispose von Hippel–Lindau disease patients to develop tumors at multiple sites, including retinal angiomas, hemangioblastomas of the central nervous system, pheochromocytomas, renal cell carcinomas, and pancreatic cancers (reviewed in refs. 1 and 2). In addition, somatic VHL inactivation through deletion of one allele coupled with either mutation or hypermethylation of the remaining allele has been detected in approximately 80% of sporadic clear cell renal carcinomas examined (2), supporting a tumor suppressor function for VHL.
The deduced amino acid sequence of the VHL protein gives no indication of the protein’s function. Studies designed to assign VHL function through the identification of physically associating proteins found that VHL stably binds the B and C subunits of the elongin complex (3–5). Elongin, an RNA polymerase II transcription elongation factor, contains a catalytic A subunit that is stabilized and activated by the B and C regulatory subunits. In addition, the heterotrimeric VHL-elongin B/C complex (VBC) stably interacts with the human CUL-2 homolog, a member of a conserved gene family involved in cell cycle and growth control in lower eukaryotes (6). The cellular function(s) of the VBC and/or VBC-CUL-2 complexes have yet to be elucidated.
To analyze the role of VHL in normal cell growth and differentiation, we used targeted homologous recombination to develop a mouse line that is deleted for one VHL allele. Heterozygous VHL mice (+/−) have survived thus far to beyond 15 months without evidence of spontaneous disease. However, VHL-deficient mice (−/−) develop placental lesions at 9.5 to 10.5 days of gestation (E9.5 to E10.5) and die in utero between E10.5 to E12.5 due to the absence of placental embryonal vasculogenesis and subsequent hemorrhage and necrosis.
MATERIALS AND METHODS
Construction of a Murine VHL-Targeting Vector.
Two genomic DNA clones corresponding to the VHL gene were isolated from a 129 mouse-derived P1 library (Genome Systems, St. Louis). Physical mapping showed that three BamHI restriction fragments of 5 kb, 6 kb, and 4 kb contained exons 1, 2, and 3, respectively. These fragments were subcloned into pBluescript II (Stratagene), and partial sequence analysis showed that the human and murine VHL genes had a conserved intron/exon structure (see also ref. 7). The plasmid pPNT, containing pgk-neo and a pgk-HSV thymidine kinase selectable markers, was used to construct the VHL targeting vector (pPNT-VHL). A 3-kb KpnI–NotI fragment containing the VHL promoter and first 24 codons of exon 1 was cloned into pPNT (the NotI site of the insert was filled in with DNA polymerase before cloning, thus destroying that site and maintaining, as unique, the single NotI site in pPNT). In addition, a 3-kb NcoI–BamHI fragment containing 3′ untranslated sequences of the third VHL exon (the NcoI site is seven nucleotides 3′ of the translation termination codon) was also cloned into pPNT, such that the exon 1 and exon 3 sequences flanked the neomycin-resistance gene, and the herpes simplex virus tk gene was outside of the regions of VHL homology. At the targeted allele, neo would be transcribed from the opposite strand as VHL.
Targeted Disruption of VHL in Embryonic Stem (ES) Cells.
The VHL targeting vector, pPNT-VHL, was linearized with NotI, and 20 μg of DNA was electroporated (Bio-Rad, 400 volts, 25 μF) into J1 ES cells. After electroporation 2.5 × 105 cells per dish were plated in 60-mm2 tissue culture dishes containing a monolayer of G418-resistant embryonic fibroblasts. Selection was initiated 24 hr after plating using 400 μg/ml G418 (GIBCO/BRL) and 2 μM gancyclovir (Syntex, Palo Alto, CA). Double selection resulted in an approximate 10-fold enrichment, and resistant colonies were isolated and expanded after 5 to 6 days of selection.
Generation of Chimaeric Mice and VHL +/− Mice.
Two ES cell clones, G15 and G35, were introduced into blastocysts of C57BL/6 embryos and injected into pseudopregnant foster mothers to complete development. Chimaeric offspring were identified based on the contribution of agouti coat color and mated with C57BL/6 males and females. Agouti offspring then were analyzed for VHL disruption as described below.
Genotyping of Targeted ES Cells, VHL Mice, and Embryos.
