Abstract
CD4-expressing T cells in lymphoid organs are infected by the primary strains of HIV and represent one of the main sources of virus replication. Gene therapy strategies are being developed that allow the transfer of exogenous genes into CD4+ T lymphocytes whose expression might prevent viral infection or replication. Insights into the mechanisms that govern virus entry into the target cells can be exploited for this purpose. Major determinants of the tropism of infection are the CD4 molecules on the surface of the target cells and the viral envelope glycoproteins at the viral surface. The best characterized and most widely used gene transfer vectors are derived from Moloney murine leukemia virus (MuLV). To generate MuLV-based retroviral gene transfer vector particles with specificity of infection for CD4-expressing cells, we attempted to produce viral pseudotypes, consisting of MuLV capsid particles and the surface (SU) and transmembrane (TM) envelope glycoproteins gp120-SU and gp41-TM of HIV type 1 (HIV-1). Full-length HIV-1 envelope glycoproteins were expressed in the MuLV env-negative packaging cell line TELCeB6. Formation of infectious pseudotype particles was not observed. However, using a truncated variant of the transmembrane protein, lacking sequences of the carboxyl-terminal cytoplasmic domain, pseudotyped retroviruses were generated. Removal of the carboxyl-terminal domain of the transmembrane envelope protein of HIV-1 was therefore absolutely required for the generation of the viral pseudotypes. The virus was shown to infect CD4-expressing cell lines, and infection was prevented by antisera specific for gp120-SU. This retroviral vector should prove useful for the study of HIV infection events mediated by HIV-1 envelope glycoproteins, and for the targeting of CD4+ cells during gene therapy of AIDS.
Keywords: gene therapy, env gene, transmembrane protein truncation
Peripheral blood lymphocytes are well suited as target cells in gene therapeutic protocols, particularly for disorders of the immune system. CD4-expressing cells can be enriched, cultured, and transduced with a gene transfer vector in vitro and reintroduced into a patient. This ex vivo approach to gene therapy has been used for the treatment of adenosine deaminase deficiency and AIDS (1). The most efficient way to stably introduce genes into target cells uses retroviral vectors. Most ongoing clinical trials are based on procedures using modified versions of the Moloney murine leukemia virus (MuLV) for gene transfer. These MuLV-based viral vector particles package viral RNA from which the genes required for viral replication have been removed and replaced by therapeutic genes and selection markers. The particles carrying the vector genome are termed retroviral vector particles. The viral RNAs retain long terminal repeat sequences and the packaging signal, allowing the transcription and packaging into viral particles and the reverse transcription of the RNA upon the infection of the target cell. Functions necessary for virus formation, RNA packaging, and entry into target cells are provided by the packaging cells in trans. These cells are designed to provide the viral proteins Gag, Pol, and Env, which are necessary for the assembly of viral particles and which are encoded by RNAs that do not contain packaging signals. This results in the production of helper-free, replication-incompetent, recombinant retroviral vector particles able to transfer therapeutic genes encoded by suitable retroviral vectors.
The host range of a retrovirus (i.e., its specificity of infection) is determined by the viral surface envelope protein and by the receptors expressed by target cells. Infection is initiated by the interaction of the envelope protein with a receptor molecule on the surface of the target cell (2). In contrast to HIV, which selectively infects CD4-expressing human cells, retroviral particles derived from amphotropic MuLV infect human cells without cell-type specificity. Because the HIV envelope surface (SU) glycoprotein gp120-SU mediates the specific infection of CD4+ lymphocytes, and because capsid particles derived from MuLV can efficiently package and transfer therapeutic genes, it seemed desirable to derive a gene transfer vector combining both properties. The formation of a viral pseudotype comprising the envelope glycoproteins of HIV and the core particle of MuLV could result in a gene transfer vector with infection specificity for CD4+ cells.
The concept of retroviral pseudotypes is based on the ability to assemble envelope glycoproteins on the surface of core particles from a different strain. It has been shown that MuLV core particles can be pseudotyped with envelope proteins of simian sarcoma associated virus (3), feline leukemia virus subgroup B (4), and feline endogenous virus RD114 (5), for example. Incorporation of the vesicular stomatitis virus G protein into virions formed upon coexpression of the gag-pol gene region of MuLV also has been demonstrated (6). Retroviral vectors with an expanded host range and increased stability were generated. MuLV pseudotypes were further produced by rescue of a MuLV vector in a single cell clone stably transfected with a plasmid comprising for the humane T cell leukemia virus type 1 env gene (7). In contrast, pseudotyping of MuLV-derived capsid particles by the envelope glycoproteins of D-type retroviruses or HIV-1 has not yet been observed (8).
