Abstract
The til-1 locus was identified as a common retroviral integration site in virus-accelerated lymphomas of CD2-myc transgenic mice. We now show that viral insertions at til-1 lead to transcriptional activation of PEBP2αA (CBFA1), a transcription factor related to the Drosophila segmentation gene product, Runt. Insertions are upstream and in the opposite orientation to the gene and appear to activate a variant promoter that is normally silent in T cells. Activity of this promoter was detected in rodent osteogenic sarcoma cells and primary osteoblasts, implicating bone as the normal site of promoter activity. The isoforms encoded by the activated gene all encompass the conserved runt DNA-binding domain and share a novel N terminus different from the previously reported PEBP2αA products. Minor products include isoforms with internal deletions due to exon skipping and a novel C-terminal domain unrelated to known runt domain factors. The major isoform expressed from the activated til-1 locus (G1) was found to account for virtually all of the core binding factor activity in nuclear extracts from its corresponding lymphoma cell line. Another member of this gene family, AML1(CBFA2), is well known for its involvement in human hemopoietic tumors. These results provide evidence of a direct oncogenic role for PEBP2αA and indicate that the Myc and Runt family genes can cooperate in oncogenesis.
The use of retroviral gene tagging in oncogene transgenic mice has provided a powerful methodology to resolve the complex genetics of cancer (1). Although the number of genes identified in this way continues to grow, the known retroviral insertion loci in the most thoroughly studied system (murine lymphoma) fall into one of three complementation groups that appear to comprise either genetically linked loci or functionally redundant genes. The first group is represented by the Myc family (c-myc, N-myc, L-myc), a second by the Pim family (pim-1, pim-2), and a third large group is defined by the bmi-1/bla-1 and eis-1/gfi-1/evi-5/pal-1 insertion clusters where the bmi-1 and gfi-1 genes appear to be the critical targets for viral activation (2, 3).
Although these three gene groups are involved in a remarkably high percentage of tumors of Eμ-myc and Eμ-pim-1 mice (4–6), there is emerging evidence that Myc is promiscuous with respect to its oncogenic collaborators and that a different pattern of complementing genes can be revealed by altering transcriptional control of the transgene. For example, in virus-accelerated T cell tumors of mouse mammary tumor virus long terminal repeat (MMTVd-LTR)-myc transgenic mice, the preferred target was recently characterized as the murine homologue of Notch1, a gene known for its involvement in translocations in human T cell leukemias (7). In Moloney murine leukemia virus-infected CD2-myc mice, which develop exclusively T cell tumors, we found very few insertions at the known myc collaborating genes, but a high frequency (33%) at a novel locus, til-1, on mouse chromosome 17 (8, 9).
We have now identified a gene affected by these insertions as PEBP2αA (CBFA1), one of a three-member family of mammalian transcription factors related to the Drosophila segmentation gene product Runt. The three CBFα chains bind their target sites in DNA directly, but affinity is increased significantly by interaction with a common β-chain (CBFβ) (10). Although a close relative of PEBP2αA, AML1 (CBFA2) is known to be involved widely in chromosomal translocations in myeloid leukemia and other hemopoietic malignancies (reviewed in refs. 11 and 12). These results provide direct evidence for oncogenic involvement of PEBP2αA and indicate that the Myc and runt domain gene families can cooperate in oncogenesis. Moreover, the overexpression of PEBP2αA in CD2-myc lymphoma cells reveals isoforms with alternative N- and C-terminal domains that may have distinct physiological roles.
METHODS
Cells.
The lymphoma cell lines T1i, T22i, T27i, and T47i were established from thymic lymphomas induced following Moloney murine leukemia virus infection of CD2-myc mice. These mice were heterozygous for an inactivated p53 allele (13). The Tn94 lymphoma cell line was established from a spontaneous thymic lymphoma in a p53−/− mouse (14). The EL4 and BW5147 cell lines were obtained from the European Collection of Animal Cell Cultures (Salisbury, U.K.). Other lines used were NIH/3T3 and its derivatives Ψ2 (15) and E86 (16), and the rat osteogenic sarcoma cell lines ROS and UMR-106 (ATCC CRL-1661). ROS cells and rat primary long-bone osteoblasts were kindly provided by L. Lanyon and R. Suswillo (University of London).
RNA Extraction and Blot Hybridization.