Genomic DNA was extracted from cultured ES cells, mouse tail biopsies, and embryo yolk sacs as previously described (8). For Southern analysis, DNA was digested with either BamHI or HindIII, electrophoresed on 1% agarose gels, and capillary blotted to nylon membranes. Probes flanking the targeted VHL sequences were labeled with a random primer labeling kit (Stratagene) and used in hybridizations. Wild-type and mutated alleles were identified by predicted restriction fragment size differences. Eight of 160 ES clones examined showed homologous recombination of pPNT-VHL with 5′ and 3′ flanking probes. Digestion of ES cell DNA with enzymes that did not cut within the targeting vector and Southern blotting with a neo probe showed only a single hybridizing fragment, indicating the there was only a single site of vector insertion in the targeted ES cell clones. In some cases mouse tail biopsy DNA was digested with NcoI instead of HindIII. It was found that the wild-type VHL locus could not be identified in these samples after HindIII digestion, perhaps because of loss of recognition as a result of methylation at the site. PCR also was used to genotype DNA isolated from mouse tail biopsies or embryo yolk sacs. Oligonucleotide primer pairs included those specific for wild-type VHL exon 3, 5′-CCTGAAAGAGCGGTGCCTTCAGGT-3′ (forward) and 5′-GGCTGAAACTCAGGAACACT-3′ (reverse), and wild-type VHL exon 1 primers, 5′-CGAGCCCTCTCAGGTCATCT-3′ (forward) and 5′-CGGTGCCCGGTGGTAAGATC-3′ (reverse). An oligonucleotide primer specific for the targeted allele was from pgk-neo, flanking the NcoI site in VHL exon 3 (5′-CTACCCGGTAGAATTGACCT-3′) and was used in combination with the exon 3 reverse primer shown above. Oligonucleotide primers corresponding to internal neo sequences were, 5′-ATGATTGAACAAGATGGATTGCACG-3′ (forward) and 5′-CGGCACTTCGCCCAATAGCAGCC-3′ (reverse).
Microdissection of Abnormal Embryos and Genotyping.
Tissue from abnormal embryos was isolated using laser capture microdissection (9). This technique allowed for the procurement of viable tissue from abnormal embryos, without contamination from necrotic embryonal tissues or maternally derived tissues. DNA was extracted as described (9), and PCR analysis was performed in triplicate using the primers described above in the presence of [32P]α-dCTP. Products were electrophoresed on polyacrylamide/urea gels and visualized by autoradiography.
Histologic Analysis of Embryos.
Mouse placentas and embryos from E8.5 to E16.5 were fixed in the uterus in neutral buffered formalin or 4% paraformaldehyde. After fixation, embryos were carefully dissected to grossly examine the embryo and placenta, embedded in paraffin, and sectioned at 5 μm. Sections were stained with hematoxylin and eosin. Immunohistochemistry was performed after paraformaldehyde, ethanol, or formalin fixation using antibodies directed against VHL or VEGF (Santa Cruz Biotechnology), avidin–biotin complex development, and hematoxylin counterstaining.
In Situ Hybridization.
Paraformaldehyde or formalin-fixed, paraffin-embedded sections of placentas from VHL and control mice were used for in situ hybridization to determine VHL mRNA expression. A 3-kb NcoI–BamHI insert corresponding to the 3′ untranslated region of the VHL mRNA (see above) was cloned into pBluescript and pGem 7Z plasmids and used to generate radiolabeled antisense and sense strand RNA probes. RNA probes were diluted to a specific activity of 2 × 106 dpm/μl and hydrolyzed with alkali before hybridization. In situ hybridization was performed as previously described (10, 11). Briefly, histologic sections were dewaxed and hydrated, blocked in acetic anhydride and succinic anhydride, and hybridized overnight. After stringent washes and RNase digestion, the slides were coated with Kodak NTB2 emulsion and exposed for 4 days. Slides were developed in diluted D-19, fixed, and viewed with transmitted and darkfield illumination.
RESULTS
Generation of VHL Mice.
