Abstract
Biological speciation ultimately results in prezygotic isolation—the inability of incipient species to mate with one another–but little is understood about the selection pressures and genetic changes that generate this outcome. The genus Chlamydomonas comprises numerous species of unicellular green algae, including numerous geographic isolates of the species C. reinhardtii. This diverse collection has allowed us to analyze the evolution of two sex-related genes: the mid gene of C. reinhardtii, which determines whether a gamete is mating-type plus or minus, and the fus1 gene, which dictates a cell surface glycoprotein utilized by C. reinhardtii plus gametes to recognize minus gametes. Low stringency Southern analyses failed to detect any fus1 homologs in other Chlamydomonas species and detected only one mid homolog, documenting that both genes have diverged extensively during the evolution of the lineage. The one mid homolog was found in C. incerta, the species in culture that is most closely related to C. reinhardtii. Its mid gene carries numerous nonsynonymous and synonymous codon changes compared with the C. reinhardtii mid gene. In contrast, very high sequence conservation of both the mid and fus1 sequences is found in natural isolates of C. reinhardtii, indicating that the genes are not free to drift within a species but do diverge dramatically between species. Striking divergence of sex determination and mate recognition genes also has been encountered in a number of other eukaryotic phyla, suggesting that unique, and as yet unidentified, selection pressures act on these classes of genes during the speciation process.
Sexual eukaryotes carry two classes of sex-related genes: sex determination genes act in individual organisms to determine their gender or mating type, and mate recognition genes encode traits that assure that mating occurs between the correct gender/mating type of the correct species. We have cloned and characterized a gene of each class in the unicellular green alga Chlamydomonas reinhardtii. The sex determination mid (minus dominance) gene encodes a regulatory protein that is necessary to express the mating type minus sexual differentiation program and to switch off the mating type plus program (1). The mate recognition fus1 gene encodes a cell surface glycoprotein called fringe that is necessary for plus gametes to adhere to minus gametes and subsequently to fuse to form zygotes (2). The mid gene is located in a highly rearranged region, the R domain, of the mating-type minus (mt−) locus and is unique to the mt− chromosome; the fus1 gene is located in the R domain of the mt+ locus and is unique to the mt+ chromosome (3).
This study reports the results of experiments designed to identify, by low stringency hybridization, mid and fus1 homologs in other members of the Volvocales, focusing in particular on species that have been found, by cladistic analysis (4), to be near relatives of C. reinhardtii. No fus1 homologs were detected, and only one mid homolog was detected, indicating that both genes are evolving very rapidly between species. In contrast, several geographic isolates of C. reinhardtii carry mid and fus1 genes that are nearly identical in coding sequence, indicating strong within-species conservation of these genes.
The one detected interspecies mid homolog is found in C. incerta, a species shown here and by others (5, 6), to be the closest C. reinhardtii relative in culture although they fail to mate. The mid genes of these two species prove to carry nonsynonymous codon differences at 35/148 positions (24%) and synonymous codon differences at 32/148 positions (22%). Despite this divergence, C. incerta mid transgenes can direct mt+ C. reinhardtii cells to undergo minus gametic differentiation.
These results are discussed in the context of several additional studies that collectively document that the genes governing both sex determination and mate recognition are evolving very rapidly throughout the eukaryotic kingdom, apparently in conjunction with speciation.
MATERIALS AND METHODS
General.
All of the species and C. reinhardtii isolates listed in Table 1 were cultured on solid TAP medium (7) except for C. cribrum, C. starrii, and C. sp. J., which failed to grow on Tris/acetate/phosphate but grew successfully on Volvox medium (8). Volvox DNA was kindly provided by D. Kirk (Washington University). Electron microscopy was performed as described in Goodenough et al. (9).
Table 1.
Strains and sources
| Organism | Source/comments |
|---|---|
| C. callosa | UTEX 624 |
| C. cribrum | UTEX 1341 |
| C. debaryana | UTEX 579 (also called C. komma) |
| C. debaryana | CCAP 11/56A (mating partner with 11/56B) |
| C. debaryana | CCAP 11/56B |
| C. debaryana | CC-1741 [=UTEX 344, probably corre sponds to CCAP 11/56A or B; see (6)] |
| C. eugametos mt+ | UTEX 9 |
| C. eugametos mt− | UTEX 10 |
| C. globosa | CC-1872, UTEX 2608 |
| C. incerta | CC-1870, UTEX 2607 |
| C. iyengarii | UTEX 221 |
| C. mexicana mt− | UTEX 729 |
| C. mexicana mt+ | UTEX 730 |
| C. monoica | UTEX 220 (also called C. noctigama) |
| C. reinhardtii mt+ | CC-620 (Mass.) |
| C. reinhardtii mt− | CC-621 (Mass.) |
| C. reinhardtii mt+ | CC-1373 (Mass., formerly called C. smithii) |
| C. reinhardtii mt− | CC-1952 (Minn.) |
| C. reinhardtii mt− | CC-2342 (Penn.) |
| C. reinhardtii mt− | CC-2931 (NC) |
| C. starrii | SAG 3.73, obtained from M. Turmel |
| C. sp. J. | obtained from A. Coleman, (see ref. 5) |
| C. zebra | UTEX 1904 |
| Gonium pectorale mt+ | Brian, Alaska #1, from A. Coleman |
| G. pectorale mt− | Brian, Alaska #2, from A. Coleman |
| Volvox carteri | nagariensis male, from D. Kirk |
| V. carteri | nagariensis female, from D. Kirk |
Strains labeled UTEX derive from the University of Texas Algal Collection; CCAP, the Culture Centre of Algae and Protozoa; CC, the Chlamydomonas Genetics Center; and SAG, the Sammlung von Algenkulturen.