Total RNA was extracted from tumor cell lines using RNAzolB (Biogenesis, Bournemouth, U.K.). Poly(A)+ RNA was purified using the polyATract mRNA isolation system (Promega). RNAs were separated on 1% agarose gels containing 2.2 M formaldehyde and transferred to Hybond N membrane (Amersham) in 20× SSC. Blots were hybridized and washed as previously described (17) and exposed to x-ray film. Probes used were til-1E, a 877-bp PstI-EcoRI fragment from the til-1 locus as shown in Figs. 1a and 2a; PEBP2αA 3′UT, a 1.9-kb NcoI-HindIII fragment spanning the 3′ untranslated sequences of the G1 cDNA clone; and GAPDH, glyceraldehyde 3-phosphate dehydrogenase gene.
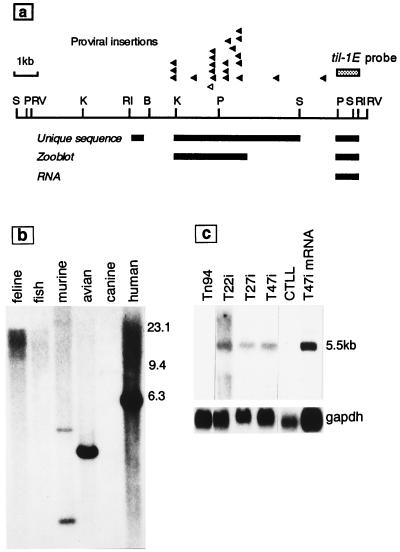
Figure 1.
(a) Physical map of the til-1 insertion locus showing the clustered proviral insertion sites (arrowheads) in T cell lymphomas of Moloney murine leukemia virus-infected CD2-myc mice. The empty arrowhead shows the site of insertion in lymphoma cell line T47i. The locations of single-copy, conserved, and transcribed sequences are indicated by bars underneath. Restriction enzyme abbreviations: RI, EcoRI; RV, EcoRV; B, BamHI; K, KpnI; P, PstI; S, SstI. (b) Evolutionary conservation of til-1E. Human, murine, canine, feline, avian, and fish DNA samples were digested with SstI and hybridized with the til-1E probe. The blot was washed at low stringency (twice in 1× SSC, 0.5% SDS, 50°C). (c) Transcripts related to til-1E are expressed in lymphoma cell lines with proviral insertions at the til-1 locus. Total RNA samples (20 μg) or poly(A)+ mRNA (1 μg) were separated on a 2.2 M formaldehyde gel, and the blot was probed with til-1E before washing at high stringency (0.1× SSC, 60°C). The T22i, T47i, and T27i cell lines carry proviral insertions at til-1. The T cell line CTLL and the lymphoma cell line Tn94 are unrearranged at til-1.
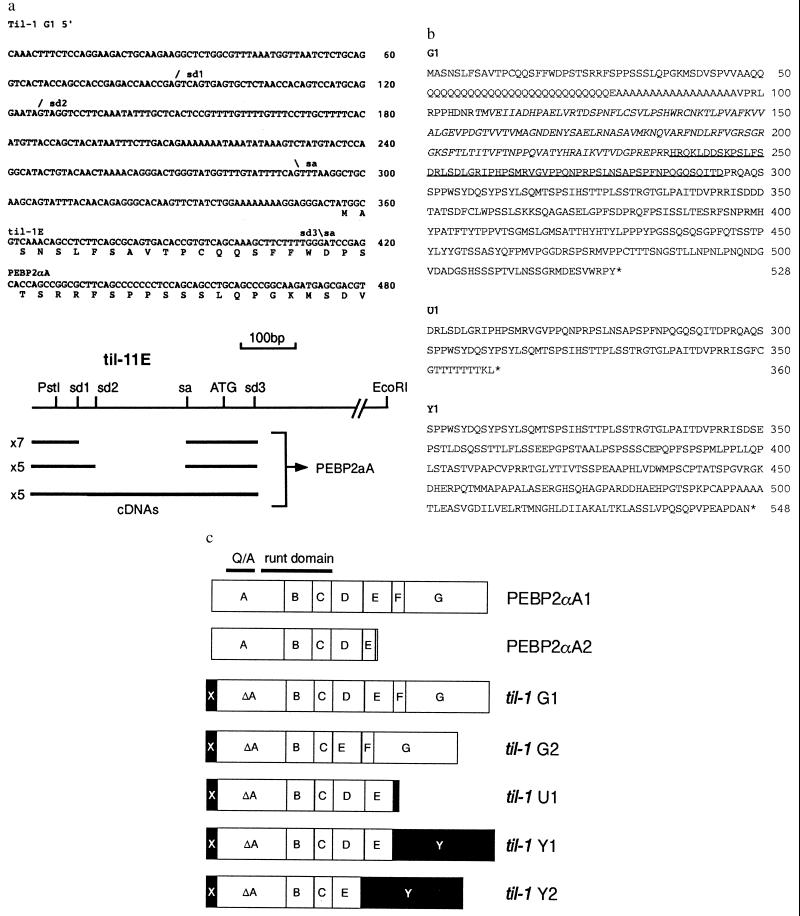
Figure 2.