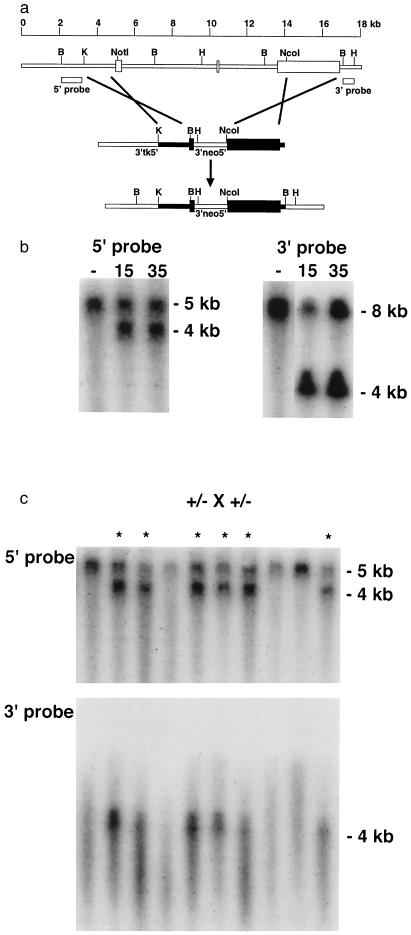
The murine VHL genomic locus was molecularly cloned and mapped, and a targeting vector was constructed. The targeting vector was designed such that the sequences between the exon 1 NotI site and the exon 3 NcoI site were replaced with the neomycin-resistance gene (Fig. 1a). ES cells were electroporated with the linearized VHL targeting vector and were subjected to G418 and gancyclovir double selection. Southern blotting analysis showed that 8 of 160 double-resistant clones tested had targeted disruption of one VHL locus (Fig. 1b). Although the VHL promoter, the first 23 codons of the 181 amino acid coding sequence (7), and the 3′ untranslated region would be retained at the disrupted locus, expression analysis using coupled reverse transcription and PCR failed to detect a VHL transcript from the targeted locus in ES cells (data not shown). Southern blotting using the neo gene as a probe showed that the targeted ES cells contained only a single insertion of the targeting vector (data not shown). Two ES cell clones, G15 and G35, were injected into C57BL/6 recipient blastocysts and produced a total of nine chimaeric mice. Seven G35 chimaeras had strong ES cell contribution based on agouti coat color and were bred to C57BL/6 mice. All seven achieved germ-line transmission of the disrupted VHL allele at a very high frequency. Two G15 chimeras had poor ES cell contribution and did not transmit the disrupted VHL allele.
Figure 1.
Targeted disruption of the murine VHL gene in ES cells and mice. (a) Schematic representation of the VHL locus and the VHL targeting vector. Boxes represent the three VHL exons. The open boxes in the targeting vector represent the pgk-neo and pgk-tk selectable markers. The filled boxes in the targeting vector represent a 3-kb KpnI–NotI restriction fragment from the 5′ end of the VHL gene and a 3-kb NcoI–BamHI restriction fragment from the 3′ end of the gene. The 5′ and 3′ flanking probes used in Southern blotting are indicated. (b) Detection of wild-type and targeted alleles in ES clones 15 and 35 and a representative, untargeted ES clone (−) by Southern blotting. After homologous recombination of the targeting vector in ES cells, BamHI digestion and Southern blotting with the 5′ flanking probe detected in addition to the 5-kb fragment from the wild-type locus, a 4-kb fragment from the targeted locus due to the introduction of a new BamHI site in the targeting vector. Similarly, HindIII digestion and Southern blotting with the 3′ flanking probe detected the wild-type 8-kb fragment and a 4-kb fragment at the targeted locus, due to the insertion of a HindIII site in the targeting vector. (c) Southern analysis of BamHI-digested (Upper) or HindIII (Lower) mouse tail biopsy DNA from a representative intercross litter, showing absence of VHL homozygous mice. The wild-type, 8-kb locus could not be identified in HindIII digests of DNA extracted from mouse tail biopsies, perhaps because of loss of recognition due to methylation interference.
Genotyping VHL Mice and Embryos.
Heterozygous VHL (+/−) offspring appeared phenotypically normal, and the oldest have survived beyond 15 months of age without evidence of significant spontaneous disease. Genotypic evaluation of weaned offspring from VHL heterozygous intercrosses (+/− × +/−), and Southern blot analysis of a representative litter is shown in Fig. 1c. Overall, genotyping of VHL heterozygous intercrosses revealed an approximate 2:1 ratio of VHL heterozygotes to +/+ mice and the absence of VHL-deficient (−/−) mice (Table 1). Because there was no evidence of perinatal death during the first 3 weeks of age (at which point the pups were genotyped), this indicated that VHL homozygotes died in utero. Embryos from heterozygous intercrosses were analyzed, and gross analysis revealed numerous abnormal placental sites containing hemorrhagic lesions and in the process of resorption (Fig. 2). From 10 litters examined between E8.5 and E12.5, 43 embryos (78%) appeared phenotypically normal and 12 (22%) appeared phenotypically abnormal (Table 1). Three abnormal placental sites were identified as late in gestation as E14.5, and none were seen at E16.5 or E18.5 (Table 1). Genotyping of the normal, live embryos by PCR identified 28 VHL +/− embryos and 15 VHL +/+ embryos in these litters, a ratio similar to that observed with the weanlings (Table 1). None of the phenotypically normal embryos were VHL −/− (Table 1). However, genotyping of the phenotypically abnormal embryos in the process of resorption proved extremely difficult, due to the presence of necrotic tissue and the inability to isolate yolk sacs without contaminating maternal blood. Therefore, the technique of laser capture microdissection (9) was used to analyze additional litters. Serial sections from normal or abnormal embryos were visualized microscopically, and maternal or embryonic tissues were isolated from 14 abnormal placental sites from six litters. Analysis of laser capture microdissection-procured DNA showed that 12 of 14 abnormal placentas and/or embryos tested contained the targeted locus but failed to amplify wild-type VHL exon 2 sequences (Fig. 3 and data not shown), indicating that these placentas and embryos had VHL −/− genotypes. All of the maternal samples tested showed +/− genotypes (Fig. 3 and data not shown). The remaining 2 of 14 abnormal placentas and embryos tested also showed +/− genotypes (data not shown). Whereas some VHL +/− placentas and embryos may exhibit an abnormal phenotype and embryonic lethality, spontaneous death and resorption of mouse embryos is quite common, and these two embryos may have developed abnormalities unrelated to VHL disruption (see also below). In addition, we cannot rule out the possibility that some maternally derived tissue was procured in the laser capture microdissection process, thereby, inappropriately assigning a +/− genotype based on positive amplification of VHL exon 2 sequences. However, it is clear that the high rate of embryonic lethality in VHL-deficient mice was likely due to homozygous VHL disruption.