Molecular Biology Techniques.
The preparation of genomic DNA and protocols for Southern blotting and hybridization have been described (10). High stringency hybridizations were carried out at 65°C, with subsequent washes also at 65°C (11). Low stringency hybridizations were carried out variously at 37°C, 42°C, or 50°C and washed at the hybridization temperature. The tubulin hybridization probe was the whole pcf9–12 plasmid, which contains a C. reinhardtii β1-tubulin cDNA (12). The fus1 probe was a 1.7-kb EcoRV–BamHI fragment from cDNA17 (2), which consists entirely of coding sequence. The C. reinhardtii mid probe was a 0.5-kb XhoI–PstI fragment from cDNA1 (1), which contains the entire coding region. Transformation of the C. reinhardtii nuclear genome used the glass bead/vortexing protocol (13), with minor modifications (14).
For library construction, genomic DNA from C. incerta (CC-1870) and C. reinhardtii strain CC-1373 was digested to completion with EcoRI and separately ligated into the EcoRI site of λEMBL3 (Promega). The ligated DNA was packaged in vitro (Promega) and plated on Escherichia coli strain LE392. Protocols for making plaque lifts on nitrocellulose, screening by hybridization, and purifying phage DNA were essentially as in Maniatis et al. (15).
Fragments for DNA sequencing were cloned into pUC118 or 119, and their sequence was determined from single-stranded or double-stranded plasmid using the Sequenase Kit (United States Biochemical). The mid genes from CC-1952, -2342, and -2931 were amplified from genomic DNA by PCR using primers and conditions described elsewhere (1). Their sequence was determined either directly from the purified PCR products or after cloning into pUC118. Each strain was amplified and sequenced twice with the same result.
The cDNA clones of the C. incerta mid and ypt1 genes were generated by performing reverse transcriptase PCR using the reverse transcriptase PCR Kit (Stratagene) on poly(A)+ RNA isolated from nitrogen-starved C. incerta cells. The PCR reaction amplifying mid used the primers GCATCATGGCCTGCTTGC (which contains the initiating ATG) and GATGCCAGCTGCTGCAC (which ends 57 bp 3′ of the stop codon) and the following conditions: 94°C for 1 min, 65°C for 1 min, and 72°C for 1 min for 35 cycles. The resulting product was purified from a low melt agarose gel, ligated into HincII-digested pUC118, and sequenced. The amplification of ypt1 was performed by hot start PCR with the first three cycles at lower stringency (1 min at 94°C, 1 min at 50°C, and 1 min at 72°C) and the next 35 cycles at enhanced stringency (1 min at 94°C, 1 min at 53°C, and 1 min at 72°C). The primers used were yptC1 upstream and yptC1 downstream (16). The obtained PCR product was purified and sequenced directly by cycle sequencing (6).
Observations on Several Chlamydomonas Species.
We and others (5) have determined that a mixup has occurred between C. incerta and C. globosa. C. incerta was collected by F. Hindak in Cuba in 1965. C. globosa was collected by H. W. Kroes in The Netherlands in 1967. A comparison of BamHI-digested chloroplast DNA of C. incerta (CC-1870) and C. globosa (CC-1872) from the Chlamydomonas Genetics Center at Duke had suggested that these two strains were similar or perhaps identical (17). We prepared genomic DNA from CC-1870 and -1872, strains that had been obtained from the Sammlung von Algenkulturen, Gottingen, Germany (SAG), by the Chlamydomonas Genetics Center in 1985 (C. incerta = SAG 7.73, C. globosa = SAG 81.72), and from UTEX 2607 and 2608, strains that had been obtained from the SAG by the University of Texas Algal Collection in 1994. All four strains were digested with HindIII and PstI and, the resulting Southern blot was hybridized successively with the β-tubulin (at 65°C) and mid (at 55°C) cDNAs. The hybridizing fragments were of identical size in all four samples. In addition, introns VI and VII of the ypt4 gene were sequenced after PCR amplification as described (6) from both CC-1870 and -1872. Intron ypt4-VII (349 bp) was completely identical between the strains; intron ypt4-VI (451–453 bp) showed two 1-bp insertions in CC-1872 compared with CC-1870 (= 99.6% identity). This level of similarity is higher than found in the closest-related geographic Chlamydomonas isolates analyzed so far (6), and we conclude therefore that a strain mixup has resulted in the loss of either C. incerta or C. globosa from the culture collections. Schlösser et al. (18) found that both species are susceptible to the C. reinhardtii vegetative wall lysin, and although it is possible that the original C. incerta and C. globosa were both susceptible, these results more likely indicate that the mixup had occurred prior to the onset of their studies. It is impossible to know whether the extant species is the Cuban or the Dutch isolate, but since most of our work has been with CC-1870, we refer to the strain as C. incerta.
