Abstract
This study aimed to exploit bacterial artificial chromosomes (BAC) as large antigen-capacity DNA vaccines (BAC-VAC) against complex pathogens, such as herpes simplex virus 1 (HSV-1). The 152-kbp HSV-1 genome recently has been cloned as an F-plasmid-based BAC in Escherichia coli (fHSV), which can efficiently produce infectious virus progeny upon transfection into mammalian cells. A safe modification of fHSV, fHSVΔpac, does not give rise to progeny virus because the signals necessary to package DNA into virions have been excluded. However, in mammalian cells fHSVΔpac DNA can still replicate, express the HSV-1 genes, cause cytotoxic effects, and produce virus-like particles. Because these functions mimic the lytic cycle of the HSV-1 infection, fHSVΔpac was expected to stimulate the immune system as efficiently as a modified live virus vaccine. To test this hypothesis, mice were immunized with fHSVΔpac DNA applied intradermally by gold-particle bombardment, and the immune responses were compared with those induced by infection with disabled infectious single cycle HSV-1. Immunization with either fHSVΔpac or disabled infectious single cycle HSV-1 induced the priming of HSV-1-specific cytotoxic T cells and the production of virus-specific antibodies and conferred protection against intracerebral injection of wild-type HSV-1 at a dose of 200 LD50. Protection probably was cell-mediated, as transfer of serum from immunized mice did not protect naive animals. We conclude that BAC-VACs per se, or in combination with genetic elements that support replicative amplification of the DNA in the cell nucleus, represent a useful new generation of DNA-based vaccination strategies for many viral and nonviral antigens.
The immune system reacts to most systemic infections with broad and vigorous cytokine, cellular, and humoral responses that restrict spread of the infectious agent within the organism. For example, within days after primary infection with herpes simplex virus 1 (HSV-1), the immune system induces a variety of cytokines, chemokines, adhesion molecules, natural killer cells, macrophages, γ/δ T cells, and CD4+ and CD8+ T cells and produces neutralizing antibodies (1–10). Antiviral vaccines, which have been around for at least two centuries, are modified target agents designed to reduce pathogenicity but conserve immunogenicity (11). Modified live virus (mlv) vaccines are based on either attenuated or replication-defective viruses and induce an immune response comparable to that induced by the parent viruses. Although biologically much safer, inactivated virus-vaccines and subunit vaccines are much less effective in inducing immune responses than mlv vaccines, indicating a link between some elements of virus replication and immunogenicity (12–14). In recent years, the use of naked DNA for immunization stirred great hopes for the development of safe and efficient vaccines (15, 16). As opposed to the purified proteins used as subunit vaccines, immunization with antigen-encoding DNA supports protein synthesis in host cells, thereby activating the cellular arm of the immune system (17, 18). However, the plasmid vectors previously used for vaccinations did not have the capacity to hold the wide range of antigens needed to induce broad immune responses against complex (viral) pathogens (19–21). Live virus vaccines, on the other hand, are not entirely safe and, moreover, are not easily manipulated to counter the complicated interplay of the virus with the host’s defense system.
Based on bacterial artificial chromosomes (BACs), we have established a vaccination strategy (BAC-VAC) using HSV-1 as an example. BAC-VAC allows replicative amplification of the vaccine DNA in the host cell nucleus and combines safety and simplicity of DNA vaccines with the high immunogenicity of live-virus vaccines. BACs have been widely used in genome research because of their capacity to accommodate large inserts (>300 kb) and their genetic stability in bacteria (22, 23). BACs are also suitable for cloning the large genomes of DNA viruses, as first demonstrated by Luckow et al. (24) with baculovirus (130 kb). Recently, the genomes of several herpesviruses, including those of murine cytomegalovirus (230 kb), Epstein–Barr virus (170 kb), and HSV-1 (152 kb) have been cloned successfully in Escherichia coli where they are stably maintained as supercoiled plasmid DNA and accessible to the prokaryotic tools for modification (25–28). Upon transfection into mammalian cells, these plasmids can efficiently mediate the production of infectious virus progeny. To explore the potential usefulness of BACs for vaccination protocols, we have chosen a plasmid that contains a replication-competent but packaging-defective HSV-1 genome (fHSVΔpac) developed by Saeki et al. (27). The investigators have excluded the HSV-1 DNA cleavage/packaging signals (pac), which are essential for cleavage of the concatemeric products of viral DNA replication into unit-length genomes and their subsequent packaging into virions, to prevent the formation of HSV-1 progeny from the BAC DNA (27). Although packaging-defective in mammalian cells, fHSVΔpac can still replicate, express the HSV-1 genes, cause cytotoxic effects, produce noninfectious, virus-like particles, and support the packaging of cotransfected HSV-1-based amplicon vectors into virions (24, 27). These functions mimic an entire lytic cycle of the HSV-1 infection and, consequently, immunization with fHSVΔpac DNA should exert all of the immunomodulatory functions considered important for efficient immune stimulation (10, 14). The set of experiments described in this report demonstrate that small amounts of the prototype BAC-VAC, fHSVΔpac, can induce broad immune responses able to protect mice from intracerebral (i.c.) challenge with wild-type (wt) HSV-1 at a dose of at least 200 LD50.
Materials and Methods
Animals, Cells, and Viruses.
Female, 7- to 10-week-old C57BL/6 (H-2b) or 129Sv/Ev (H-2b) mice were bred and maintained in specific pathogen-free conditions at the Walter and Eliza Hall Institute for Medical Research. Vero cells (American Type Culture Collection, Rockville, MD), HSV-1 glycoprotein H (gH)-expressing Vero cells (F1; refs. 26 and 29), H-2b thymoma cells (EL-4), and glycoprotein B (gB)-expressing fibroblast cells (MC57; refs. 30 and 31) were grown in complete DMEM supplemented with 10% FBS. HSV-1 strain F was obtained from B. Roizman (University of Chicago) and propagated on Vero cells (32). Disabled infectious single cycle (DISC) HSV-1, a gH deletion-mutant capable of completing a single cycle of infection, was kindly provided by J. Shields (Cantab Pharmaceuticals, Cambridge, U.K.) and was propagated on F1 cells (26, 29). HSV-1 amplicon pHSVGFP, which expresses the gene for green fluorescent protein, was packaged into HSV-1 virions by using the helper virus-free method (33–35).