Here we report the generation of a MuLV pseudotype that incorporates the HIV envelope glycoproteins and assumes specificity of infection for CD4-expressing cells. Our attempts to incorporate the full-length HIV-1 envelope protein into the viral particles were not successful. We found that only a variant HIV-1 env gene encoding the surface glycoprotein gp120-SU and a carboxyl-terminally truncated transmembrane (TM) protein was incorporated into the pseudotypes. The variant HIV-1 env gene derived from construct pLβAc/env-Tr712-neo (pTr712; refs. 9 and 10) encodes a variant TM protein lacking the carboxyl-terminal 144 amino acids, and HIV-1 variants encoding the Tr712-env gene were previously shown to replicate in permissive CD4+ T cells with kinetics comparable with those of wild-type HIV-1 (9). The Tr712-env gene was expressed in the env-negative packaging cell line TELCeB6 (5), correctly processed, assembled at the surface of cells, and incorporated into MuLV capsid particles. The viral MuLV (HIV-1 Env) pseudotypes were able to infect CD4+ cells.
MATERIALS AND METHODS
Cell Lines and Plasmids.
Plasmids were purified from transformed Escherichia coli strain DH10a or HB101. The molecular cloning of the expression constructs pLβAc/env-neo (pEnv; ref. 10) and pLβAc/env-Tr712-neo (pTr712; refs. 9 and 10) encompassing variant HIV-1 env genes and a neomycin resistance gene, the construct pCMV-env/neo encoding the env gene of human spuma retrovirus clone 2 (HSRV-2), and plasmid pCRUCA encoding the env gene of amphotropic MLV were described elsewhere (9, 11). TELCeB6 cells (5) expressing the vector MFG-nlsLacZ (12) were kindly provided by Y. Takeuchi (Institute of Cancer Research, London) and F. L. Cosset (Centre National de la Recherche Scientifique, Lyon, France), HeLa-CD4+ cells (ADP047; ref. 13) were purchased from the Medical Research Council AIDS Program Reagents Project, and 293 cells (ATCC CRL 1573; ref. 14) were purchased from American Type Culture Collection. Adherent cells were grown in DMEM (GIBCO/BRL) supplemented with 10% fetal calf serum (GIBCO/BRL). BJAB-CD4i (15), C8166, and Molt 4 cells were grown in RPMI medium 1640 containing 10% fetal calf serum. The TELCeB6 cells were transfected with plasmid pTr712 using lipofectamin (GIBCO/BRL) according to the manufacturer’s instructions. Selection of hygromycin-resistant colonies after transfection of plasmid pREP4 (Invitrogen) was carried out in medium containing 200 μg/ml of hygromycin B (Sigma), yielding the cell line TELCeB6/pTr712-K14. The cell line TELCeB6/pTr712–9 was generated by transfection of TELCeB6 cells with the plasmid pTr712 using calcium phosphate precipitation and selection in the presence of 1 mg/ml G418 (Geneticin; GIBCO/BRL).
Viral Infection, Determination of Titers, and Neutralization Experiments.
Adherent target cells were seeded in 24-multiwell plates at a density of 4 × 104 cells per well or in 6-multiwell plates at a density of 2 × 105 cells per well. C8166 and Molt 4 cells (8 × 105) were seeded in 6-multiwell plates. Before infection the cells were incubated in cell medium overnight. Infections were carried out by 3-hr incubation of the target cells with 1 ml of undiluted or diluted supernatants containing retroviral particles. Contaminating cells in the viral supernatants had been previously removed by passage through 0.45-μm filters. The cells were stained for β-galactosidase expression 2 days after infection (16). Viral titers were determined as described (17). Infection of nonadherent BJAB–CD4i cells was monitored by flow cytometric analysis with the substrate fluorescein di-β-d-galactoside, as described previously (18). The titers are expressed in colony-forming units (cfu) per ml. Neutralization of MuLV (HIV-1) infection was carried out in the presence of the mAb H902 (19) directed against the V3 loop of the HIV-1 envelope protein (9) and a serum previously shown to neutralize the infection of permissive T cells by HIV-1.
Immunostaining of Transfected Cells.
Cells were washed with PBS and incubated with ice-cold methanol for 15 min. After repeated washing, blocking buffer (PBS/2% BSA) was added for 1 hr. Cells were washed again and incubated with a 1:1,000 diluted anti-HIV-1 serum. After further washing, cells were incubated with peroxidase-conjugated protein G (Bio-Rad). Finally, antigen-containing cells were visualized by addition of substrate buffer (H2O2 plus 3-amino-9-ethylcarbazol; Sigma).
Western Blot Analysis.
Cell lysates were obtained and Western blot analyses were performed as described previously (20). Viral particles were concentrated from packaging cell supernatants by centrifugation at 45,000 rpm for 45 min at 4°C. Pellets were resuspended in SDS/PAGE sample buffer (21) and subjected to SDS/PAGE. Western blot analysis was performed with polyclonal rabbit antibodies directed against the HIV envelope glycoprotein gp120-SU or p30-MuLV-Gag and horseradish peroxidase-coupled protein A, using the enhanced chemiluminescence Western blot detection kit (Amersham).