The species C. cribrum was included in this study because it has been characterized as belonging to the same autolysin group (see Results) as C. reinhardtii (19). However, in our hands this species proved to be wall-less, so we could not confirm this identification. The sequence of the rDNA internal transcribed spacer of this species (5) does not support a close relation to C. reinhardtii.
C. starrii appears to be homothallic. Cells resuspended in nitrogen-free Volvox medium form a zygote pellicle in 24–36 hr, and during that time pairs of cells can be seen agglutinating via their flagella. Unfortunately, we have been unable to induce the resulting zygotes to germinate.
RESULTS
The mid Gene of C. incerta Has Diverged Extensively from the C. reinhardtii Gene.
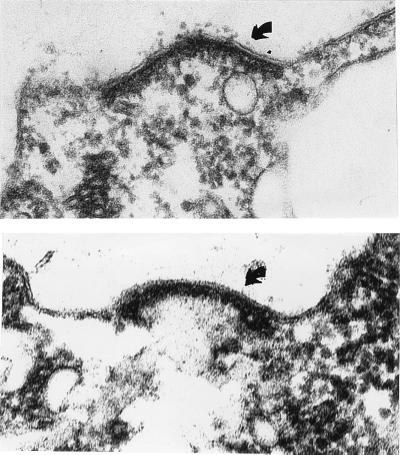
The Chlamydomonas species most closely related to C. reinhardtii is C. incerta (5, 6). To ask whether the extant strain of C. incerta is mating type plus or minus, electron microscopy was performed, and the gametes were found to produce canonical minus mating structures (Fig. 1). C. incerta genomic DNA was therefore hybridized with a mid cDNA probe from C. reinhardtii at 65°C, and a weak signal was detected. All subsequent hybridizations were performed at 55°C, which gave a signal intensity on Southern blots similar to that with C. reinhardtii mt− DNA without generating artifactual bands.
Figure 1.
Mating structures (arrows) of C. reinhardtii mt− (Upper) and C. incerta (Lower). (×96,000.)
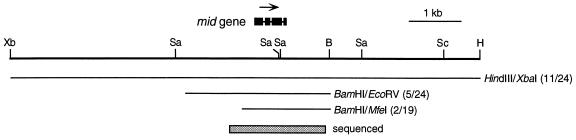
A C. incerta genomic DNA library was made in λEMBL3. This library was screened with the mid cDNA probe, and a hybridizing phage containing a 16.3 kb EcoRI fragment was purified and restriction mapped (Fig. 2). The position of the mid gene within the phage insert was determined by hybridization of Southern blots of restriction-digested phage DNA with the C. reinhardtii mid probe. The DNA sequence was then determined for 1859 bp (GenBank AF002710) encompassing the C. incerta mid gene. Part of the genomic sequence and the predicted amino acid sequence are shown in Fig. 3 and compared with that of C. reinhardtii mid. To ensure that this sequence was transcribed and that the intron locations were correctly identified, two oligonucleotide primers that correspond to positions just 5′ and 3′ of the coding region were prepared and used for reverse transcriptase PCR. Sequencing of the resulting product confirmed that it represented a spliced product of the gene.
Figure 2.
Map of the C. incerta mid gene. A restriction map of the area containing the C. incerta mid gene is shown, with the location of the mid transcript indicated above. The four blocks depict the exons; an arrow indicates the direction of transcription. The locations of the three restriction fragments used to attempt cotransformation of nic7 mt+ to a minus phenotype are shown. The number of cotransformants with at least a partial minus phenotype out of the total number of Nic+ transformants analyzed is indicated to the right of each fragment. The shaded block at the bottom of the figure indicates the extent of DNA sequencing. Restriction enzymes used: B, BamHI; H, HindIII; Sa, SalI; Sc, SacI; and Xb, XbaI.
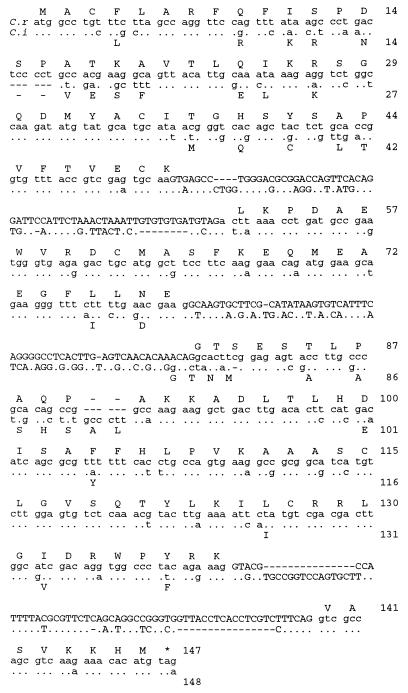
Figure 3.