Vaccine DNA.
The cloning of the 152-kb HSV-1 genome, with DNA cleavage/packaging signals (pac) excluded, as a BAC in E. coli (fHSVΔpac) has been described (27). Supercoiled fHSVΔpac DNA was isolated by alkaline lysis and Tip-500 column chromatography (Qiagen, Chatsworth, CA) and purified by cesium chloride equilibrium centrifugation. Plasmid psOVA DNA, which encodes a secreted form of chicken ovalbumin, was used as control (36). DNA preparations of fHSVΔpac and psOVA contained <100 units of endotoxin per mg as determined by the limulus amoebocyte lysate assay (37).
Immunization and Virus Challenge Protocols.
Intradermal (i.d.) injection.
Mice were immunized i.d. at the base of the tail either with 50 μg DNA in 70 μl of saline or, as a control, with 109 plaque-forming units (PFU) of DISC HSV-1 in 100 μl of saline (34, 37). Two weeks later, the animals were boosted with the same amount of DNA or virus and, 10 days later, were analyzed for the induction of cellular and humoral immune responses or challenged with wt HSV-1.
Gold-particle bombardment.
DNA was adsorbed to gold particles (1 μm) and delivered i.d. at the base of the tail by gold-particle bombardment using a gene gun as recommended by the manufacturer (Bio-Rad). The animals received two doses of 750 ng DNA each per immunization. Booster immunizations were given every 2 weeks by using the same amount of DNA. Ten days after each immunization, groups of animals were analyzed for the induction of HSV-1-specific cytotoxic T lymphocytes (CTLs) and antibody responses or challenged with wt HSV-1.
Virus challenge.
Mice were anesthetized with ether and injected i.c. with 2 × 105 PFU of HSV-1 strain F in 10 μl of PBS. The dose of HSV-1 that causes lethal infections in 50% of the animals (LD50) has been determined in age-matched C57BL/6 mice and was 102–103 PFU. The animals were examined daily for signs of disease, and the surviving animals were counted 14 days after the challenge.
Serum transfer.
Serum prepared from pooled blood of fHSVΔpac-immunized or DISC HSV-1-immunized mice was analyzed for HSV-1-specific neutralizing antibodies and transferred i.v. (100 μl) or i.p. (1 ml) into naive mice. After 4 h, the animals were challenged with HSV-1 as described above, and sera were collected from some of the mice to determine posttransfer neutralizing antibody titers.
Assessment of gB-Specific CTL Activity.
Spleen cells or lymph node cells were isolated from immunized or control animals and assayed directly, or after restimulation in vitro for 5 days by using irradiated HSV-1 gB-expressing MC57 cells, for reactivity against EL-4 cells (30, 31, 38). EL-4 target cells were pulsed for 1 h with 3 μg/ml of gB-specific H-2b restricted peptide SSIEFARL (single letter amino acid code) and analyzed by a standard 4-h 51Cr release cytotoxicity assay. An overnight (15-h) 51Cr release cytotoxicity assay was used when effector cells were analyzed directly without in vitro restimulation. Spontaneous release was consistently below 15% in a 4-h assay and approximately 25% in overnight assays.
Flow Cytometry.
Splenocytes or lymph node cells were analyzed by flow cytometry (Becton Dickinson) for the presence of CD8+ T cells (antibody 500-A2, Caltag, Burlingame, CA) carrying the T cell receptor (TCR) Vβ10 (antibody B21.5; PharMingen) (36), which is typically present in increased numbers after HSV-1 infections (31, 39).
Serology.
ELISA was performed and antibody titers were determined as described (36) by using peroxidase-conjugated polyclonal anti-mouse IgG1, IgG2a, IgG2b, and IgG3 antibodies (Southern Biotechnology Associates). HSV-1 antigen was prepared from virus-infected Vero cells and titered for optimal performance in ELISA (40). Sera collected from immunized and control animals were analyzed for HSV-1 neutralization by using standard methods.
Western Immunoblot Analysis.
Radioactively labeled cell lysates were prepared, subjected to SDS/PAGE, transferred to nitrocellulose, and immunostained essentially as described (41). Briefly, monolayers of Hep-2 cells were infected with either HSV-1 (strain F) or HSV-2 (strain G) at a multiplicity of infection of 20 PFU per cell or mock-infected and labeled with [35S]methionine from 16 to 20 h postinfection. The cells were lysed in SDS buffer, and the proteins were separated on a 9.25% SDS-polyacrylamide gel, crosslinked with N,N′-diallyltartardiamide, blotted to nitrocellulose, and visualized by autoradiography. For immunostaining, the nitrocellulose sheet was incubated for 1 h at 37°C in blocking solution (25% horse serum, 0.5% NP-40, in PBS). Serum (diluted 1:500 in a solution consisting of 1 vol of blocking solution and 2 vol of PBS) was added and incubated overnight at 4°C. Detection was carried out by using the Vectastain ABC kit and biotinylated anti-mouse IgG as a secondary antibody, avidin and peroxidase-labeled biotin, according to the instructions provided by the manufacturer (Vector Laboratories).
Results and Discussion
I.D. Injection of fHSVΔpac DNA Induces HSV-1-Specific CTL and Antibody Responses.