(a) 5′ end of a representative til-1 class 1 cDNA (see Table 1). The first 413 bases of this cDNA correspond to the til-1E domain, whereas the remainder matches closely with the published PEBP2αA1 sequence (18). The positions of heterogeneous splice variations in the 5′ noncoding sequence are indicated (sd1–3 and sa). (Bottom) The diagram shows the relationship of the til-1E genomic sequence with the cDNA clones. Seven cDNAs were found to be spliced from sd1 to sa, five from sd2 to sa, and the remaining five were unspliced within the til-1E domain. All cDNAs used sd3, which was spliced directly into the known sequence of PEBP2αA. (b) Predicted gene products of til-1 cDNA clones. Most of the clones encoded the G1 isoform in which the runt domain is shown in italics. The amino acid sequence missing from the G2 and Y2 forms is underlined. Alternative C termini for the U1 and Y1/Y2 isoforms are shown below. (c) Diagram showing the structure of proteins encoded by til-1 cDNAs. The published structures of PEBP2αA1 and αA2 (18) are shown at the top, with protein sequences boxed according to their origins from the known intron–exon structure (19). All til-1 gene products share a characteristic N-terminal domain (X), which is derived from an exon within til-1E (see a). Alternative splicing at the 3′ end gives rise to the novel U1, Y1, and Y2 isoforms (see b). Exon D is missing from cDNAs encoding G2 and Y2. Sequences distinct from those found in the previously published PEBP2αA are shown by solid boxes.
cDNA Cloning.
cDNA was prepared from 5 μg poly(A)+ RNA from the T47i lymphoma cell line using the ZAP-cDNA synthesis kit (Stratagene). The cDNAs were size fractionated, and the largest fraction was cloned into Uni-ZAP XR vector arms. An unamplified phage library was screened using the til-1E probe, and 19 positive recombinants were plaque purified and excised as pBluescript phagemids.
DNA Sequence Analysis.
cDNA clones were shotgun cloned and subjected to cycle sequence analysis using M13 IRD41-labeled primers (Hybaid, Middlesex, U.K.) and ThermoSequenase (Amersham) with separation on denaturing Long Ranger gels (FMC) and data recording on a Li-Cor (Lincoln, NB) automated sequencer. Regions of high G-C content were analyzed further using ThermoSequenase cycle sequencing with 33P-radiolabeled terminators (Amersham).
Preparation of Antiserum.
A glutathione S-transferase fusion protein was generated by insertion of a fragment encoding the C-terminal 61 amino acids of the G1 isoform (SmaI–stop) into the pGEX-5X-2 vector (Pharmacia) between the SmaI and XhoI sites. The purified fusion protein (500 μg) was injected intramuscularly into a rabbit in Freund’s complete adjuvant. Boosting was carried out 4 weeks later in Freund’s incomplete adjuvant. The anti-til1-G1 serum was collected 4 weeks later.
Electrophoretic Mobility Shift Assay.
Nuclear extracts were prepared and assayed using a commercial BandShift kit according to the manufacturer’s instructions (Pharmacia) under low-salt conditions (50 mM NaCl). The target core-binding oligonucleotide was generated by annealing of oligonucleotides 5′-GGGGATATCTGTGGTAAGCA-3′ and 5′-GGTGCTTACCACAGATATCC-3′ (20) and labeled with [α-32P]dCTP with Klenow fragment.
Reverse Transcriptase–PCR Analysis.
cDNAs were synthesized from 5-μg aliquots of total RNA primed with the oligo 5′-GCTCACGTCGCTCATCTTGC-3′ using a First-Strand cDNA Synthesis kit (Pharmacia). PCR amplification was then carried out using the same 3′ primer and a 5′ primer, 5′-AAAACAGGGACTGGGTATGG-3′. Cycling conditions were 95°C/1 min, 63°C/30 sec, 72°C/1 min for 30 cycles. Products were separated on a 4% acrylamide gel and blotted onto Hybond N (Amersham). Specific products were detected by hybridization with the til-1E probe.
RESULTS
Identification of a Transcription Unit Activated by Proviral Insertions at til-1.