Comparison of mid gene sequences. The mid coding region and the three introns from C. reinhardtii (C. r.) and C. incerta (C. i.) are compared. The alignment was performed using the bestfit program in the Genetics Computer Group Sequence Analysis Software Package (Madison, WI), using a gap weight of 5.0 and a gap length weight of 0.3. The C. reinhardtii nucleotide sequence is written out in full, with the amino acid sequence shown above. Wherever the C. incerta sequence is identical, the nucleotide is marked with a dot. Changed bases are indicated, and deletions are signified by a dash. The amino acids for the C. incerta sequence are indicated below the nucleotide sequence only when they differ from C. reinhardtii. The amino acid sequence is numbered at the right. Intron sequences are shown in capital letters. Note that the alignment suggests that the change in amino acids 80/81 in the C. incerta sequence is due to a shift in the 3′ splice junction of the second intron accompanied by a compensating frame shift in the coding sequence.
The mid sequences from C. reinhardtii and C. incerta are highly divergent: 24% of the amino acids are different, and another 22% are specified by synonymous codons. A stretch of conserved sequence is found between amino acids 45 and 75 (C. reinhardtii numbering), and the C-terminal third of the protein carries only synonymous changes or conservative amino acid substitutions. The C terminus includes a domain carrying a putative leucine zipper (positions 109–130 in C. reinhardtii) thought to be important for mid function (1).
A more quantitative estimate of divergence is provided by the algorithm of Nei and Gojobori (42). Such calculations yield a value of 13.0 ± 2.1 (on a per 100 site basis) for nonsynonymous changes (Dn) and a value of 58.6 ± 11 for synonymous changes (Ds). The ratio of these two values, Dn/Ds = 0.22, is relatively low, indicating that there has been no overall positive selection (20) for amino acid-changing mutations (see Discussion).
The three introns are found at equivalent positions in the two genes, but their sequences are highly divergent. An available algorithm for comparing intron relatedness [SI50 (6)] shows that the first two mid introns display weak relatedness just above threshold level whereas the third set already falls beyond the level of significant similarity. Despite the very small size of the mid introns, these results indicate the same degree of divergence as has been obtained from the comparison of larger intron data sets (actin, ypt4) between these species (6).
The mid gene of C. reinhardtii displays exceptionally low codon bias (B = 0.161) compared with other C. reinhardtii nuclear genes (average B value ≈0.6) (1). The numerous codon differences in the C. incerta mid gene also generate a sequence with a very low codon bias (B = 0.157). Although the overall bias of C. incerta genes has not been determined, its two sequenced genes (see below) display the same degree of GC bias found throughout the Volvocales (21), with B values of 0.709 (β-tubulin) and 0.494 (ypt1). Therefore, the mid gene will doubtless prove to display codon usage unusual for C. incerta.
Other Genomic Sequences from C. reinhardtii and C. incerta Are Highly Conserved.
A variety of data suggests that C. incerta and C. reinhardtii are very closely related. Each is sensitive to the other’s cell wall lysin, an important criterion of taxonomic relatedness in this genus (22). Moreover, studies comparing the internal transcribed spacer sequences flanking the 5.8S rRNA genes (5) and the sequences of three introns (6) have documented that C. incerta is a very close relative of C. reinhardtii. Because the foregoing studies involved the analysis of genomic regions that do not encode proteins, we analyzed two protein-encoding genes and compared their divergence with that seen for the mid genes.
β-Tubulin Sequence.
The C. incerta genomic library was hybridized with a β1-tubulin cDNA from C. reinhardtii to clone the C. incerta homolog. C. reinhardtii has two β-tubulin genes, and the pattern of fragments detected on Southern blots of C.incerta probed with the β-tubulin cDNA is consistent with its having two copies as well. A β-tubulin gene from C. incerta was sequenced (deposited as GenBank accession no. AF001379) and compared with the published sequences for C. reinhardtii (GenBank accession nos. K03281 and M10064). Only one conservative amino acid substitution (V → I) has occurred, at amino acid 30, reflecting the expected strong purifying selection on tubulin sequences. Of the 443 codons in β-tubulin, the C. incerta sequence has 29 synonymous substitutions relative to the C. reinhardtii β1 (6.5%) and 20 relative to β2 (4.5%). This would suggest that we have sequenced the C. incerta β2-tubulin. Unexpectedly, however, all three sequenced C. incerta tubulin introns showed significant similarity (SI50 1.21–1.56) to their respective C. reinhardtii β1-tubulin introns and no similarity (SI50 1.03–1.15) to their β2-tubulin counterparts. The picture is further complicated by the fact that, in C. reinhardtii, there is a very high similarity of introns 3 between β1- and β2-tubulins (SI50 = 1.78) but no similarity between introns 1 and 2. In Volvox carteri, on the other hand, the three equivalently positioned introns between β1- and β2-tubulins are unrelated to each other on the sequence level. Taken together, these results document that the pattern of intron recombination and evolution in these sets of duplicated tubulin genes is quite idiosyncratic. [A possibly analogous “swapping” of 3′ untranslated region sequences recently has been observed in a comparison of a family of cytoplasmic actin genes in several sea urchin species (23)].