Table 1.
Genotypes and gross analysis of offspring from VHL +/− intercrosses
| Days postcoitum | Total embryos (litters) | Genotype of live embryos
|
Abnormal sites | ||
|---|---|---|---|---|---|
| +/+ | +/− | −/− | |||
| Weanlings | 256 (45) | 75 | 181 | 0 | |
| 8.5 | 13 (2) | 3 | 8 | 0 | 2 |
| 10.5 | 22 (4) | 7 | 11 | 0 | 4 |
| 12.5 | 20 (4) | 5 | 9 | 0 | 6 |
| 14.5 | 27 (4) | 10 | 14 | 0 | 3 |
| 16.5 | 25 (4) | 6 | 19 | 0 | 0 |
| 18.5 | 17 (3) | 5 | 12 | 0 | 0 |
The genotypes of weanlings (approximately 3 weeks of age) or embryos were determined by either Southern blotting or PCR of DNA prepared from either tail biopsies or yolk sacs. PCR genotyping of the abnormal sites was ambiguous because of poor-quality template DNA and contaminating maternal blood in the tissues and is not reported in this table.
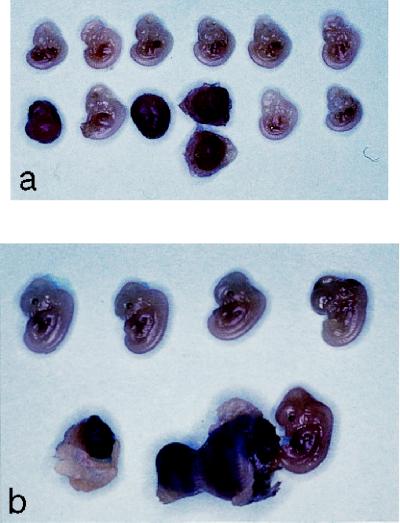
Figure 2.
Gross appearance of E12.5 embryos from intercross of VHL +/− mice. (a) Representative litter showing three abnormal embryos of 12 total. VHL −/− embryos are necrotic, and one embryo is shown with a hemorrhagic placenta. (b) Photomicrograph of another representative E12.5 litter showing one abnormal embryo and one embryo of normal appearance that is attached to a hemorrhagic placenta.
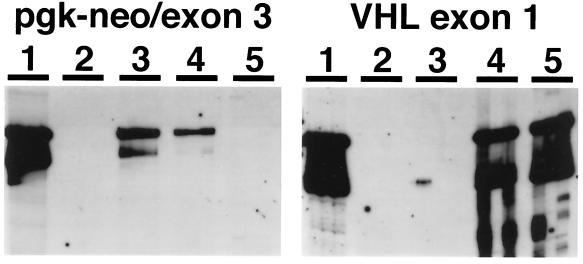
Figure 3.
Genotyping of abnormal embryos after laser capture microdissection and PCR. The PCR primers tested were specific for VHL exon 1 (reactive only with the wild-type allele), or specific for the targeted locus (the amplicon spans the pgk-neo/exon 3 boundary of the targeted locus). DNA samples were analyzed in triplicate and VHL −/− embryos were identified based on amplification with the targeted allele-specific primers and failure to amplify with VHL exon 1 primers. Group 1, positive control reaction products from PCR using VHL +/− mouse tail biopsy template DNA; group 2, negative control reaction products from PCR reactions containing no added DNA; group 3, PCR reaction products using template DNA isolated from a representative abnormal E10.5 embryo and showing a VHL −/− genotype; group 4, PCR reaction products using template DNA isolated from a portion of maternally derived placenta from a VHL +/− mother; group 5, PCR reaction products using template DNA isolated from a representative normal E10.5 embryo and showing a VHL +/+ genotype.