To search for a gene affected by insertions at til-1, we screened mouse DNA in the vicinity of the clustered integration sites (arrowheads in Fig. 1a) for sequences that are single-copy, conserved in evolution, and expressed as RNA. Sequence analysis of the 10-kb EcoRI fragment encompassing this site showed no evidence of a significant match to database sequences. However, blot hybridization with subcloned probes revealed two stretches of unique sequence that also showed evidence of cross-species hybridization. One of these, til-1E, also detected abundant transcripts in Northern blots of lymphoma cell lines carrying insertions at til-1E (T22i, 27i, 47i) but not in others of similar origin lacking til-1 rearrangements. The predominant RNA species detected was around 5.5 kb, although minor bands of lesser and greater mobility could also be seen. The hybridization of til-1E to DNA (Fig. 1b) and to Northern blots (Fig. 1c) from various species is shown.
The til-1 Transcription Unit Encodes PEBP2αA/CBFA1.
To characterize the transcripts detected by the til-1E probe in greater detail, we generated a cDNA library from polyadenylated RNA of the T47i lymphoma cell line. Screening with the probe yielded a total of 19 positive clones, of which 17 were characterized in detail by restriction mapping and complete or partial sequencing. As shown in Fig. 2a, the 5′ ends of the cDNA clones are homologous to til-1E, but the remainder of the cDNA showed a match to the published sequence of PEBP2αA (18), a gene that, like til-1, was previously mapped to the central region of mouse chromosome 17 (21). Sequence similarity to PEBP2αA commences within the published coding sequence (after codon 5), suggesting that the til-1E exons provide an alternative 5′ end and transcriptional initiator. The cDNAs reveal differential splicing within the til-1E sequence, but all share the predicted initiator ATG (as shown in Fig. 2a).
The cDNA sequences fall into five classes according to their size and coding potential (Table 1). Most of the cDNA clones encode the G1 isoform shown in Fig. 2b, which differs from the published PEBP2αA1 sequence only at the N terminus and by an extra glutamine residue within the long Q/A domain, which is characteristic of this member of the mammalian runt family. This cDNA class corresponds to the major RNA species detected by Northern blot analysis, and the G1 form therefore is likely to represent the most abundant gene product in lymphoma cells with insertions at til-1.
Table 1.
cDNA clones isolated from lymphoma cell line T47i
| cDNA class | No. of clones | Size | Priming site | Coding potential |
|---|---|---|---|---|
| 1 | 7 | 4.7–5.0 | 3′ UTR | G1: 528 aa* |
| 2 | 1 | 4.6–4.8 | 3′ UTR | G2: 470 aa |
| 3 | 1 | 1.45 | αA1-3′ | U1: 360 aa |
| 4 | 2 | 2.5 | αA2-3′ | Y1: 548 aa |
| 5 | 2 | 2.35 | αA2-3′ | Y2: 490 aa |
An additional four short cDNA clones (1.1–1.8 kb) may also encode to this form but were incomplete due to their priming within the coding sequence of exon G.
Four cDNA clones (classes 4 and 5) encode an alternative C-terminal domain (Y), which replaces the putative transactivation domain of PEBP2αA with a novel domain quite different in sequence, although similarly rich in PEST amino acids (43%) (Fig. 2b). A further variation was seen in class 2 and 5 cDNAs, which have an internal deletion apparently due to skipping of exon D. It is interesting to note that this exon is homologous to that missing from the PEBP2αB2 isoform (21).
None of the cDNAs correspond precisely to the reported short isoform PEBP2αA2 (18), but a single clone (class 3) encodes a protein of quite similar length (U1), which appears to be the result of splicing from the 3′ end of exon E to the untranslated sequences of PEBP2αA1. These structures are summarized in a diagram in Fig. 2c with the previously established exon boundaries (A-G; ref. 19) marked to indicate the origins of the coding sequences. The novel domain at the N terminus is marked X, and the alternative C-terminal domain (of unknown exon structure), by the letter Y. Filled boxes indicate sequences distinct from those seen in the published PEBP2αA isoforms.
N-Terminal Sequences of til-1 Gene Products Are Similar to Chicken runtB and Variant Isoforms of AML1.
As shown in Table 2, we noted a striking similarity between the novel N-terminal sequences of the til-1 gene products and those of AML1c, an isoform of AML1 that is expressed from an upstream promoter (22). A similar N-terminal sequence has also been found in runtB, a chicken gene that is differentially expressed during chondrocyte development (23). The til-1 sequence encoded by til-1E and the chicken runt B leader appear to be analogous to the first of two upstream exons of AML1c. We noted a further parallel in the pattern of splicing upstream of the coding sequence of AML1c (22) with that found in til-1/PEBP2αA (Fig. 2a) despite the lack of obvious sequence homology between these sequences. It appears that the til-1 locus harbors a variant upstream promoter at least 20 kb upstream of the major PEBP2αA coding exons and that this promoter is activated by the insertion of Moloney murine leukemia virus in CD2-myc mouse lymphomas.