Assuming that we have indeed cloned the C. incerta β2-tubulin gene, its 4.5% level of synonymous changes is very different from the 22% level found for the mid gene. Expressed as Ds, the tubulin value is 9.8 ± 1.9, in contrast to 58.6 for mid, a 6-fold difference.
ypt1 Sequence.
PCR primers derived from the ypt1 gene of C. reinhardtii, encodes a small G protein (16), were used to amplify the ypt1 cDNA of C. incerta from reverse-transcribed RNA. This sequence (excluding 34 bp of the 5′- and 17 bp of the 3′-primer-generated ends) was compared with the published ypt1 sequence (GenBank accession no. U13168). A single nucleotide change (T → C at nucleotide 177 of the coding region of ypt1) distinguishes the two sequences, creating a synonymous change within codon L59. This generates a Ds of 0.78 ± 0.78 for ypt1, or 75-fold less than the Ds value for mid.
The C. incerta Mid Protein Is Functional in C. reinhardtii.
Because the C. incerta mid gene is transcribed (as documented by the isolation of a spliced reverse transcriptase PCR product) and is apparently functional in C. incerta [as documented by its ability to direct the formation of a minus mating structure (Fig. 1)], its divergence from C. reinhardtii cannot be explained by proposing that it is a pseudogene. To further evaluate its function, three restriction fragments carrying the C. incerta mid gene (Fig. 2) were introduced into a C. reinhardtii nic7 mt+ recipient strain by cotransformation using pNic7.9 (14) to complement the nic7 mutation. Some of the Nic+ transformants proved to differentiate as minus gametes that agglutinate and fuse with C. reinhardtii mt+ partners, the same minus–dominance result that is obtained when the C. reinhardtii mid gene is introduced into a mt+ strain (1). [As with the C. reinhardtii mid transgene (1), some cotransformants produced a mixture of both plus and minus gametes, and others produced only minus gametes.]
Therefore, the numerous amino acid changes have not obviously altered the ability of the C. incerta Mid protein to trigger minus sexual differentiation in C. reinhardtii. Unfortunately, the reciprocal transformation experiment cannot be performed because an mt+ strain of C. incerta is not available.
fus1 and mid Homologs Are Not Detected in Most Relatives of C. reinhardtii.
Given the extensive and surprising divergence of mid genes between C. reinhardtii and C. incerta, we went on to explore the extent to which mid and fus1 sequences have diverged in more distantly related Chlamydomonas species. Genomic DNA isolated from the species listed in Table 1 was restriction digested and used to prepare Southern blots. All gave strong positive signals when hybridized with a β-tubulin cDNA probe at 65°C. In contrast, when we initially hybridized approximately one–half of the species in Table 1 with probes made from mid and fus1 cDNAs at 65°C, no signal was detected. Therefore, all of the species were hybridized at lower stringencies, varying from 37°C to 50°C. Under these conditions, multiple fragments hybridized in each species, most of which were very faint compared with the hybridization to the C. reinhardtii control on the same blot. A few more intensely hybridizing fragments matched the size of fragments that stain heavily with ethidium bromide, suggesting nonspecific hybridization to repetitive sequences. In cases in which two mating types from the same species were available, the same faint bands were detected in digests of both mating types whereas, by analogy with C. reinhardtii, these probes should only hybridize to one mating type.
In all cases, the level and pattern of hybridization suggested that most or all of the hybridization was artifactual, meaning that it will not be straightforward to identify the homologs of fus1 and mid by low stringency hybridization. For present purposes, these results are significant in that they document that both the mid and the fus1 genes are generally so divergent within the genus that homologs cannot be detected by standard protocols.
The mid and fus1 Genes Are Conserved Within C. reinhardtii.
The extensive divergence between the C. reinhardtii and C. incerta mid genes and the failure to detect any mid or fus1 homologs in more distantly related Chlamydomonas species suggest that these genes are evolving rapidly. To ascertain whether such rapid evolution is occurring within a species as well, that is, whether mid and fus1 are present in the C. reinhardtii gene pool as diverse multiple alleles that then perhaps drift or hitchhike during the speciation process, we sequenced fus1 and mid genes from C. reinhardtii isolates collected in diverse geographic locales (Table 1).