Membrane Fusion Capacity of the HIV-1 Envelope Protein.
TELCeB6/pTr712 packaging cells were cultured until they reached subconfluence. The cells were overlaid with a 5-fold excess of CD4-receptor-expressing cells or, as a control, with CD4− cells. Cocultivation was continued for 24 hr after which the cells were photographed.
RESULTS
Full-Length HIV-1 Envelope Glycoproteins Cannot Mediate the Infectivity of MuLV-Derived Vectors.
We attempted to create pseudotyped retroviral vector particles comprising the MuLV capsid and the HIV envelope glycoproteins. For this purpose we used the cell line TELCeB6 (5), which produces MuLV capsid particles, but no envelope proteins. TELCeB6 cells express a MuLV-derived gag-pol gene construct and a MFG-nlslacZ gene containing a MuLV-derived packaging signal. The transcript of the MFG-nlslacZ gene can be packaged into virus particles. The plasmid pLβAc/env-neo (pEnv; ref. 10) (Fig. 1), which encodes the full-length HIV-1 env gene under the control of the β-actin promoter was transfected into TELCeB6 cells. In a control experiment, the related envelope glycoprotein gene of the human spuma retrovirus clone 2 (HSRV-2), also was tested. The HSRV-2-derived env gene was expressed under the control of the cytomegalovirus immediate early promoter. Five micrograms of plasmid DNA comprising one of the expression constructs were transfected into 2 × 105 TELCeB6 cells. Expression of the envelope proteins in the transfected cells was demonstrated by immunostaining of the cells with antisera specific for the viral envelope proteins of HIV-1 (data not shown).
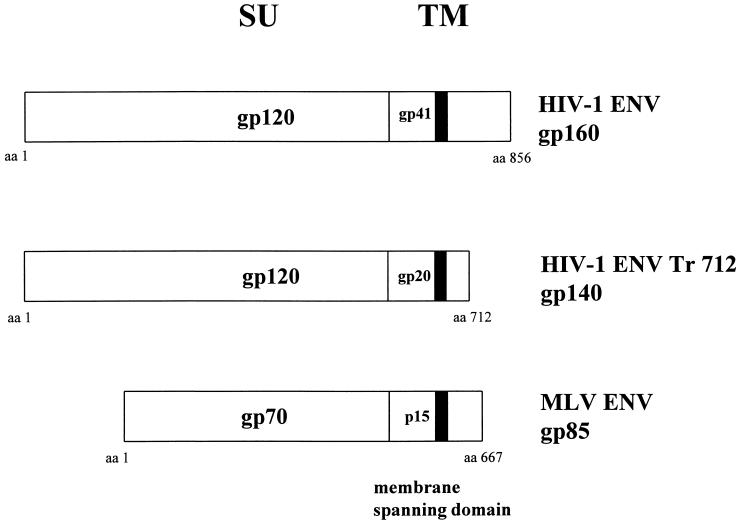
Figure 1.
Envelope glycoproteins derived from the env genes of HIV-1 and MuLV. Plasmid pEnv encodes the wild-type envelope glycoproteins of HIV-1 consisting of a total of 856 amino acids (strain BH10; ref. 22). The membrane spanning region (M) is located between amino acids 681 and 705 (23). Plasmid pTr712 encodes a truncated variant of the TM protein due to the insertion of a stop codon at amino acid position 712 by site-directed mutagenesis. The coding region of the MuLV-derived envelope proteins is shown for comparison.
Supernatants of TELCeB6 cells expressing the viral envelope glycoproteins were collected 1 to 3 days after transfection. One milliliter of supernatant was used for the infection of the human CD4-expressing cells (HeLa-CD4+) or the CD4− cells (293), which are permissive for infection by HIV-1 and human spuma retrovirus, respectively. Upon staining with 5-bromo-4-chloro-3-indolyl β-d-galactosidase (X-Gal) of the cells exposed to the cell supernatants, no blue colonies (indicative of MFG-nlslacZ gene expression) were detected, indicating the absence of detectable gene transfer (Table 1). Experiments were repeated with supernatants from clones of TELCeB6 cells, transfected with pEnv, and obtained by antibiotic selection. These clones were found to be positive for HIV env gene expression. However, infection of HeLa-CD4+ or C8166 cells with supernatants from these clones also could not be demonstrated by X-Gal assay. Evidently, the supernatants did not contain detectable amounts of infectious retroviral vector particles.
Table 1.