Table 2.
N-terminal sequences of til-1, PEBP2, AML1, and chicken runt proteins
Transcripts from the Upstream Promoter of PEBP2αA Are Undetectable in T Cell Lines Lacking Viral Insertions but Can Be Found in Normal Osteoblasts and Osteogenic Sarcoma Cell Lines.
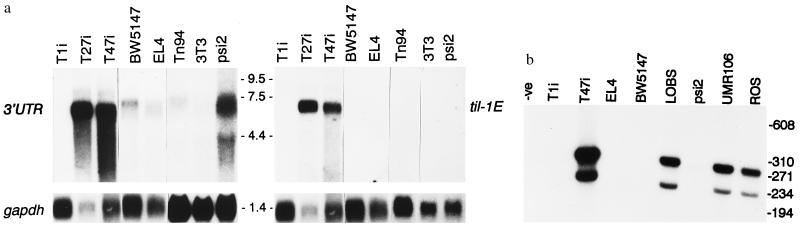
As shown in Fig. 3a, probes derived from til-1E and from the 3′ untranslated region (UTR) of til-1-G1 cDNA were used to analyze the structure of PEBP2αA transcripts in cell lines previously shown to express the gene (EL4 and BW5147 lymphoma cells, ras-transformed 3T3 fibroblasts) (18). The 3′ UTR probe revealed mRNA of the expected size in each case. In contrast, the til-1E probe detected transcripts only in lymphoma cell lines carrying proviral insertions at til-1E. The levels of RNA were significantly higher in lymphoma cells with insertions at til-1 compared with other sources, apart from Ψ2, a retroviral packaging cell line derived from 3T3 cells. We found large differences between 3T3-derived sublines in levels of PEBP2αA mRNA but with no obvious relationship to ras transformation (18). The predominant transcripts in lymphoma cell lines with til-1 insertions also appeared to be slightly smaller than from other sources, presumably due to their shorter 5′ noncoding sequences compared with the reported sequence.
Figure 3.
(a) Detection of PEBP2αA transcripts with specific probes. RNA from a variety of sources was examined with probes derived from til-1E or with a 1.9-kb NcoI-HindIII probe from the 3′ UTR of G1 cDNA. As expected, the abundant 5.5-kb transcript corresponding to the til-1-G1 form was detectable with both probes, whereas the predominant RNA species in EL4, BW5147, and 3T3-derived cells (18) were detectable only with the 3′ UTR probe. Rehybridization with the GAPDH gene was used as a control for RNA loading. Analyses were performed with 5 μg of poly(A)+ RNA (T1i, 27i, 47i, BW5147, El4), or 20 μg of total cellular RNA (Tn94, 3T3, Ψ2). (b) Analysis of expression from the til-1E promoter by reverse transcriptase–PCR. As detailed in Methods, primers spanning the splice junction from til-1E to the exon A of PEBP2αA were used to analyze cDNA generated from the cell lines indicated. A fragment of the expected size (228 bp) was detected in the T47i lymphoma cell line, in rat primary long-bone osteoblasts (LOBS), and in osteosarcoma cell lines UMR106 and ROS. The larger band is single-stranded DNA. In contrast, T cell and fibroblast cell lines previously shown to express PEBP2αA were negative by this criterion, as were CD2-myc lymphoma cells lacking proviral insertions at til-1 (e.g., T1i).
The til-1E probe did not reveal any transcripts in poly(A)-enriched RNA from a wide range of neonatal mouse tissues. However, the 3′ UTR probe also failed to show any detectable transcripts in the same tissues, and we could not reproduce the faint hybridizing bands of 4.7 kb and 2.1 kb reported in thymus and testis poly(A)+ RNA (24), suggesting that expression of PEBP2αA in normal thymus is very low and/or restricted to specific subsets (not shown).
In view of the expression of the chicken runtB gene in developing bone tissues (23) and reports of an AML1-related gene product that regulates the osteocalcin promoter in mouse and rat osteoblasts (25, 26), we tested bone-derived cell lines for the presence of transcripts related to til-1E. Northern blot analysis with the til-1E probe revealed faint signals at 5.5 kb from UMR106, a rat osteogenic sarcoma line (ATCC CRL-1661) (not shown). A more sensitive reverse transcriptase–PCR assay was devised to detect these transcripts (Fig. 3b), and this revealed expression in T47i lymphoma, UMR106, and another rat osteosarcoma cell line, ROS. The apparently larger product seen in Fig. 3b was found to be sensitive to S1-nuclease but not restriction endonucleases (not shown) and, hence, may be the result of asymmetric PCR amplification.