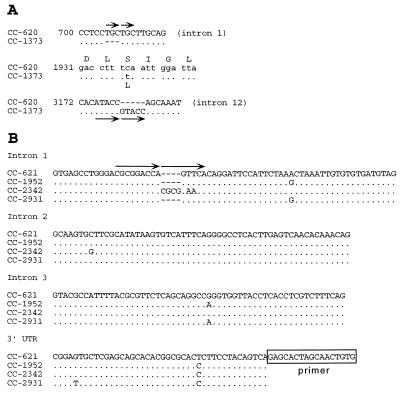
A 4.7-kb EcoRI genomic fragment carrying the fus1 gene from CC-1373 (Mass.) was identified in a phage clone and subsequently subcloned into plasmids. A region of 3812 bp covering the entire coding region and the 12 introns was sequenced, and three changes were noted (Fig. 4A): an insertion/deletion in introns 1 and 12 and a nonsynonymous change (S → L) in codon 423.
Figure 4.
Comparison of C. reinhardtii fus1 and mid genes. (A) Three differences were noted in the sequence of the fus1 gene between the two indicated mt+ strains. Amino acid 423 has changed from serine to leucine in CC-1373, and an insertion/deletion has occurred in both introns 1 and 12. The conventions for comparing the sequences are as in Fig. 2. (B) The mid genes were compared among four C. reinhardtii mt− isolates. Most of the changes were in the intron sequences and the 3′ UTR, which are shown. Not shown are two synonymous changes in codons A88 and P136 in strain CC-2342 (see text). The pairs of arrows in A and B show short duplications of identical or nearly identical sequence in the CC-620 fus1 intron 1, the CC-1373 fus1 intron 12, and the CC-2342 mid intron 1. Such short duplications are characteristic of the excision footprints of some transposons (41), which could have been the mechanism for these three changes.
The mid gene was sequenced from three additional C. reinhardtii mt− strains: -CC-1952 (Minn.), -2342 (Penn.), and -2931 (NC). Several changes were noted in the introns and the 3′ untranslated region (Fig. 4B). Only the CC-2342 coding region contained any changes, both of which were single-base substitutions creating synonymous codons. The intron level of divergence (ranging from 0 to 6 × 10−2 exchanges per position) is comparable to that seen in the introns of actin and ypt4 of the same strains [average range from 6 × 10−3 to 5 × 10−2 exchanges per position (6)], although the particular interstrain differences deviate (in actin and ypt4 introns, CC-2931 and CC-2342 are much more closely related than both are to CC-1952 whereas mid introns are identical between CC-1952 and CC-2931, setting CC-2342 apart).
These results indicate that the mid and fus1 genes are subject to very little within-species divergence in the natural isolates of C. reinhardtii that are available for analysis.
DISCUSSION
We document in this report that two genes that play central roles in sexual differentiation in Chlamydomonas have evolved rapidly between species. Homologs of the fus1 gene of C. reinhardtii cannot be detected in other members of the Volvocales even though mating structures per se are found not only throughout the Volvocales but also in other members of the Chlorophyta (24). The one detectable homolog of the mid gene, found in C. incerta, is highly divergent even though, by several taxonomic and genomic criteria, C. incerta and C. reinhardtii are sibling species that have diverged only very recently (refs. 5 and 6 and present study). Within the C. reinhardtii group, on the other hand, there is strict conservation of both sequences, even between those estimated to have been separated for over 1 million years (6). This suggests that the genes are subject to purifying selection within a species and that they diversify at the time of speciation.
As reviewed in detail elsewhere (U.G., unpublished work), these same patterns are found throughout the sexual eukaryotes. Studies on the mate recognition genes of mollusks (25), echinoderms (26), and fungi (27) show dramatic differences between closely related species, as do the sex determination genes of insects (28), worms (29, 30), fungi (31), and mammals (32, 33).
Whereas the need to generate novel sets of mate recognition traits at the time of speciation has long been recognized as a key feature of the speciation process (34), the sequence of events that generates and selects these new traits is poorly understood and is the subject of much controversy (35). Not appreciated until recently is the parallel generation of novel sex determination gene sequences in conjunction with speciation; intuitively, it would be expected that such genes would be subject to the same level of stringent conservation found for genes that govern other pathways of somatic differentiation (36). A satisfactory explanation for this evolutionary pattern has not yet been offered.
Two recent studies of mate recognition genes in abalone (25) and sea urchins (26) document a marked excess of amino acid substitutions over synonymous changes, yielding Dn/Ds values > 1 (in the sea urchin case, this is seen only in certain domains of the protein). Two studies of the SRY sex determination gene in primates (32) and rodents (33) also report high Dn/Ds values in certain domains and conservation in others. In all of these studies, the data are interpreted to show that the rapid evolution of sex genes is being driven by positive selection (although, as noted above, the traits being selected are unclear). This same argument could be made for certain domains of the mid gene, notably in the amino-terminal half of the protein (Fig. 3), but, to be convincing, such an argument must be based on more than two gene sequences.