In a first set of experiments, mice were injected i.d. with fHSVΔpac (Fig. 1) or psOVA DNA as described in Materials and Methods. Two weeks after each injection, splenocytes or lymph node cells were isolated to analyze CTL activity, and blood was collected to determine HSV-1-specific antibodies. The isolated splenocytes were restimulated for 5 days by using irradiated HSV-1 gB-expressing MC57 cells, and cytotoxic activity was analyzed by 51Cr release by using peptide-loaded EL-4 target cells. HSV-1 gB was chosen for this assay because it includes the dominant CTL epitope for the H-2b haplotype of C57BL/6 mice (42). Fig. 2A represents one of six experiments and shows specific lysis of the target cells by the restimulated effector cells from fHSVΔpac DNA, but not from psOVA control DNA-immunized mice. Direct CTL activity without restimulation of the isolated splenocytes or lymph node cells could not be detected. A low, but significant, amount of HSV-1-specific antibody was produced after two DNA immunizations, as determined by ELISA (data not shown). The animals were not protected from lethal i.c. HSV-1 infection, even after three immunizations with fHSVΔpac (data not shown).
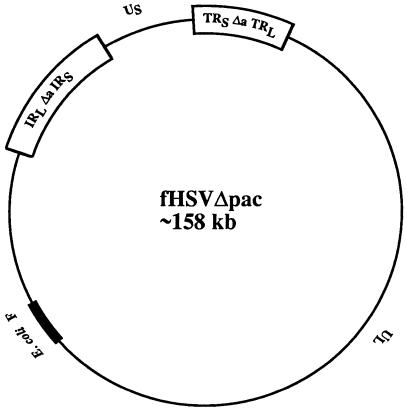
Figure 1.
Structure of a BAC containing a replication-competent, packaging-defective HSV-1 genome (fHSVΔpac). The modified HSV-1 genome (HSVΔpac) is composed of unique long (UL) and unique short (US) segments, which are both flanked by inverted repeats: IRS, internal repeat of the short segment; TRS, terminal repeat of the short segment; TRL, terminal repeat of the long segment; IRL, internal repeat of the long segment. The DNA cleavage/packaging signals contained in the a sequences located at both junctions between the S and the L component have been excluded (Δa). The backbone of the E. coli F-plasmid is indicated.
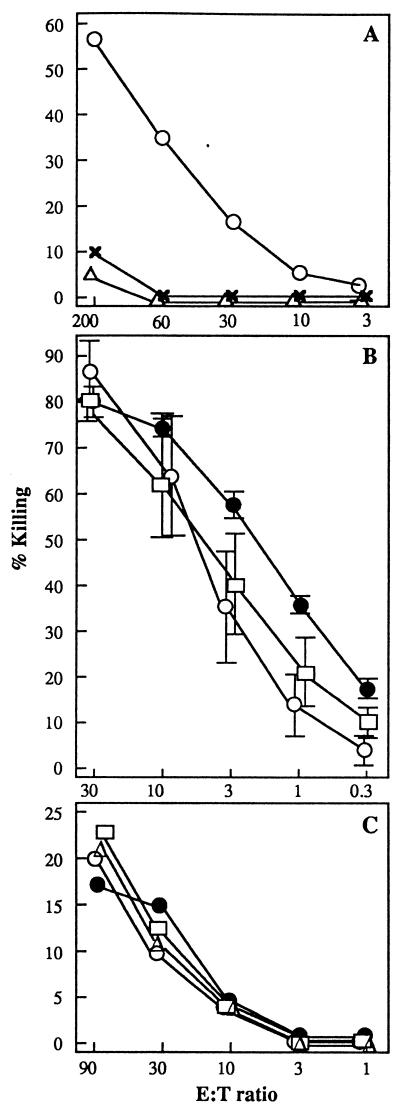
Figure 2.
Graphs showing gB-specific CTL activities of splenocytes (A and B) or lymph node cells (C). (A) Mice were injected i.d. with 50 μg fHSVΔpac DNA (○), 50 μg psOVA control DNA (x), or 106 transducing units of HSV-1 amplicon vector pHSVGFP (▵) and, 2 weeks later, boosted with the same amount of DNA or vector. Ten days after the booster immunizations, splenocytes were isolated, restimulated in vitro, and analyzed for HSV-1-specific CTL activity in a 4-h 51Cr release assay. Results are expressed as the specific lysis at various effector to target (E:T) ratios. (B) Two groups of three mice were immunized once with 1.5 μg of fHSVΔpac DNA by gold-particle bombardment (○, □), and a third group of three mice was injected with 109 PFU of DISC HSV-1 (●). Two weeks later, splenocytes from the three mice in each group were isolated, restimulated separately, and tested individually for CTL activity as described in A. The mean (+/− SD) of three assays is shown. (C) Three groups of three mice were immunized with 1.5 μg fHSVΔpac DNA by gold-particle bombardment (○, □), and a fourth group of three mice was immunized with 109 PFU of DISC HSV-1 by injection (■). The animals were boosted 2 weeks later with the same reagents. Ten days after the booster immunization, the draining lymph node cells of the mice of each group were pooled and analyzed in a direct overnight 51Cr release assay. Lysis of nonpeptide loaded target cells is subtracted from the data shown.
Other investigators have demonstrated that administration of DNA by gold-particle bombardment is 10- to 100-fold more effective for the induction of immune responses than direct i.d. injection (16, 43). The method of DNA transfer may be even more critical for large DNA molecules, such as the 158-kb plasmid fHSVΔpac. The following experiment was performed to address this point.
Immunizing Mice Once with fHSVΔpac DNA by Gold-Particle Bombardment Induces Strong CTL Activity After in Vitro Restimulation.