Normal rat osteoblasts (LOBS) were also positive by this assay, whereas T cell lines and fibroblast cells expressing the previously known form of PEBP2αA were negative. These results indicate that proviral insertions at til-1 induce ectopic expression of bone-specific isoforms of PEBP2αA.
Til-1/PEBP2αA Is the Major Core Binding Activity in T47i Lymphoma Cells.
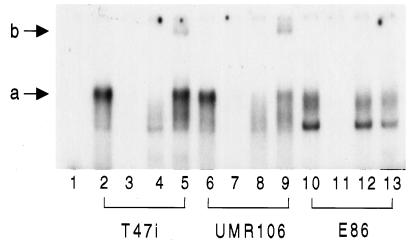
The runt family factors bind to a distinctive motif (R/TACCRCA, R = purine), and this activity can be used to monitor the functional binding proteins in nuclear protein extracts (18, 20). We used a similar approach to analyze binding activity in the T47i cell line. As shown in Fig. 4, electrophoretic mobility shift assay with a core oligonucleotide revealed a strong and sequence-specific band shift pattern in T47i nuclear extracts (a). An antiserum raised to the C-terminal 61 amino acids recognized a predominant 70-kDa protein in cells expressing the gene (not shown). This antiserum appeared to inhibit formation of the complex or rendered it too large to enter the gel (lane 4), but the shift pattern was restored by the addition of excess glutathione S-transferase fusion protein, where a partial supershift was also induced (lane 5, b). In contrast, the complex band shift pattern in E86 cells was unaffected by the antiserum and is presumably due to the binding of other CBF family members (lanes 10–13). These results demonstrate that the major core binding factor in T47i cells is PEBP2αA.
Figure 4.
Electrophoretic mobility shift analysis of core binding factor activity in cell extracts. Nuclear extracts were derived from T47i lymphoma cells (lanes 2–5), UMR 106 rat osteogenic sarcoma cells (lanes 6–9), or E86 mouse fibroblasts (lanes 10–13). No extract was added to lane 1. All reactions included nonspecific competitor poly-dI-dC, whereas those in lanes 3, 7, and 11 included an excess (100×) of unlabeled target oligonucleotide. Antiserum to the C terminus of the G1 isoform (see Fig. 2) was added in lanes 4, 8, and 12, along with an excess of the corresponding glutathione S-transferase fusion protein in lanes 5, 9, and 13. The letters a and b indicate the major bandshift and supershift complexes observed with the T47i and UMR106 extracts.
A similar-size complex was formed with nuclear extracts from the UMR106 cell line, and this could also be specifically inhibited by the antiserum to PEBP2αA (lanes 6–9), indicating that this member of the family is the predominant core binding activity in this bone-derived cell line.
DISCUSSION
We previously identified til-1 as a novel insertion locus in retrovirus-accelerated tumors of CD2-myc mice. We have now shown that these insertions activate the PEBP2αA gene, a close relative of the AML1 oncogene. From the position of the insertions upstream and in the opposite orientation to the gene, it appears that activation is mediated by the action of long terminal repeat enhancers on a cellular promoter element. Furthermore, these insertions reveal the presence of a novel coding exon for PEBP2αA that resembles the 5′ coding sequences of other Runt domain family members, which are expressed in a tissue-specific manner. The til-1-specific upstream promoter is not detectably active in T cell lines previously shown to express PEBP2αA, suggesting that insertions at til-1 induce the ectopic expression of PEBP2αA isoforms that are normally absent from T cells.
We found no evidence of activity of the til-1 promoter in a range of neonatal mouse tissues, including thymus and other hemopoietic tissues. Moreover, very few tissues and cell lines expressed detectable levels of transcripts of PEBP2αA. These observations are consistent with studies of core binding factor activity in a variety of hemopoietic cell lines which showed that the protein product of CBFA1/PEBP2αA was detectable only in a single myeloid cell line, 32D.3, and in Buffalo rat liver cells (27). Because homozygous inactivation of the PEBP2αA gene has revealed a critical role in bone formation (M. Owen, personal communication), developing bone is another likely site of normal gene expression. Moreover, the presence of a til-1E-like N-terminal sequence in a chicken runt family isoform expressed in chondrocytes (23) suggests that the upstream variant promoter may be involved in this process. In support of this hypothesis, we found expression of a til-1E-related transcript in normal rat osteoblasts and in osteogenic sarcoma cell lines, including UMR106, where PEBP2αA proved to be the major core binding activity. Taken together, these results strongly suggest that PEBP2αA encodes the previously described AML1-related factor that binds to the CBF motif in the osteocalcin promoter (25, 26).