Titers of pseudotyped vectors determined by X-Gal staining
| env gene | Target cell | Titer, cfu/ml |
|---|---|---|
| Ampho MuLV | 293 | 6 × 105 |
| HeLa | 5 × 105 | |
| HeL–CD4+ | 4 × 105 | |
| C8166 | n.d. | |
| HIV-1+ | 293 | <1 |
| HeLa | <1 | |
| HeLa–CD4+ | <1 | |
| C8166 | n.d. | |
| HIV-1 truncated‡ | 293* | <1 |
| HeLa | <1 | |
| HeLa–CD4+ | 9 × 104 | |
| C8166* | 3 × 104 | |
| BJAB | <1 | |
| BJAB–CD4i | 3 × 103 | |
| BJAB–CD4i | 1.7 × 104 | |
| Jurkat | 2 × 104 |
X-Gal, 5-bromo-4-chloro-3-indolyl β-d-galactosidase. n.d., not determined.
Pool of hygromycin resistant colonies.
‡ Infections carried out using supernatants from cell clone TELCeB6/pTr712-9. Cells marked with * were infected using supernatants from clone TELCeB6/pTr712-K14.
A Carboxyl-Terminally Truncated Variant of the Transmembrane Protein of HIV-1 Is Incorporated by MuLV Pseudotype Particles.
It previously has been shown that the full-length surface glycoprotein gp120-SU and a carboxyl-terminally truncated variant of the TM protein of HIV-1, of which most of the cytoplasmic domain had been removed, can be expressed, processed, and integrated into HIV particles (9). The variant TM protein encoded by plasmid pTr712 (Fig. 1) has 712 amino acids, including only seven amino acids in its intracellular domain. Whether or not TELCeB6 cells, upon transfection with the plasmid pTr712, expressed envelope glycoprotein on the cell surface and supported the formation of MuLV pseudotypes was investigated.
The expression of the surface HIV-1 envelope glycoprotein was studied by cocultivation of TELCeB6/pTr712 cells with HeLa-CD4+ cells or dexamethasone-induced BJAB-CD4i cells (15) and examination of syncytia formation (Fig. 2 B and D). Syncytia are presumably formed through the association of the SU envelope glycoprotein encoded by pTr712 on the surface of the packaging cell line and the CD4 receptors present on the surface of the HeLa-CD4 cells. No syncytia were formed when TELCeB6/pTr712 cells were cocultivated with HeLa cells or with noninduced BJAB-CD4i cells (Fig. 2 A and C). TELCeB6 cells expressing the amphotropic MuLV envelope protein or ecotropic envelope proteins did not form syncytia with CD4-expressing cells (data not shown).
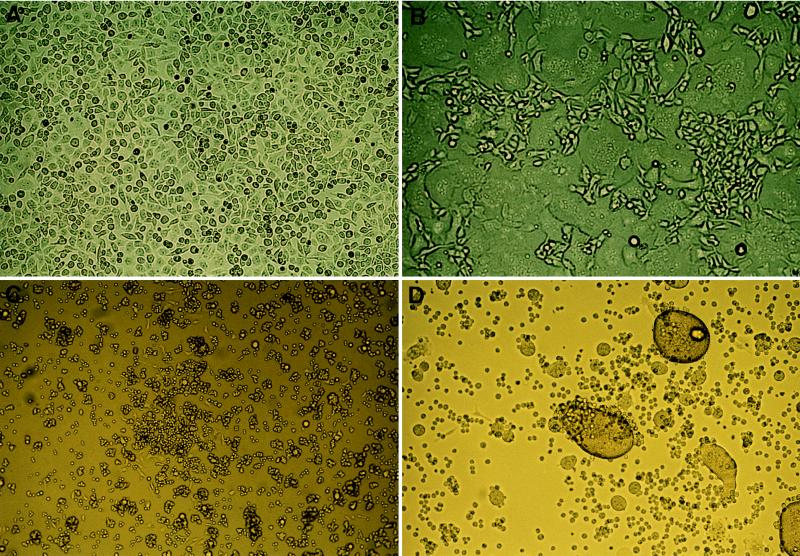
Figure 2.
Demonstration of the functional expression of the HIV-1 surface glycoprotein by syncytia formation with CD4 receptor-expressing cells. The TELCeB6/pTr712 cultures were overlaid with a 5-fold excess of CD4 receptor-expressing cells or as a control, CD4− cells. Cocultivation was continued for 24 hr before they were photographed. (A) TELCeB6/pTr712 cells + HeLa cells. (B) TELCeB6/pTr712 cells + HeLa-CD4 cells. (C) TELCeB6/pTr712 cells + BJAB-CD4i cells. (D) TELCeB6/pTr712 cells + BJAB-CD4i cells treated for 48 hr with 25 nM dexamethasone.