Histologic Evaluation of VHL Embryos and Placentas.
The histology of embryos from VHL heterozygous intercrosses was studied in detail. In total, 63 placental sites from seven litters from gestational days 9.5 to 12.5 were examined. Seventeen of the 63 placental sites (27%) showed placental and embryonic lesions, and 14 were evaluable by PCR genotyping (see above). Both placenta and embryo appeared to develop normally until about E9.5. At this gestational age (just after chorioallantoic fusion) the placental labyrinth is beginning to develop (12). A common feature of VHL −/− embryos was a lack of embryonic endothelium and embryonic blood vessels in the placental labyrinth. Embryonic vessels with nucleated erythrocytes were identified on the edge of the labyrinth, but no vessels were found to invade the labyrinth (Fig. 4b). The trophoblasts appeared normal and viable until about E9.5, but failed to progress to development of syncytiotrophoblasts. Therefore, VHL −/− embryos show defective vasculogenesis within the placental labyrinth, but not at other sites.
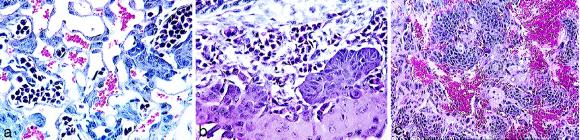
Figure 4.
Histopathology of normal and VHL −/− embryos. (a) Hematoxylin and eosin staining of a normal E10.5 placental labyrinth showing trophoblasts and nucleated erythrocytes in the embryonic vessels and non-nucleated maternal erythrocytes (×300). (b) Hematoxylin and eosin staining of a VHL −/− E10.5 placental labyrinth lacking embryonal vasculogenesis (×300). Note the presence of embryonic vessels with nucleated erythrocytes on the edge of the thin labyrinth that have not invaded the labyrinth (arrows). (c) Hematoxylin and eosin staining of a VHL −/− E11.5 placental showing hemorrhage into the abnormal labyrinth (×150).
By E11.5 to E12.5 the placental labyrinth showed great disruption, loss of normal structure, and hemorrhage (Fig. 4c). These abnormalities ultimately led to the development of a hemorrhagic and necrotic placental site. The VHL −/− embryos were generally normal histologically until the placental lesion developed, at which point the embryo became necrotic. As a control, phenotypically abnormal embryos and placentas from normal C57BL/6 mouse crosses were evaluated. Two of 36 embryos (6%) from four litters between E8.5 and E12.5 were determined to be abnormal by gross examination. Whereas embryonic abnormalities were noted histologically in these embryos, none displayed the characteristic placental lesions seen in the VHL mice. Therefore, it is clear that the embryonic lethality seen in VHL-deficient mice is due to a lack of placental vasculogenesis and subsequent placental failure.
VHL and Vascular Endothelial Cell Growth Factor (VEGF) Expression in the Placenta.
VHL expression was analyzed in the placental labyrinth of normal and abnormal embryos. High levels of VHL mRNA expression were detected by in situ hybridization in the placental labyrinth of normal embryos at E10.5 to E12.5 (Fig. 5a). Lower levels of VHL expression were detected in the yolk sac and other embryonic tissues (data not shown). No VHL mRNA was detected in VHL −/− embryos (Fig. 5b). In addition, VHL was detected by immunohistochemical staining in the labyrinth trophoblasts, the allantoic mesoderm, and some placental embryonic endothelium of normal placentas and embryos (Fig. 5 c and d). Therefore, VHL mRNA and protein are normally expressed in the placental labyrinth and mesoderm, and loss of expression in VHL-deficient embryos is associated with the lack of vasculogenesis at that site.
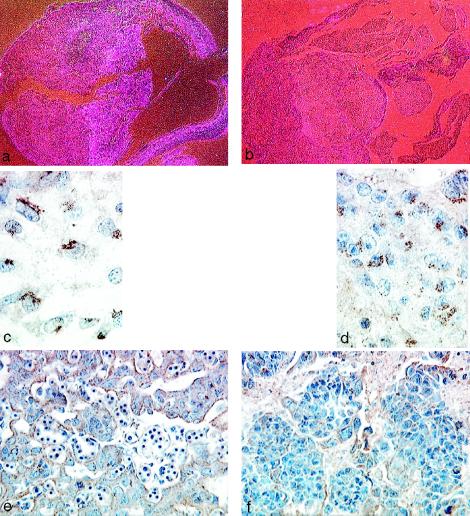
Figure 5.