Four of the cDNAs cloned from the T47i lymphoma cell line encode a novel C terminus (Y domain) unrelated to that of any known runt-domain family member. The significance of this observation for oncogenesis is unknown because these are likely to be minor products of the overexpressed gene. However, a role for this domain in normal development must be considered. A transactivation domain has been identified in the C terminus of AML1 (21), and the homologous portion of PEBP2αA is completely replaced by the alternative Y domain. The importance of the C-terminal domains of these factors is also clear from recent work on Drosophila runt, where it was shown that the replacement of the last three amino acids with a heterologous activation domain causes severe perturbations in development (28). Requirement for the C-terminal domain of AML1 for functional cooperation with other transcriptional regulators such as the Ets and Myb oncoproteins further underlines its importance (29, 30).
How does ectopic expression of PEBP2αA isoforms contribute to thymic lymphoma development? As genes of this family can act as positive transcriptional regulators (19, 30, 31), the unscheduled expression of target genes is a strong possibility. Regulatory sites for the core binding factors are found in a range of cellular gene promoters including T cell antigen receptor (TCR) α, β, γ, δ, CD3ɛ, granulocyte/macrophage colony-stimulating factor, and interleukin 3, as well as in viral enhancers (31–34). Ectopic expression of growth factors is an attractive model for synergy with Myc, which can induce apoptosis in starved cells (35). However, the most notable feature of the list of candidate CBF target genes is the number of TCR complex genes. Up-regulation of TCR/CD3 expression is a critical event in T cell development (36), and transcriptional stimulation of the component genes might conceivably promote cell transit through growth/differentiation checkpoints or allow survival of cells carrying TCRs with suboptimal or excessive affinity for major histocompatibility complex. The collaboration of TCR complex signaling with Myc would also provide a parallel for our earlier observation of retroviral transduction of both c-myc and TCR-β in a naturally occurring T cell lymphoma (37) and could account for the highly consistent TCR+CD3+ phenotype of CD2-myc lymphomas (8).
However, other models must also be considered. For example, AML1 has been reported to transform 3T3 cells, suggesting a possible direct role of these genes in controlling cell proliferation (38). Also, antagonistic interactions may arise through competition of α-chain isoforms for their common CBF-β binding cofactor or DNA target sites (21, 39). This type of dominant interference mechanism has been postulated for the AML1 fusion proteins that are generated by chromosomal translocations in myeloid and pre-B cell leukemias (27, 40, 41). Moreover, the observation that the AML1 gene promoter includes binding sites for PEBP2/CBF factors (42) suggests the possibility of cross-regulation between isoforms and family members at the transcriptional level.
Further experiments will be aimed at establishing the selective advantage conferred by PEBP2αA activation and its relationship to the other-known pathways of myc-induced oncogenesis. We will also examine the involvement of the other runt domain family members in the tumors of CD2-myc mice that lack insertions at til-1.
Acknowledgments
We are grateful to the Cancer Research Campaign and the Leukemia Research Fund for their support of our research. We also thank Prof. L. Lanyon and R. Suswillo for the kind gift of primary osteoblast cells.
ABBREVIATIONS
- UTR
untranslated region
- TCR
T cell antigen receptor
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF005936).