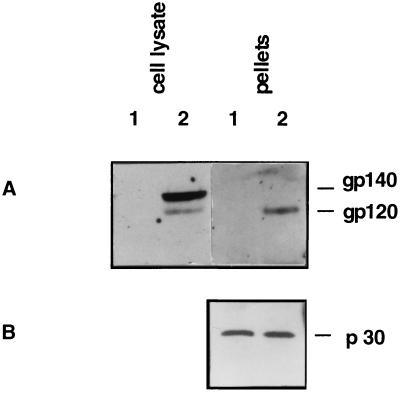
Western blot analysis was performed to investigate the expression of HIV-1 envelope protein in transfected TELCeB6/pTr712 cells and in virus particles isolated from the supernatant of these cells. The precursor (gp140) and the processed surface envelope glycoprotein (gp120-SU) were detected with a polyclonal antiserum specific for gp160 in cell lysates of TELCeB6/pTr712 cells (Fig. 3A, lane 2) whereas untransfected TELCeB6 cells did not react (Fig. 3A, lane 1). In addition, the surface glycoprotein gp120-SU of HIV-1 was detected in pelleted virus particles (Fig. 3A, lane 2), indicating that the surface envelope glycoprotein gp120-SU can become incorporated into MuLV particles if coexpressed with the truncated variant of the HIV-1 envelope TM protein. The presence of proteins derived from the MuLV core particles was demonstrated by Western blot analysis using an antiserum specific for the MuLV p30-Gag protein (Fig. 3B, lanes 1 and 2).
Figure 3.
Detection of HIV-1 Env expression in TELCeB6/pTr712 cells and concentrated supernatants. Nontransfected TELCeB6 cells were used as a negative control. Western blot analysis was performed with polyclonal rabbit antibodies directed against the HIV envelope glycoprotein gp160 (A) or p30-MLV-Gag (B) and horseradish peroxidase-coupled protein A, using the ECL system (Amersham) for detection. Lane 1, TELCeB6; lane 2, TELCeB6/pTr712.
Incorporation of Variant HIV-1 Envelope Glycoproteins into Pseudotypes Results in Virus Particles with Specificity of Infection for CD4+ Cells.
After exposing HeLa-CD4+ cells to supernatants from TELCeB6/pTr712 cells and monitoring expression of β-galactosidase, between 2 × 102 and 1 × 103 blue colonies per ml of supernatant were detected (Fig. 4F). No blue colonies were observed after infection of the parental, CD4−-HeLa cells (Fig. 4E).
Figure 4.
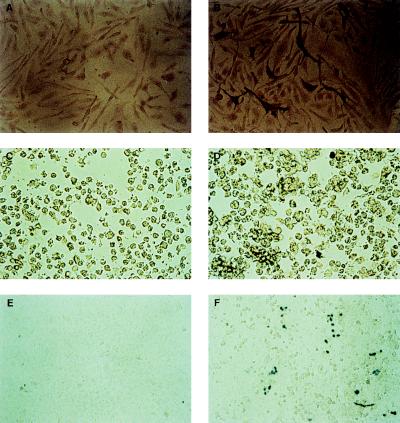
Detection of HIV-1 Env expression in TELCeB6 cells stably transfected by the HIV-1 env expression gene pTr712 and gene transfer into CD4+ human cell lines using MuLV (HIV-1 Tr712) vectors. TELCeB6 cells were mock-transfected (A) or transfected with env expression plasmid pTr712 (B) and immunohistologically stained using an anti-HIV-1 serum. The dark cells represent transfected cells expressing mutant Tr712-env gene. After incubation of CD4+ C8166 T cells with supernatants of cell line TELCeB6/pTr712 (D) and env-negative TELCeB6 cells (C; negative control), 5-bromo-4-chloro-3-indolyl β-d-galactosidase (X-Gal) staining demonstrated successful gene transfer (blue cells in D) by the pseudotyped vector particles derived from the TELCeB6/pTr712 cell clones. Similarly, gene transfer was detected in the CD4+ cell line HeLa-CD4+ (F), but not in parental CD4-HeLa cells (E) using pseudotype vector-containing supernatants from the cell lines TELCeB6/pTr712.
To increase the viral yield, stable clones of TELCeB6 packaging cells expressing the truncated envelope gene pTr712 were selected after cotransfection with the selectable marker gene pREP4(hygr). The expression of the HIV-1 env gene was tested in 50 hygromycin B-resistant cell colonies by immunostaining. Forty-seven of the colonies showed at least 10% HIV-1 env-positive cells (Fig. 4B; see Fig. 4A for the negative control). Several clones were investigated, and the titers of the viral particles released were determined by infection of HeLa-CD4+ cells. Cell clone TELCeB6/pTr712−K14 released about 4 × 104 cfu/ml and clone TELCeB6/pTr712−9 about 9 × 104 cfu/ml.