Analysis of VHL and VEGF expression in normal and VHL −/− E10.5 embryos. (a) Darkfield photomicrograph of VHL in situ hybridization of a normal embryo using an antisense probe (×30). High levels of VHL expression are seen in the placental labyrinth. (b) Darkfield photomicrograph of VHL in situ hybridization of a VHL −/− embryo using an antisense probe showing no significant hybridization (×30). (c and d) Avidin–biotin complex immunohistochemistry of a normal VHL +/+ E10.5 placenta showing VHL expression in the allantoic mesenchyme (c, ×750) and placental labyrinth trophoblasts (d, ×300). (e) Avidin–biotin complex immunohistochemistry of a normal E10.5 placenta showing VEGF expression in the placental labyrinth trophoblasts (×300). (f) Avidin–biotin complex immunohistochemistry of a VHL −/− E10.5 placenta showing greatly reduced VEGF protein levels in the placental labyrinth trophoblasts (×300).
Because VEGF expression is negatively regulated by VHL in renal carcinoma cell lines (13–15) and VEGF is known to regulate vasculogenesis (16–19), placentas from normal and VHL-deficient embryos were analyzed for VEGF protein. High levels of VEGF protein were found in the placental labyrinth trophoblasts of normal E10.5 embryos (Fig. 5e), demonstrating that both VHL and VEGF are simultaneously expressed in the placental labyrinth trophoblasts. However, contrary to what would have been predicted from the above studies, VEGF protein levels were greatly reduced in the labyrinth trophoblasts of VHL-deficient placentas (Fig. 5f). These results imply that VHL regulation of VEGF expression may differ in placental labyrinth trophoblasts as compared with renal carcinoma cells.
DISCUSSION
Among the earliest differentiation events In the mammalian embryo are those that direct cells to develop into the extraembryonic tissues, the placenta, the yolk sac, and the amnion (20). The placenta is derived from trophoblast cells and mesodermal cells from the inner cell mass, and these cell types appear histologically normal in VHL-deficient mice. However, loss of normal VHL expression in the trophoblasts and mesodermal cells appears to account for the lack of vasculogenesis in the placental labyrinth, leading to placental failure. On the other hand, the yolk sac is derived from endodermal cells of the inner cell mass, and mesenchymal cells in the wall of the yolk sac condense to form blood islands (20). Blood islands differentiate to form vessel endothelial cells and primitive blood cells. VHL expression is detected in the yolk sac endoderm, but not the endothelium of normal embryos (data not shown). In VHL-deficient mice, the majority of yolk sacs examined appeared normal, and primitive blood cells could easily be discerned. However, in some embryos the yolk sacs appeared atrophic and had decreased numbers of blood islands (data not shown). Thus, whereas VHL may play some role in development of the yolk sac and blood islands, the major cause of embryonic lethality in VHL-deficient mice is placental failure.
Control of VEGF expression has been shown to occur both at the level of transcription initiation, and in response to hypoxia, through mRNA stability (21–24). Negative regulation of VEGF expression by VHL was previously demonstrated in renal carcinoma cell lines containing inactivated endogenous VHL and expressing high levels of VEGF (13–15). Re-expression of wild-type VHL after gene transfer results in decreased VEGF levels in those cells due to decreased VEGF mRNA stability rather than transcription (14, 15). However, in this study VEGF expression was found to be decreased in VHL-deficient placental labyrinth trophoblasts. Whereas the placental labyrinth trophoblasts seen in VHL-deficient placentas appeared normal and viable, further investigation will be required to determine whether this decrease is directly related to VHL inactivation (indicating an alternative form of regulation in placental labyrinth trophoblasts than seen in renal carcinoma cells) or a secondary effect and an early indicator of placental failure.
Whereas a number of factors are known to stimulate or inhibit endothelial cell proliferation and vessel formation (25), VEGF is the only known factor shown thus far to be involved in vasculogenesis (16–19). VEGF is the ligand for a family of receptor tyrosine kinases that include Flt-1/VEGF-R1, Flk-1/VEGF-R2, and Flt-4/VEGF-R3 (26, 27). Mice deficient for VEGF, Flt-1, or Flk-1 have been developed and show embryonic lethality by E9.5 to E11.5 (16–19), similar to the VHL-deficient embryos described in this report. Lethality in VEGF and VEGF receptor mice appears to involve defects in vasculogenesis as well as blood island development (hematopoiesis). Interestingly, VEGF heterozygous mice also show embryonic lethality by E10.5 to E11.5 (18–19), indicating that gene dosage is important for VEGF. These results imply that the decreased VEGF levels seen in VHL homozygous embryos may play a contributory role in embryonic lethality.