Mice were primed with 1.5 μg of fHSVΔpac DNA delivered i.d. by gold-particle bombardment, and the immune responses were compared with those induced by i.d. injection of 50 μg of fHSVΔpac DNA (see above), or infection with either 104–109 PFU of DISC HSV-1 or 106 transducing units of helper virus-free HSV-1 amplicon vector pHSVGFP. DISC HSV-1 can complete one cycle of replication without producing infectious progeny virus (29) and was used as standard modified live virus in all forthcoming experiments. HSV-1 amplicon vectors are replication-defective, do not express any viral genes, but contain all structural components of HSV-1 particles (33).
One or two weeks after a single vaccination, splenocytes were harvested and analyzed for HSV-1 gB-specific CTL activity after in vitro restimulation. The results are shown in Fig. 2B and can be summarized as follows: A single immunization with 1.5 μg of fHSVΔpac DNA by gold-particle bombardment was far more effective in inducing CTL activity than two i.d. injections with 50 μg of DNA from the same batch. A similarly high gB-specific CTL activity was induced after immunization with 109 PFU of DISC HSV-1, which decreased with smaller amounts of virus in a dose-dependent manner (not shown). Immunization with psOVA DNA or with 106 transducing units of HSV-1 amplicon vector did not result in detectable CTL activity against gB (Fig. 2 A and B). However, restimulated splenocytes from psOVA-immunized mice revealed cytotoxic activity when assayed against the OVA peptide 257–264 (44) (data not shown).
The next experiments aimed to further characterize the fHSVΔpac-induced CTL response and to determine whether CTL activity was detectable without in vitro restimulation of splenocytes or lymph node cells. DNA immunizations were performed by gold-particle bombardment in all further experiments.
CTLs from fHSVΔpac DNA-Immunized Mice Have an Increased Frequency of the TCR Vβ10.
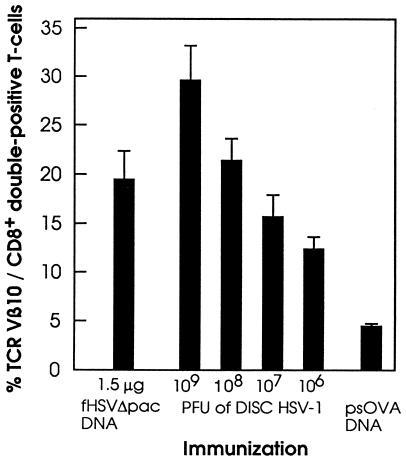
A single H-2b restricted peptide of gB is a major target for CTL cell lines or T cells freshly isolated from draining lymph nodes of HSV-1-infected C57BL/6 mice (30). These T cells are CD8+ and express a highly conserved TCR Vβ10/junctional sequence combination that can be recognized by a TCR Vβ10-specific mAb (31, 39, 45). Fluorescence-activated cell sorting analysis of the spleen cells from fHSVΔpac DNA-immunized mice showed that 20–30% of the cells expressed TCR Vβ10 after restimulation (Fig. 3). In vitro restimulated splenocytes from mice immunized with 106–109 PFU of DISC HSV-1 also contained significant numbers of TCR Vβ10 expressing CD8+ T cells (Fig. 3). Similar results also were obtained in 129Sv/Ev (H-2b) mice immunized with either fHSVΔpac DNA or DISC HSV-1 (data not shown). The presence of TCR Vβ10 and CD8+ double-positive T cells specific against a single H-2b restricted peptide of gB after immunization with fHSVΔpac DNA indicates an immunodominance of gB over the other HSV-1 (glyco)proteins that resembles infection with wt HSV-1 or DISC HSV-1. The i.d. injection of 106 transducing units of HSV-1-amplicon vector, which was used as a control because it does not express any HSV-1 genes, did not induce detectable levels of CTL activity (Fig. 2B) and no HSV-1-specifc antibodies (data not shown). Moreover, the population of TCR Vβ10 and CD8+ double-positive T cells among the restimulated splenocytes was approximately 5%, similar to splenocytes from control mice (Fig. 3). These results are consistent with earlier experiments that had indicated that infusion of helper virus-free HSV-1 amplicon vector into mouse liver supported the long-term expression of the transgene without causing apparent inflammatory reactions (46).
Figure 3.
Graph showing TCR Vβ10/CD8+ double-positive T cells. Mice (three per group) were immunized with 1.5 μg fHSVΔpac DNA by gold-particle bombardment, or with 109, 108, 107, or 106 PFU of DISC HSV-1 by infection. Ten days after the immunizations, splenocytes were harvested, restimulated in vitro for 5 days, and then analyzed by fluorescence-activated cell sorting. The numbers indicate the ratios of TCR Vβ10 and CD8+ double-positive T cells to the total numbers of CD8+ T cells × 100%. Restimulated splenocytes from psOVA DNA- immunized mice served as control.
Primary CTL Response by Lymph Node Cells from Mice Immunized Twice with fHSVΔpac DNA.
Lymph node cells and spleen cells from mice vaccinated with a single dose of fHSVΔpac DNA were analyzed for direct CTL activity on gB peptide-loaded EL-4 target cells. CTL activity could not be detected in both cell populations without in vitro restimulation, and the number of TCR Vβ10 and CD8+ double-positive T cells was similar to that of the control animals (data not shown). To determine whether primary CTL activity is detectable after two immunizations, mice were primed with either 1.5 μg of fHSVΔpac DNA or 109 PFU of DISC HSV-1 and, after 14 days, boosted with the same amount of DNA or DISC virus. Ten days after the booster immunizations, significant primary CTL activity was found in lymph node cells draining the site of DNA or virus inoculations (Fig. 2C). In addition, the number of TCR Vβ10 and CD8+ double-positive T cells in the lymph node cell population was 5–10% higher than that of control mice (data not shown). Hence, mice immunized with fHSVΔpac DNA or DISC HSV-1 responded with CD8+ T cells expressing TCR Vβ10, similar to animals infected with replicating wt HSV-1 (31, 39, 45).