Supernatants of the TELCeB6/pTr712 packaging cell clones were able to infect HeLa-CD4+ cells, but not the parental CD4−-HeLa cells (Table 1). The specificity of infection also was investigated using the cell line BJAB-CD4i, which can be induced to express CD4 upon treatment with dexamethasone (15). However, residual CD4 expression can be observed in the absence of dexamethasone and a corresponding low level of infectivity was detected. After dexamethasone treatment, a 10-fold increase in infection was observed with virus from TELCeB6/pTr712 supernatants whereas untransfected BJAB cells could not be infected. The specificity of infection of CD4+ cells by the viral pseudotypes also was confirmed in experiments with C8166 T cells (Fig. 4D; see Fig. 4C for the negative control), which demonstrated 5 × 103 to 3 × 104 cfu/ml of supernatant (Table 1).
In a control experiment, TELCeB6 packaging cells were transfected with a gene encoding the envelope glycoproteins of amphotropic MuLV (pCRUCA) and pREP4 (hygr). Upon selection, supernatants from these cells were used to infect 293, HeLa, and HeLa-CD4+ cells (Table 1). The titers observed with the amphotropic MuLV were about 10-fold higher than those measured in the supernatants of cell clones TELCeB6/pTr712-K14 or TELCeB6/pTr712–9.
The specificity of infection was confirmed in blocking experiments in which an antiserum specific for HIV gp120-SU was added to the viral pseudotypes and the infectivity on HeLa-CD4+ cells then was measured. Whereas infection of HeLa-CD4 cells was inhibited by this antibody, efficient infection in the absence of serum or in the presence of a human control serum was achieved (Table 2). Inhibition also was observed with the mAb H902, directed against the V3 loop of the HIV-1 envelope glycoprotein gp120-SU (Table 2).
Table 2.
Neutralization of pseudotyped vectors by incubation with anti-HIV-1 serum
| Serum* | Titer, cfu/ml† |
|---|---|
| None | 1 × 103 |
| Normal donor | 5 × 102 |
| HIV-1-infected donor | <1 |
| None‡ | 3 × 104 |
| Hybridoma H902 supernatant‡ | 1 × 103 |
Supernatants of TELCeB6 cells transiently transfected with plasmid pTr712 were harvested and incubated in the presence or absence of respective sera for 1 hr at room temperaature.
† Titration was performed in HeLa-CD4+ cells.
‡ Pseudotyped vectors were derived from the cell clone TELCeB6/pTr712-9.
DISCUSSION
We describe here the generation of a retroviral gene transfer vector particle with specificity for human CD4-expressing cells. This specificity was conferred upon the MuLV-based retrovirus by supplying the surface envelope glycoprotein gp120-SU and a truncated variant of the envelope TM protein in a packaging reaction and incorporation of the envelope glycoproteins into viral pseudotypes. However, incorporation of the wild-type HIV-1 env gene products into pseudotype particles and formation of infectious virus was not observed, consistent with previous studies in which MuLV (HIV-1 ENV) pseudotype formation could not be demonstrated (8). Why the short form of the envelope TM protein is able to become part of the virus particle and the long form is not is a matter for speculation. Mono- or multimers of the envelope protein precursor are proteolytically cleaved into the SU- and the TM-glycoproteins, which remain associated or covalently linked by disulfide bridges during transport to the cell surface, although further modification or cleavage may occur. Finally, during the budding process, a multimer of the associated or linked Env glycoproteins is incorporated into the capsid particles. Compared with the cytoplasmic domains of other retroviral TM proteins, which generally comprise 22–38 amino acids, that of HIV-1 Env, with 150 amino acids, is somewhat exceptional (24). Because the cytoplasmic residues are ultimately located in the interior of the virion, it is likely that the long cytoplasmic domain of the HIV-1-derived TM protein cannot be accommodated by the MuLV core and interferes with particle formation. If so, the pTr712 short form of the HIV-1 TM might not interfere with particle formation, and incorporation into budding virions would allow the formation of infectious pseudotypes. In agreement with this hypothesis, a failure to accommodate a long heterologous C-terminus also has been demonstrated in the case of HIV-1. In this case, a HIV-1 glycoprotein with a 118-aa-long cytoplasmic region from a cellular protein (CD22), although functionally present at the cell surface, could not be incorporated into particles (25). It also was shown that heterologous proteins with short intracellular domains can be incorporated by MuLV-derived capsid particles if expressed at high levels (26). Therefore, the absence of release of infectious virus from TELCeB6 cells (TELCeB6/pEnv) transfected by the wild-type HIV-1 env gene most likely resulted from absence of wild-type Env incorporation by MuLV capsid particles due to spacial inhibition.