Embryonic lethality as a result of abnormal placental development also has been seen in Mash-2 (28) and VCAM1 mice (29). Mash-2 is a basic-helix–loop–helix transcription factor and is normally expressed in the trophoblasts (28). Mash-2-deficient mice have an absence of diploid trophoblast precursors at E8.5 and die due to placental failure. Chimaeric rescue of the placental defect resulted in Mash-2 mutants that were viable and apparently normal, indicating a noncritical role for Mash-2 in embryonic development. In VCAM1 mice there is a failure of the allantois to fuse with the chorion at E8.5, resulting in abnormal placental development and embryonic death by E9.5 to E11.5 (29). This defect is not complete, however. In some VCAM1-deficient embryos, the allantois fuses with the chorion, but the allantoic mesoderm develops abnormally. Also, a minority of VCAM1-deficient mice have survived and are fertile.
The results presented in this report demonstrate that VHL is required for normal extraembryonic vascular development. Embryonic lethality in VHL-deficient mice at a similar stage of gestation has also been found by another group (N. Kley, personal communication). The determination of any additional developmental role(s) for VHL must await attempts to rescue developing embryos from placental failure. Heterozygous VHL mice have lived beyond 15 months without evidence of significant disease, indicating that VHL haploinsufficiency may not be a critical feature for normal VHL function. Genotyping has shown that most, if not all, VHL tumors tested show deletional inactivation of the inherited wild-type VHL allele and retention of the inherited inactivated allele, as predicted for a tumor suppressor gene. The development of tumors in VHL mice might occur through a similar mechanism, but may take in excess of 15 months to develop. Further analysis of VHL mice should provide us with a better understanding of the role of VHL in development and tumorigenesis.
Acknowledgments
We gratefully acknowledge Dr. N. Kley for discussions on VHL- deficient mice, Drs. F. Latif and M. Lerman for sharing murine VHL cDNA sequence information before publication, Dr. C. Fox and C. Rehm, Molecular Histology Laboratories, Gaithersburg, MD for performing the in situ hybridization studies, B. Kasprzak for immunohistochemistry, and C. F. Dudley and S.-P. Huang for technical assistance. This work was supported in part by a Public Health Service contract to Science Applications International Corporation, Frederick, MD.
ABBREVIATIONS
- ES
embryonic stem
- E
gestational day
- VEGF
vascular endothelial cell growth factor
References
- 1.Linehan W M, Lerman M I, Zbar B. J Am Med Assoc. 1995;273:564–570. [PubMed] [Google Scholar]
- 2.Gnarra J R, Duan D R, Weng Y, Humphrey J S, Chen D Y T, Lee S, Pause A, Dudley C F, Latif F, Kuzmin I, Schmidt L, Duh F-M, Lerman M I, Zbar B, Klausner R D, Linehan W M. Biochim Biophys Acta. 1996;1242:201–210. doi: 10.1016/0304-419x(95)00012-5. [DOI] [PubMed] [Google Scholar]
- 3.Duan D R, Pause A, Burgess W H, Aso T, Chen D Y T, Garrett K P, Conaway R C, Conaway J W, Linehan W M, Klausner R D. Science. 1995;269:1402–1406. doi: 10.1126/science.7660122. [DOI] [PubMed] [Google Scholar]
- 4.Aso T, Lane W S, Conaway J W, Conaway R C. Science. 1995;269:1439–1443. doi: 10.1126/science.7660129. [DOI] [PubMed] [Google Scholar]
- 5.Kibel A, Iliopoulos O, DeCaprio J A, Kaelin W G. Science. 1995;269:1444–1446. doi: 10.1126/science.7660130. [DOI] [PubMed] [Google Scholar]