Immunization with fHSVΔpac DNA Induces the Production of Antibodies of All IgG Isotypes and Against a Variety of HSV-1-Specific Proteins.
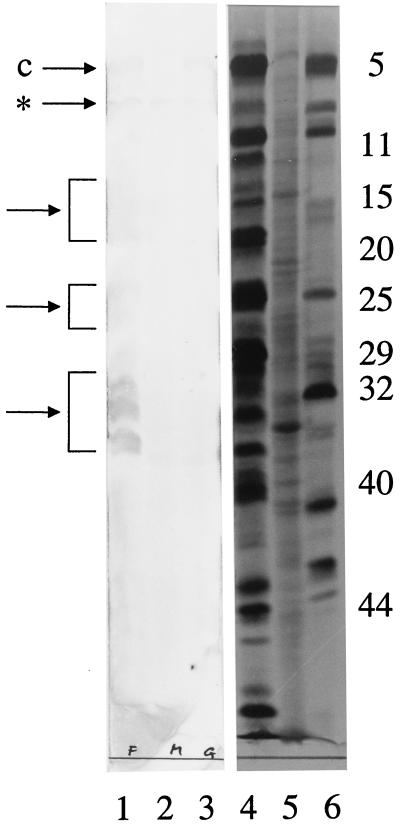
C57BL/6 mice were immunized once with 1.5 μg of fHSVΔpac DNA by gold-particle bombardment or with 109 PFU of DISC HSV-1 by infection. ELISA data of the sera analyzed 14 days later showed that DNA immunization resulted in significant HSV-1-specific antibody titers that were, however, 20- to 30-fold lower than those obtained after DISC HSV-1 infection (Table 1). HSV-1-specific antibodies of the IgG2a and IgG2b isotypes were detected in the sera after priming with DNA or virus. Ten days after a second immunization with fHSVΔpac DNA, the HSV-1-specific antibody titer increased 10-fold, whereas a second dose of DISC HSV-1 resulted only in a 2- to 3-fold increase. This increase in the titer of HSV-1-specific antibodies upon second immunizations with fHSVΔpac or DISC HSV-1 indicates an induction of immune memory. Neutralizing antibodies were detected in sera of mice immunized twice with fHSVΔpac DNA or once with DISC HSV-1 (Table 1). A second immunization with DISC HSV-1 increased the titer 8-fold. Serum from fHSVΔpac-immunized mice recognized a wide range of HSV-1-specific proteins, as determined by Western immunoblot analysis of cells infected with either HSV-1 or HSV-2 (Fig. 4). One of the proteins detected in both HSV-1-and HSV-2-infected cells represents the major capsid protein (ICP5).
Table 1.
Serology
| Immunogen | Mouse strain | No. of immunizations | IgG isotype
|
Neutralization | |||
|---|---|---|---|---|---|---|---|
| IgG1 | IgG2a | IgG2b | IgG3 | ||||
| fHSVΔpac DNA | C57BL/6 | 1 | 0.1 | 0.2 | 1.5 | <0.1 | <2 |
| 2 | 0.5 | 2.5 | 12.1 | 1 | 8 | ||
| 129Sv/Ev | 1 | 0.1 | <0.1 | <0.1 | <0.1 | n.d. | |
| 2 | 2.5 | 0.5 | 0.5 | <0.1 | n.d. | ||
| 3 | 6.7 | 12.4 | 2.5 | 0.5 | 8 | ||
| DISC HSV-1 | C57BL/6 | 1 | 2.5 | 11.1 | 32.5 | 1 | 8 |
| 2 | 1.6 | 12.5 | 61.3 | 2.5 | 64 | ||
| 129Sv/Ev | 1 | 1.5 | 14.1 | 16.7 | 2.1 | n.d. | |
| 2 | 3.2 | 18.5 | 11.3 | 8.3 | n.d. | ||
| 3 | 2.2 | 16.5 | 33.1 | 3.2 | 64 | ||
Groups of four mice were immunized with either 1.5 μg fHSVΔpac DNA by gold-particle bombardment or 109 PFU of DISC HSV-1 by infection. Mice were boosted at biweekly intervals, and blood was collected immediately before boosting. Sera were analyzed for different IgG isotypes by ELISA (the mean titer is given as serum dilution × 10−3) or for the capacity to neutralize HSV-1 (titer = 1/serum dilution). n.d., not done.
Figure 4.
Immunoblot of HSV-1- and HSV-2-infected cell proteins. Sera from fHSVΔpac DNA-immunized mice were pooled and analyzed for the presence of anti-HSV-1-specific and anti-HSV-2-specific antibodies by Western immunoblot analysis of infected cells (lanes 1–3). Lanes 4–6 represent the autoradiographic images of the cell lysates shown in lanes 1–3. Radioactively labeled cell extracts were prepared, subjected to SDS/PAGE, transferred to nitrocellulose, and immunostained as described in Materials and Methods. Lanes 1 and 4: lysates of cells infected with HSV-1 (strain F); lanes 2 and 5: mock-infected cells (M); lanes 3 and 6: cells infected with HSV-2 (strain G). The numbers on the right refer to infected cell proteins (ICP) specified by HSV-1 (see ref. 49). The arrows point to the major capsid proteins (c) of both HSV-1 (ICP5) and HSV-2 or to clusters of immunologically stained HSV-1-specific proteins. * indicates a band, which was stained in the infected as well as in the mock-infected cell lysates.
Immunization of mice of a different strain, 129Sv/Ev, which is MHC compatible to C57BL/6, also resulted in the production of HSV-1-specific antibodies of all IgG isotypes upon two immunizations with fHSVΔpac or a single immunization with DISC HSV-1. Interestingly, a second immunization with DISC HSV-1 did not increase the antibody titer in these mice. Preliminary data indicate that after three immunizations with fHSVΔpac, the antibody titer is maintained for more than 3 months without further boosting the mice.