The Mid protein of C. incerta is able to function in C. reinhardtii, suggesting that the two conserved domains of the protein (roughly residues 45–75 and 91–147; Fig. 3), including the putative leucine zipper (1), are critical to the sex determination function of Mid. A similar conservation of function in widely divergent sex determination proteins has been observed for the transformer gene product of Drosophila by O’Neil and Belote (28), and the DNA binding motif of the SRY protein is conserved throughout the mammals (32, 33). The entire mid gene is apparently conserved within species but only portions of the gene are conserved between species, so this again suggests that there is positive selection for diversification in the nonconserved domains in conjunction with speciation. It cannot, however, be readily argued that the nonconserved domains are “unimportant to function” and hence “free to drift” because, in this case, one would expect to find within-species allelism in these domains, and this was not observed.
A striking finding was the large number of synonymous codon changes in the mid genes of C. reinhardtii vs. C. incerta. The mid Ds value of 58.6 was far higher than that of the two other structural genes analyzed (9.8 for β-tubulin and 0.78 for ypt1) and, indeed, higher than has been seen for other sex-related genes in genera that are far more divergent than C. reinhardtii and C. incerta: for example, the Ds value for the SRY gene is 22.1 in mouse vs. rat (33) and 27.6 in human vs. marmoset (32). The strong codon bias found in all Chlamydomonas genes (37) is absent from the mid (and fus1) genes (1, 2), so it can be argued that synonymous changes are not selected against and therefore might tend to accumulate (38). Such an argument, however, only raises the question of why these genes are uniquely devoid of codon bias in the first place. The most parsimonious answer to this question is that the weak selection pressures postulated to maintain codon bias (39) are not able to override the large number of synonymous mutations that, for some reason, accumulate in these two genes. One way to escape the circularity of these arguments is to propose that there is positive selection for amino acid-changing mutations that arise in the mid gene at the time of speciation and that synonymous mutations hitchhike along. This postulate returns attention to the question of why amino acid changes in this gene might be under positive selection.
An additional feature of the mid and fus1 genes that may be relevant to their evolution is that each gene is without a homolog on the opposite chromosome (3), meaning that neither gene is a substrate for recombinational repair during the diploid phase of the life cycle. It follows that purifying selection alone, without the assistance of recombinational repair, is acting to maintain the monomorphism of each gene within the C. reinhardtii species. It further follows that, if selection pressures were to reverse such that there were positive selection for variants, recombinational repair would not act to homogenize the locus and variants would be more likely to accumulate in the gene and in the population. This dynamic would operate as well for sex-related genes located in dimorphic chromosomes (e.g, the mammalian Y chromosome), but it cannot explain sex–gene divergence in general because, for example, the sex determination genes of worms and flies have homologs.
Taken together, these observations indicate that something unexplained, but very interesting, happens to the selection pressures on sex-related genes in conjunction with speciation. Possibly relevant is the unexplained observation that lineages that are especially prone to speciation tend also to be prone to extinction (40). Perhaps being prone to extinction is the hallmark of a sexual system that is vulnerable to selection for variation and hence is prone to generate new species at the expense of the old.
Acknowledgments
We thank A. Coleman and M. Turmel for providing strains, M. LeDizet for the calculation of B values, W. Swanson for the Nei and Gojobori calculations, both M. LeDizet and W. Swanson for stimulating conversations about evolution, and L. Ellis, F. Bourgeois, A. Fearncombe, L. Small, and M. Liss for technical assistance. This research was supported by Grant MCB-18817 from the National Science Foundation and Grant GM-26150 from the National Institutes of Health.
Footnotes
References
- 1.Ferris, P. J. & Goodenough, U. W. (1997) Genetics, in press. [DOI] [PMC free article] [PubMed]
- 2.Ferris P J, Woessner J P, Goodenough U W. Mol Biol Cell. 1996;7:1235–1248. doi: 10.1091/mbc.7.8.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ferris P J, Goodenough U W. Cell. 1994;76:1135–1145. doi: 10.1016/0092-8674(94)90389-1. [DOI] [PubMed] [Google Scholar]
- 4.Buchheim M A, Lemieux C, Otis C, Gutell R R, Chapman R L, Turmel M. Mol Phylogenet Evol. 1996;5:391–402. doi: 10.1006/mpev.1996.0034. [DOI] [PubMed] [Google Scholar]
- 5.Coleman, A. W. & Mai, J. C. (1997), J. Mol. Evol., in press.