- 6.Pause A, Lee S, Worrell R A, Chen D Y T, Burgess W H, Linehan W M, Klausner R D. Proc Natl Acad Sci USA. 1997;94:2156–2161. doi: 10.1073/pnas.94.6.2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gao J, Naglich J G, Laidlaw J, Whaley J M, Seizinger B R, Kley N. Cancer Res. 1995;55:743–747. [PubMed] [Google Scholar]
- 8.Laird P W, Zijderveld A, Linders K, Rudnicki M A, Jaenisch R, Berns A. Nucleic Acids Res. 1991;19:4293. doi: 10.1093/nar/19.15.4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Emmert-Buck M R, Bonner R F, Smith P D, Chuaqui R F, Zhuang Z, Goldstein S R, Weiss R A, Liotta L A. Science. 1996;274:998–1001. doi: 10.1126/science.274.5289.998. [DOI] [PubMed] [Google Scholar]
- 10.Fox C H, Cottler-Fox M. Microsc Res Tech. 1993;25:78–84. doi: 10.1002/jemt.1070250111. [DOI] [PubMed] [Google Scholar]
- 11.Fox C H, Cottler-Fox M. In: Protocols in Immunology. Coligan J, Kruisbeek A, Margulies D, Shevach E, Strober W, editors. New York: Wiley; 1993. pp. 12.8.1–12.8.21. [Google Scholar]
- 12.Rossant J, Croy B A. J Embryol Exp Morphol. 1985;86:177–189. [PubMed] [Google Scholar]
- 13.Siemeister G, Weindel K, Mohrs K, Barleon B, Martiny-Baron G, Marme D. Cancer Res. 1996;56:2299–2301. [PubMed] [Google Scholar]
- 14.Gnarra J R, Zhou S, Merrill M J, Wagner J R, Krumm A, Papavassilou E, Oldfield E H, Klausner R D, Linehan W M. Proc Natl Acad Sci USA. 1996;93:10595–10599. doi: 10.1073/pnas.93.20.10589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iliopoulos O, Levy A P, Jiang C, Kaelin W G J, Goldberg M A. Proc Natl Acad Sci USA. 1996;93:10595–10599. doi: 10.1073/pnas.93.20.10595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shalaby F, Rossant J, Yamaguchi T P, Gertsenstein M, Wu X F, Breitman M L, Schuh A C. Nature (London) 1995;376:62–66. doi: 10.1038/376062a0. [DOI] [PubMed] [Google Scholar]
- 17.Fong G H, Rossant J, Gertsenstein M, Breitman M L. Nature (London) 1995;376:66–70. doi: 10.1038/376066a0. [DOI] [PubMed] [Google Scholar]
- 18.Carmeliet P, Ferreira V, Breier G, Pollefeyt S, Kieckens L, Gertsenstein M, Fahrig M, Vandenhoeck A, Harpal K, Eberhardt C, Declercq C, Pawling J, Moons L, Collen D, Risau W, Nagy A. Nature (London) 1996;380:435–439. doi: 10.1038/380435a0. [DOI] [PubMed] [Google Scholar]
- 19.Ferrara N, Carver-Moore K, Chen H, Dowd M, Lu L, O’Shea K S, Powell-Braxton L, Hillan K J, Moore M W. Nature (London) 1996;380:439–442. doi: 10.1038/380439a0. [DOI] [PubMed] [Google Scholar]
- 20.Gilbert S F. Developmental Biology. Sunderland, MA: Sinauer; 1994. pp. 234–242. [Google Scholar]
- 21.Shweiki D, Itin A, Soffer D, Keshet E. Nature (London) 1992;359:843–845. doi: 10.1038/359843a0. [DOI] [PubMed] [Google Scholar]
- 22.Ikeda E, Achen M G, Breier G, Risau W. J Biol Chem. 1995;270:19761–19766. doi: 10.1074/jbc.270.34.19761. [DOI] [PubMed] [Google Scholar]
- 23.Finkenzeller G, Tachnau A, Marme D. Biochem Biophys Res Commun. 1995;208:432–439. doi: 10.1006/bbrc.1995.1356. [DOI] [PubMed] [Google Scholar]
- 24.Levy A P, Levy N S, Goldberg M A. J Biol Chem. 1996;271:2746–2753. doi: 10.1074/jbc.271.5.2746. [DOI] [PubMed] [Google Scholar]
- 25.Hanahan D, Folkman J. Cell. 1996;86:353–364. doi: 10.1016/s0092-8674(00)80108-7. [DOI] [PubMed] [Google Scholar]
- 26.Mustonen T, Alitalo K. J Cell Biol. 1995;129:895–898. doi: 10.1083/jcb.129.4.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Merenmies J, Parada L F, Henkemeyer M. Cell Growth Differ. 1997;8:3–10. [PubMed] [Google Scholar]
- 28.Guillemot F, Nagy A, Auerbach A, Rossant J, Joyner A L. Nature (London) 1994;371:333–336. doi: 10.1038/371333a0. [DOI] [PubMed] [Google Scholar]
- 29.Gurtner G C, Davis V, Li H, McCoy M J, Sharpe A, Cybulsky M I. Genes Dev. 1995;9:1–14. doi: 10.1101/gad.9.1.1. [DOI] [PubMed] [Google Scholar]