Immunization with fHSVΔpac DNA Protects Mice from Lethal HSV-1 Infection.
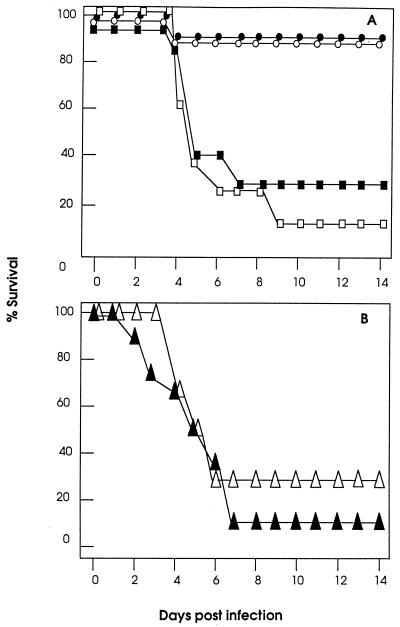
Next, we determined whether the immune responses induced by immunization with fHSVΔpac DNA was protective against lethal HSV-1 infection. The i.c. route was chosen for the challenge experiments because mice are relatively resistant to HSV-1 infections by peripheral routes but highly susceptible to i.c. inoculation (10, 47, 48). Ten days after a second immunization, groups of eight mice were challenged i.c. with 2 × 105 PFU (200 LD50) of wt HSV-1. The results show clear protection of fHSVΔpac-immunized and DISC HSV-1-immunized mice, but not of control animals. Seven of eight mice immunized with fHSVΔpac DNA or DISC HSV-1 survived the challenge without showing any signs of disease. Animals that were immunized with a single dose of fHSVΔpac DNA or with control DNA were not protected and showed signs of disease, including incoordination, ragged fur, and hunched position. Seven of the eight animals died within 9 days (Fig. 5A). These data indicate that immunization with fHSVΔpac DNA (twice 1.5 μg) confers protective immunity as effectively as infection with DISC HSV-1 (twice 109 PFU). Of note, 109 PFU of the DISC HSV-1 vaccine was chosen for the protection experiments because the CTL activity was comparable to that induced by 1.5 μg of fHSVΔpac DNA (Fig. 2 A and B).
Figure 5.
Graph showing survival of mice after lethal challenge with wt HSV-1. (A) Mice (eight per group) were immunized once (□) or twice (○) with 1.5 μg fHSVΔpac DNA, once with 1.5 μg psOVA DNA (■) by gold-particle bombardment, or twice with 109 PFU of DISC HSV-1 by infection (●). Ten days after the last immunization, the animals were challenged i.c. with 2 × 105 PFU of wt HSV-1 strain F (200 LD50). (B) Animals (eight per group) were i.v. inoculated with 100 μl of pooled sera from fHSVΔpac DNA (▵) or DISC HSV-1 (▴) immunized mice. Four hours later, the animals were challenged with wt HSV-1 as described in A.
Serum transfer experiments were performed to determine whether the humoral arm of the immune system was sufficient to protect mice from lethal i.c. HSV-1 infection. Mice were i.v. inoculated with 100 μl of pooled sera from fHSVΔpac DNA- or DISC HSV-1-immunized mice, which had a neutralization titer of 32. Posttransfer serum collected 4 h later had a neutralization titer of 16. The animals (eight mice per group) challenged 4 h after serum transfer by i.c. infection with 2 × 105 PFU of wt HSV-1 showed signs of disease within a few days, and only three of the 16 mice survived longer than 7 days (Fig. 5B). Furthermore, all four animals injected i.p. with 1 ml of immune-serum with a neutralization titer of 64 died within 7 days after i.c. challenge with 2 × 105 PFU of wt HSV-1 (data not shown). These data are consistent with previous results, which had established that cellular immune responses are required to clear infectious HSV-1 from the brain (48).
Summary and Perspectives.
The prototype BAC-VAC described in this report contains a 150-kb, modified HSV-1 genome that is replication-competent in mammalian cells and expresses at least all of the ≈36 viral genes that are essential for HSV-1 replication, but does not produce infectious progeny virus (fHSVΔpac; ref. 27). Immunization with fHSVΔpac DNA induced broad cellular and humoral immune responses and protected the animals from lethal HSV-1 infection as effectively as vaccination with an equivalent amount of a modified live virus (DISC HSV-1). The results also show that the humoral immune responses induced by fHSVΔpac or DISC HSV-1 were not sufficient to confer protection against the lethal challenge. However, exact mechanisms and duration of protection need to be elucidated for further evaluation of fHSVΔpac as a potential vaccine against HSV-1. Moreover, it will be important to determine whether immunization with fHSVΔpac DNA has an influence on the capability of wt HSV-1 to establish latency after experimental infections via eye, ear, or footpad. The i.c. challenge route chosen for this study because of its stringency is not suitable to address the issue of latency. The focus of this study was to demonstrate the usefulness of BACs to carry large, antigen-encoding DNA fragments, such as modified viral genomes. However, similar strategies could be established on other large-capacity shuttle vectors, such as yeast artificial chromosomes (YAC-VAC), P1-derived artificial chromosomes (PAC-VAC), or cosmids (COS-VAC). BAC-VAC allows to combine the high immunogenicity of modified live virus vaccines with the safety and simplicity of DNA vaccines. Moreover, as opposed to live virus vaccines, the E. coli propagated BAC-VAC can easily and extensively be modified to (i) study the complex interactions between host and parasite, (ii) eliminate specific components of the target pathogen to increase safety and/or counter the capability to evade detection and elimination by the host‘s immune system, (iii) combine heterologous genetic elements, such as antigen-encoding DNA and sequences that support replicative amplification, to enhance or manipulate the immune response, and (iv) develop novel vaccination strategies against many different target agents, including viruses, parasites, and tumors.