Other examples of viral pseudotypes comprising HIV-1 components have been described. Rabies virus was pseudotyped with the HIV-1-derived envelope glycoproteins (27) with titers of up to 4 × 103 cfu/ml. In this case, pseudotyping was possible only when a hybrid TM protein was used in which the cytoplasmic domain was replaced with that of the rabies virus G protein. Pseudotype formation was not detected when wild-type HIV-1 envelope proteins or mutants with a truncated cytoplasmic domain were used. Expression of the HIV-1 SU envelope protein together with a truncated TM protein, as in the example with the MuLV pseudotype virus described in this paper, is not sufficient for pseudotyping rabies virus. Earlier reports have described the incorporation of HIV-1 envelope protein into vesicular stomatitis virus particles. Similar to the situation with rabies virus, pseudotype particles were formed only when the cytoplasmic domain of the vesicular stomatitis virus G protein was fused to the extracellular and transmembrane domain of the HIV-1 TM protein (28) and titers up to 3 × 103 cfu/ml were achieved.
Several important applications of the MuLV (HIV-1) vector particles produced and characterized in this paper can be envisaged. Two possibilities are the delivery of therapeutic genes to CD4-expressing cells and the study of the molecular events of HIV infection. The primary target cells of HIV are CD4+ lymphocytes, and several gene therapy strategies have been described that aim to introduce protective genes into these cells, which then interfere with infection or the viral replication process. Such “intracellular immunization” can be based upon the transfer of transdominant mutant genes encoding HIV proteins (29), the intracellular expression of HIV specific antibodies (30), the sequestration of viral nucleic acid binding regulatory proteins through overexpression of their cognate RNAs (RNA decoys) (31), ribozymes (32), or antisense constructs (33). It is conceivable that in vivo or ex vivo transduction of CD4+ lymphocytes with retroviral vectors as described above might help to reduce the virus load in the peripheral blood and/or lymph nodes of infected patients.
Applications might go beyond the fight against HIV infection and AIDS. Gene therapy protocols for adenosine deaminase deficiency, for example, depend on the ex vivo gene transfer into CD4-enriched T lymphocytes (1). High titers of the CD4-cell-specific viral vector might allow periodic in vivo application, which would greatly simplify the procedure. MuLV (HIV-1) pseudotypes also will be useful as a safe and rapid assay system for the analysis of molecules that affect HIV-1 entry and permit the development of drugs that interfere with the HIV-1 infection process.
Acknowledgments
We would like to thank D. Kahlenberg and T. Kearns for excellent technical assistance, M. Selbert for expert automatic DNA sequencing, and S. Norley, M. Baier, S. Ottmann, and M. Grez for constructive discussions. We are grateful to M. Grez for the donation of HeLa and NIH 3T3 cells, to A. Rethwilm for the donation of plasmid pCMVenv/neo, to F. Cosset for the donation of TELCeB6 cells, and to J. Schmidt for the donation of anti-p30 (Gag) antibodies. H902 hybridoma cells were obtained from Dr. B. Chesebro through the AIDS Research and Reference Reagent Programme. This work was supported by Grant 01KV9501/9 of the Bundesministerium für Bildung, Wissenschaft, Forschung und Technologie to B.G. and Grants 01 Kl 9406-Teilprojekt 1 and 01 KV 9550 of the Bundesministerium für Bildung, Wissenschaft, Forschung und Technologie to K.C.
ABBREVIATIONS
- HIV-1
HIV type 1
- cfu
colony-forming units
- MuLV
murine leukemia virus
- SU
surface
- TM
transmembrane
References
- 1.Morgan R A, Anderson W F. Annu Rev Biochem. 1993;62:191–217. doi: 10.1146/annurev.bi.62.070193.001203. [DOI] [PubMed] [Google Scholar]
- 2.Weiss R A, Tailor C S. Cell. 1995;82:531–533. doi: 10.1016/0092-8674(95)90024-1. [DOI] [PubMed] [Google Scholar]
- 3.Takeuchi Y, Simpson G, Vile R G, Weiss R A, Collins M K. Virology. 1992;186:792–794. doi: 10.1016/0042-6822(92)90049-u. [DOI] [PubMed] [Google Scholar]
- 4.Porter C D, Collins M K, Tailor C S, Parkar M H, Cosset F L, Weiss R A, Takeuchi Y. Hum Gene Ther. 1996;7:913–919. doi: 10.1089/hum.1996.7.8-913. [DOI] [PubMed] [Google Scholar]