- 6.Liss M, Kirk D L, Beyser K, Fabry S. Curr Genet. 1997;31:214–227. doi: 10.1007/s002940050198. [DOI] [PubMed] [Google Scholar]
- 7.Harris E H. The Chlamydomonas Sourcebook. San Diego: Academic; 1989. [Google Scholar]
- 8.Kirk D L, Kirk M M. Dev Biol. 1983;96:493–506. doi: 10.1016/0012-1606(83)90186-0. [DOI] [PubMed] [Google Scholar]
- 9.Goodenough U W, Detmers P A, Hwang C. J Cell Biol. 1982;92:378–386. doi: 10.1083/jcb.92.2.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferris P J. Genetics. 1989;122:363–377. doi: 10.1093/genetics/122.2.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Church G M, Gilbert W. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Youngblom J, Schloss J A, Silflow C D. Mol Cell Biol. 1984;4:2686–2696. doi: 10.1128/mcb.4.12.2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kindle K L. Proc Natl Acad Sci USA. 1990;87:1228–1232. doi: 10.1073/pnas.87.3.1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferris P J. Genetics. 1995;141:543–549. doi: 10.1093/genetics/141.2.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maniatis T, Fritsch E F, Sambrook J. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1982. [Google Scholar]
- 16.Dietmaier W, Fabry S, Huber H, Schmitt R. Gene. 1995;158:41–50. doi: 10.1016/0378-1119(95)00052-8. [DOI] [PubMed] [Google Scholar]
- 17.Harris E H, Boynton J E, Gillham N W, Burkhart B D, Newman S M. Arch Protistenkd. 1991;139:183–192. [Google Scholar]
- 18.Schlösser U G, Sachs H, Robinson D G. Protoplasma. 1976;88:51–64. doi: 10.1007/BF01280359. [DOI] [PubMed] [Google Scholar]
- 19.Matsuda Y, Musgrave A, van den Ende H, Roberts K. Bot Mag Tokyo. 1987;100:373–384. [Google Scholar]
- 20.Hughes A L, Nei M. Nature (London) 1988;335:167–170. doi: 10.1038/335167a0. [DOI] [PubMed] [Google Scholar]
- 21.Schmitt R, Fabry S, Kirk D L. Int Rev Cytol. 1992;139:189–265. doi: 10.1016/s0074-7696(08)61413-8. [DOI] [PubMed] [Google Scholar]
- 22.Schlösser U G. In: Systematics of the Green Algae. Irvine D E G, John D M, editors. London: Academic; 1984. pp. 409–418. [Google Scholar]
- 23.Kissinger J C, Hahn J-H, Raff R A. Mol Biol Evol. 1997;14:654–665. doi: 10.1093/oxfordjournals.molbev.a025805. [DOI] [PubMed] [Google Scholar]
- 24.Goodenough U W. In: Microbial Cell-Cell Interactions. Dworkin M, editor. Washington, DC: Am. Soc. Microbiol.; 1991. [Google Scholar]
- 25.Swanson W J, Vacquier V D. Proc Natl Acad Sci USA. 1995;92:4957–4961. doi: 10.1073/pnas.92.11.4957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Metz E C, Palumbi S R. Mol Biol Evol. 1996;13:397–406. doi: 10.1093/oxfordjournals.molbev.a025598. [DOI] [PubMed] [Google Scholar]
- 27.Bakkeren G, Kronstad J W. Proc Natl Acad Sci USA. 1994;91:7085–7089. doi: 10.1073/pnas.91.15.7085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O’Neil M T, Belote J M. Genetics. 1992;131:113–123. doi: 10.1093/genetics/131.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Bono M, Hodgkin J. Genetics. 1996;144:587–595. doi: 10.1093/genetics/144.2.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuwabara P E. Genetics. 1996;144:597–607. doi: 10.1093/genetics/144.2.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Turgeon B G, Böhlman H, Ciuffetti L M, Christiansen S K, Yang G, Schafer W, Yoder O C. Mol Gen Genet. 1993;238:270–284. doi: 10.1007/BF00279556. [DOI] [PubMed] [Google Scholar]
- 32.Whitfield L S, Lovell-Badge R, Goodfellow P N. Nature (London) 1993;364:713–715. doi: 10.1038/364713a0. [DOI] [PubMed] [Google Scholar]
- 33.Tucker P K, Lundrigan B L. Nature (London) 1993;364:715–717. doi: 10.1038/364715a0. [DOI] [PubMed] [Google Scholar]
- 34.Paterson H E H. Evolution and the Recognition Concept of Species. Baltimore: Johns Hopkins Univ. Press; 1993. [Google Scholar]
- 35.Gibbons A, Morell V. Science. 1996;273:1496–1502. [Google Scholar]
- 36.Hunter C P, Kenyon C. Nature (London) 1995;377:229–232. doi: 10.1038/377229a0. [DOI] [PubMed] [Google Scholar]
- 37.LeDizet M, Piperno G. Mol Biol Cell. 1995;6:697–711. doi: 10.1091/mbc.6.6.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kliman R M, Hey J. Mol Biol Evol. 1993;10:1239–1258. doi: 10.1093/oxfordjournals.molbev.a040074. [DOI] [PubMed] [Google Scholar]
- 39.Akashi H. Genetics. 1995;139:1067–1076. doi: 10.1093/genetics/139.2.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stanley S M. In: Causes of Evolution: A Paleontological Perspective. Ross R M, Allmon W D, editors. Chicago: Univ. of Chicago Press; 1990. pp. 103–127. [Google Scholar]
- 41.Federoff N V. In: Mobile DNA. Howe M M, Berg D E, editors. Washington, DC: Am. Soc. Microbiol.; 1989. pp. 375–411. [Google Scholar]
- 42.Nei M, Gojobori T. Mol Biol Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]