Acknowledgments
We thank Drs. F. Carbone, M. Jones (Monash Medical School, Melbourne), W. Heath (Walter and Eliza Hall Institute), Y. Saeki (Osaka University), and Marco W. J. Schreurs (University Nijmegen) for generously sharing ideas and material, and Drs. P. Cameron (Melbourne University) and P. Chaplin (Commonwealth Scientific and Industrial Research Organization, Melbourne) for their assistance with gold-particle bombardment. M.S.’s sabbatical stay at the Walter and Eliza Hall Institute was made possible by members of the institute, in particular K.S. and Dr. F. Battey. We are grateful to Daniella Campania (Burnet Centre Research Fund, Melbourne) and Irma Heid and Beat Scheier (Institute of Virology, University of Zurich) for their skilled technical assistance. This work was supported in part by fellowships to M.S. from the International Union Against Cancer, National Institutes of Health Australia, the Swiss National Science Foundation, and the Swiss Federal Office for Education and Research (European Union Network No. ERBFMRXCT960053).
Footnotes
Abbreviations: BAC, bacterial artificial chromosome; CTL, cytotoxic T lymphocyte; HSV-1, herpes simplex virus 1; DISC, disabled infectious single cycle; wt, wild type; gB, glycoprotein B; i.c., intracerebral; i.d., intradermal; PFU, plaque-forming unit; TCR, T cell receptor.
References
- 1.Kapoor A K, Buckmaster A, Nash A A, Field H J, Wildy P. Immunol Lett. 1982;5:259–265. doi: 10.1016/0165-2478(82)90109-2. [DOI] [PubMed] [Google Scholar]
- 2.Jennings S R, Bonneau R H, Smith P M, Wolcott R M, Chervenak R. Cell Immunol. 1991;133:234–252. doi: 10.1016/0008-8749(91)90194-g. [DOI] [PubMed] [Google Scholar]
- 3.Valyi-Nagy T, Deshmane S L, Raengsakulrach B, Nicosia M, Gesser R M, Wysocka M, Dillner A, Fraser N W. J Virol. 1992;66:7336–7345. doi: 10.1128/jvi.66.12.7336-7345.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lopez Z, Arvin A M, Ashley R. In: The Human Herpesviruses. Roizman B, Whitley R J, Lopez C, editors. New York: Raven; 1993. pp. 397–425. [Google Scholar]
- 5.Smith P M, Wolcott R M, Chervenak R, Jennings S R. Virology. 1994;202:76–88. doi: 10.1006/viro.1994.1324. [DOI] [PubMed] [Google Scholar]
- 6.Manickan E, Rouse B T. J Virol. 1995;69:8178–8179. doi: 10.1128/jvi.69.12.8178-8179.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu T, Tang Q, Hendricks R L. J Virol. 1996;70:264–271. doi: 10.1128/jvi.70.1.264-271.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hendricks R L. Cornea. 1997;16:503–506. [PubMed] [Google Scholar]
- 9.Posavad C M, Koelle D M, Corey L. Nat Med. 1998;4:381–382. doi: 10.1038/nm0498-381. [DOI] [PubMed] [Google Scholar]
- 10.Grob P, Schijns V E C J, van den Broek M F, Cox P J S, Ackermann M, Suter M. J Virol. 1999;73:4748–4754. doi: 10.1128/jvi.73.6.4748-4754.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jenner, E. (1798) reprinted in Camac, C. N. B., ed. (1959) Classics of Medicine and Surgery (Dover, NY), pp. 213–240.
- 12.Morrison L A, Knipe D M. J Virol. 1994;68:689–696. doi: 10.1128/jvi.68.2.689-696.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ghiasi H, Nesburn A B, Wechsler S L. Vaccine. 1996;14:107–112. doi: 10.1016/0264-410x(95)00169-2. [DOI] [PubMed] [Google Scholar]
- 14.Siegrist C A, Saddallah F, Tougne C, Martinez X, Kovarik J, Lambert P H. Vaccine. 1998;16:1473–1478. doi: 10.1016/s0264-410x(98)00111-x. [DOI] [PubMed] [Google Scholar]
- 15.Fynan E F, Webster R G, Fuller D H, Haynes J R, Santoro J C, Robinson H L. Proc Natl Acad Sci USA. 1993;90:11478–11482. doi: 10.1073/pnas.90.24.11478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Robinson H L, Torres C A. Semin Immunol. 1997;9:271–283. doi: 10.1006/smim.1997.0083. [DOI] [PubMed] [Google Scholar]
- 17.Porgador A, Irvine K R, Iwasaki A, Barber B H, Restifo N P, Germain R N. J Exp Med. 1998;188:1075–1082. doi: 10.1084/jem.188.6.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Akbari O, Panjwani N, Garcia S, Tascon R, Lowrie D, Stockinger B. J Exp Med. 1999;189:169–178. doi: 10.1084/jem.189.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Manickan E, Rouse R J, Yu Z, Wire W S, Rouse B T. J Immunol. 1995;155:259–265. [PubMed] [Google Scholar]
- 20.Ghiasi H, Cai S, Slanina S, Nesburn A B, Wechsler S L. Antiviral Res. 1995;28:147–157. doi: 10.1016/0166-3542(95)00045-n. [DOI] [PubMed] [Google Scholar]
- 21.Hariharan M J, Driver D A, Townsend K, Brumm D, Polo J M, Belli B A, Catton D J, Hsu D, Mittelstaedt D, McCormack J E, et al. J Virol. 1998;72:950–958. doi: 10.1128/jvi.72.2.950-958.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O’Connor M, Peifer M, Bender W. Science. 1989;244:1307–1312. doi: 10.1126/science.2660262. [DOI] [PubMed] [Google Scholar]