- 5.Cosset F L, Takeuchi Y, Battini J L, Weiss R A, Collins M K. J Virol. 1995;69:7430–7436. doi: 10.1128/jvi.69.12.7430-7436.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burns J C, Friedmann T, Driever W, Burrascano M, Yee J-K. Proc Natl Acad Sci USA. 1993;90:8033–8037. doi: 10.1073/pnas.90.17.8033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vile R G, Schulz T F, Danos O F, Collins M K, Weiss R A. Virology. 1991;180:420–424. doi: 10.1016/0042-6822(91)90050-l. [DOI] [PubMed] [Google Scholar]
- 8.Wilson C, Reitz M S, Okayama H, Eiden M V. J Virol. 1989;63:2374–2378. doi: 10.1128/jvi.63.5.2374-2378.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wilk T, Pfeiffer T, Bosch V. Virology. 1992;189:167–177. doi: 10.1016/0042-6822(92)90692-i. [DOI] [PubMed] [Google Scholar]
- 10.Kräusslich H-G, Ochsenbauer C, Traenckner A-M, Mergener K, Fäcke M, Gelderblom H R, Bosch V. Virology. 1993;192:605–617. doi: 10.1006/viro.1993.1077. [DOI] [PubMed] [Google Scholar]
- 11.Battini J-L, Heard J-M, Danos O. J Virol. 1992;66:1468–1475. doi: 10.1128/jvi.66.3.1468-1475.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takeuchi Y, Cosset F L, Lachmann H, Okada H, Weiss R A, Collins M K. J Virol. 1994;68:8001–8007. doi: 10.1128/jvi.68.12.8001-8007.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chesebro B, Wehrly K, Maury W, Trono D, Baltimore D. J Virol. 1990;9:4155–4160. [Google Scholar]
- 14.Graham F L, Smiley J, Russell W C, Nairn R. J Gen Virol. 1977;36:59–74. doi: 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- 15.Krüger U, Pfeiffer T, Bosch V. AIDS Res Hum Retroviruses. 1996;12:783–792. doi: 10.1089/aid.1996.12.783. [DOI] [PubMed] [Google Scholar]
- 16.Sanes J R, Rubenstein J L R, Nicolas J F. EMBO J. 1986;5:3133–3142. doi: 10.1002/j.1460-2075.1986.tb04620.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pear W S, Nolan G P, Scott M L, Baltimore D. Proc Natl Acad Sci USA. 1993;90:8392–8396. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fiering S N, Roederer M, Nolan G P, Micklem D R, Parks D R, Herzenberg L A. Cytometry. 1991;12:291–301. doi: 10.1002/cyto.990120402. [DOI] [PubMed] [Google Scholar]
- 19.Chesebro B, Wehrly K. J Virol. 1988;198:3070–3075. [Google Scholar]
- 20.Schnierle B S, Moritz D, Jeschke M, Groner B. Gene Ther. 1996;3:334–342. [PubMed] [Google Scholar]
- 21.Laemli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 22.Ratner L, Haseltine W, Patarca R, Livak K J, Starcich B, Josephs S F, Doran E R, Rafalski J A, Whitehorn E A, Baumeister K, Ivanoff L, Petteway S R, Jr, Pearson M L, Lautenberger J A, Papas T S, Ghrayeb J, Chang N T, Gallo R C, Wong-Staal F. Nature (London) 1985;313:277–284. doi: 10.1038/313277a0. [DOI] [PubMed] [Google Scholar]
- 23.Becker Y. Virus Genes. 1991;5:287–312. doi: 10.1007/BF00271529. [DOI] [PubMed] [Google Scholar]
- 24.Hunter E, Swanstrom R. Curr Top Microbiol Immunol. 1990;157:187–253. doi: 10.1007/978-3-642-75218-6_7. [DOI] [PubMed] [Google Scholar]
- 25.Wilk T, Pfeiffer T, Bukovsky A, Moldenhauer G, Bosch V. Virology. 1996;218:269–274. doi: 10.1006/viro.1996.0190. [DOI] [PubMed] [Google Scholar]
- 26.Suomalainen M, Garoff H. J Virol. 1994;68:4879–4889. doi: 10.1128/jvi.68.8.4879-4889.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mebatsion T, Conzelmann K-K. Proc Natl Acad Sci USA. 1996;93:11366–11370. doi: 10.1073/pnas.93.21.11366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Owens R J, Rose J K. J Virol. 1993;67:360–365. doi: 10.1128/jvi.67.1.360-365.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Malim M H, Freimuth W W, Liu J, Boyle T J, Lyerly H K, Cullen B R, Nabel G J. J Exp Med. 1992;176:1197–1201. doi: 10.1084/jem.176.4.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marasco W A, Haseltine W A, Chen S Y. Proc Natl Acad Sci USA. 1993;90:7889–7893. doi: 10.1073/pnas.90.16.7889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee T C, Sullenger B A, Gallardo H F, Ungers G E, Gilboa E. New Biol. 1992;4:66–74. [PubMed] [Google Scholar]
- 32.Sarver N, Cantin E M, Chang P S, Zaia J A, Ladne P A, Stephens D A, Rossi J J. Science. 1990;247:1222–1225. doi: 10.1126/science.2107573. [DOI] [PubMed] [Google Scholar]
- 33.Chatterjee S, Johnson P R, Wong K K. Science. 1992;258:1485–1488. doi: 10.1126/science.1359646. [DOI] [PubMed] [Google Scholar]