- 23.Shizuya H, Birren B, Kim U J, Mancino V, Slepak T, Tachiiri Y, Simon M. Proc Natl Acad Sci USA. 1992;89:8794–8797. doi: 10.1073/pnas.89.18.8794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luckow V A, Lee S C, Barry G F, Olins P O. J Virol. 1993;67:4566–4579. doi: 10.1128/jvi.67.8.4566-4579.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Messerle M, Crnkovic I, Hammerschmidt W, Ziegler H, Koszinowski U H. Proc Natl Acad Sci USA. 1997;94:14759–14763. doi: 10.1073/pnas.94.26.14759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Delecluse H J, Hilsendegen T, Pich D, Zeidler R, Hammerschmidt W. Proc Natl Acad Sci USA. 1998;95:8245–8250. doi: 10.1073/pnas.95.14.8245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saeki Y, Ichikawa T, Saeki A, Chiocca E A, Tobler K, Ackermann M, Breakefield X O, Fraefel C. Hum Gene Ther. 1998;9:2787–2794. doi: 10.1089/hum.1998.9.18-2787. [DOI] [PubMed] [Google Scholar]
- 28.Stavropoulos T A, Strathdee C A. J Virol. 1998;72:7137–7143. doi: 10.1128/jvi.72.9.7137-7143.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McLean C S, Erturk M, Jennings R, Challanain D N, Minson A C, Duncan I, Boursnell M E, Inglis S C. J Infect Dis. 1994;170:1100–1109. doi: 10.1093/infdis/170.5.1100. [DOI] [PubMed] [Google Scholar]
- 30.Vasilakos J P, Michael J G. J Immunol. 1993;150:2346–2355. [PubMed] [Google Scholar]
- 31.Cose S C, Kelly J M, Carbone F R. J Virol. 1995;69:5849–5852. doi: 10.1128/jvi.69.9.5849-5852.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ejercito P M, Kieff E D, Roizman B. J Gen Virol. 1968;2:357–364. doi: 10.1099/0022-1317-2-3-357. [DOI] [PubMed] [Google Scholar]
- 33.Fraefel C, Song S, Lim F, Lang P, Yu L, Wang Y, Wild P, Geller A I. J Virol. 1996;70:7190–7197. doi: 10.1128/jvi.70.10.7190-7197.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aboody-Guterman K S, Pechan P A, Rainov N G, Sena-Esteves M, Jacobs A, Snyder E Y, Wild P, Schraner E, Tobler K, Breakefield X O, Fraefel C. NeuroReport. 1997;8:3801–3808. doi: 10.1097/00001756-199712010-00029. [DOI] [PubMed] [Google Scholar]
- 35.Fraefel C. In: Current Protocols in Neuroscience. Crawley J, Gerfen C, McKay R, Rogawski M, Sibley P, Skolnik P, editors. New York: Wiley; 1999. pp. 4.14.1–4.14.16. [Google Scholar]
- 36.Boyle J S, Koniaras C, Lew A M. Int Immunol. 1997;9:1897–1906. doi: 10.1093/intimm/9.12.1897. [DOI] [PubMed] [Google Scholar]
- 37.Boyle J S, Silva A, Brady J L, Lew A M. Proc Natl Acad Sci USA. 1997;94:14626–14631. doi: 10.1073/pnas.94.26.14626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Salvucci L A, Bonneau R H, Tevethia S S. J Virol. 1995;69:1122–1131. doi: 10.1128/jvi.69.2.1122-1131.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jones C M, Cose S C, Carbone F R. Int Immunol. 1997;9:1319–1328. doi: 10.1093/intimm/9.9.1319. [DOI] [PubMed] [Google Scholar]
- 40.Knuchel M, Ackermann M, Muller H K, Kihm U. Vet Microbiol. 1992;32:117–134. doi: 10.1016/0378-1135(92)90100-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ackermann M, Braun D K, Pereira L, Roizman B. J Virol. 1984;52:108–118. doi: 10.1128/jvi.52.1.108-118.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bonneau R H, Salvucci L A, Johnson D C, Tevethia S S. Virology. 1993;195:62–70. doi: 10.1006/viro.1993.1346. [DOI] [PubMed] [Google Scholar]
- 43.Pertmer T M, Eisenbraun M D, McCabe D, Prayaga S K, Fuller D H, Haynes J R. Vaccine. 1995;13:1427–1430. doi: 10.1016/0264-410x(95)00069-d. [DOI] [PubMed] [Google Scholar]
- 44.Hogquist K A, Jameson S C, Heath W R, Howard J L, Bevan M J, Carbone F R. Cell. 1994;76:17–27. doi: 10.1016/0092-8674(94)90169-4. [DOI] [PubMed] [Google Scholar]
- 45.Cose S C, Jones C M, Wallace M E, Heath W R, Carbone F R. Eur J Immunol. 1997;27:2310–2316. doi: 10.1002/eji.1830270927. [DOI] [PubMed] [Google Scholar]
- 46.Fraefel C, Jacoby D R, Lage C, Hilderbrand H, Chou J Y, Alt F W, Breakefield X O, Majzoub J A. Mol Med. 1997;3:813–825. [PMC free article] [PubMed] [Google Scholar]
- 47.Meignier B, Longnecker R, Mavromara-Nazos P, Sears A E, Roizman B. Virology. 1988;162:251–254. doi: 10.1016/0042-6822(88)90417-5. [DOI] [PubMed] [Google Scholar]
- 48.Lewandowski G. Brain Behav Immun. 1997;11:264–272. doi: 10.1006/brbi.1997.0497. [DOI] [PubMed] [Google Scholar]
- 49.Morse L S, Pereira L, Roizman B, Schaffer P A. J Virol. 1978;26:389–410. doi: 10.1128/jvi.26.2.389-410.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]