References
- 1.Berns A. J Cell Biochem. 1994;47:130–135. doi: 10.1002/jcb.240470206. [DOI] [PubMed] [Google Scholar]
- 2.Jonkers J, Berns A. Biochem Biophys Acta. 1996;1287:29–57. doi: 10.1016/0304-419x(95)00020-g. [DOI] [PubMed] [Google Scholar]
- 3.Grimes H L, Chan T O, Zweidler-McKay P A, Tong B, Tsichlis P N. Mol Cell Biol. 1996;16:6263–6272. doi: 10.1128/mcb.16.11.6263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van Lohuizen M, Verbeek S, Scheijen B, Wientjens E, van der Gulden H, Berns A. Cell. 1991;65:737–752. doi: 10.1016/0092-8674(91)90382-9. [DOI] [PubMed] [Google Scholar]
- 5.van der Lugt N M T, Domen J, Verhoeven E, Linders K, van der Gulden H, Allen J, Berns A. EMBO J. 1995;14:2536–2544. doi: 10.1002/j.1460-2075.1995.tb07251.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schmidt T, Zornig M, Beneke R, Moroy T. Nucleic Acids Res. 1996;24:2528–2534. doi: 10.1093/nar/24.13.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Girard L, Hanna Z, Beaulieu N, Hoemann C D, Simard C, Kozak C A, Jolicoeur P. Genes Dev. 1996;10:1930–1944. doi: 10.1101/gad.10.15.1930. [DOI] [PubMed] [Google Scholar]
- 8.Stewart M A, Cameron E, Toth S, Campbell M, McFarlane R, Onions D, Neil J C. Int J Cancer. 1993;53:1023–1030. doi: 10.1002/ijc.2910530628. [DOI] [PubMed] [Google Scholar]
- 9.Stewart M A, Terry A, O’Hara M, Cameron E R, Onions D E, Neil J C. J Gen Virol. 1996;77:443–446. doi: 10.1099/0022-1317-77-3-443. [DOI] [PubMed] [Google Scholar]
- 10.Ogawa E, Inuzuka M, Maruyama M, Satake M, Naito-Fujimoto M, Ito Y, Shigesada K. Virology. 1993;194:314–331. doi: 10.1006/viro.1993.1262. [DOI] [PubMed] [Google Scholar]
- 11.Zent C, Kim N, Hiebert S, Zhang D E, Tenen D G, Rowley J D. Curr Top Microbiol Immunol. 1996;211:243–252. doi: 10.1007/978-3-642-85232-9_24. [DOI] [PubMed] [Google Scholar]
- 12.Meyers S, Hiebert S W. Crit Rev Eukaryotic Gene Expression. 1995;5:365–383. doi: 10.1615/critreveukargeneexpr.v5.i3-4.70. [DOI] [PubMed] [Google Scholar]
- 13.Donehower L A, Harvey M, Slagle B L, McArthur M J, Montgomery C A, Butel J S, Bradley A. Nature (London) 1992;356:215–221. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- 14.Baxter E W, Blyth K, Donehower L A, Cameron E R, Onions D E, Neil J C. J Virol. 1996;70:2095–2100. doi: 10.1128/jvi.70.4.2095-2100.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mann R, Mulligan R C, Baltimore D. Cell. 1983;33:153–159. doi: 10.1016/0092-8674(83)90344-6. [DOI] [PubMed] [Google Scholar]
- 16.Markowitz D G, Goff S P, Bank A. Virology. 1988;167:400–406. [PubMed] [Google Scholar]
- 17.Neil J C, Hughes D, McFarlane R, Wilkie N M, Onions D E, Lees G, Jarrett O. Nature (London) 1984;308:814–820. doi: 10.1038/308814a0. [DOI] [PubMed] [Google Scholar]
- 18.Ogawa E, Maruyama M, Kagoshima H, Inuzuka M, Lu J, Satake M, Shigesada K, Ito Y. Proc Natl Acad Sci USA. 1993;90:6859–6863. doi: 10.1073/pnas.90.14.6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ahn M-Y, Bae S-C, Maruyama M, Ito Y. Gene. 1996;168:279–280. doi: 10.1016/0378-1119(95)00751-2. [DOI] [PubMed] [Google Scholar]
- 20.Wang S, Speck N A. Mol Cell Biol. 1992;12:89–102. doi: 10.1128/mcb.12.1.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bae S-C, Ogawa E, Murayama M, Oka H, Satake M, Shigesada K, Jenkins N A, Gilbert D J, Copeland N G, Ito Y. Mol Cell Biol. 1994;14:3242–3252. doi: 10.1128/mcb.14.5.3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miyoshi H, Ohira M, Shimizu K, Mitani K, Hirai H, Imai T, Yokoyama K, Soeda E, Ohki M. Nucleic Acids Res. 1995;23:2762–2769. doi: 10.1093/nar/23.14.2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Castagnola P, Gennari M, Gaggero A, Daga A, Corsetti M T, Calabi F, Cancedda R. Exp Cell Res. 1996;223:215–226. doi: 10.1006/excr.1996.0075. [DOI] [PubMed] [Google Scholar]
- 24.Satake M, Nomura S, Yamaguchi-Iwai Y, Takahama Y, Hashimoto Y, Niki M, Kitamura Y, Ito Y. Mol Cell Biol. 1995;15:1662–1670. doi: 10.1128/mcb.15.3.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Geoffrey V, Ducy P, Karsenty G. J Biol Chem. 1995;270:30973–30979. doi: 10.1074/jbc.270.52.30973. [DOI] [PubMed] [Google Scholar]
- 26.Merriman H L, Van Wijnen A J, Hiebert S, Bidwell J P, Fey E, Lian J, Stein J, Stein G S. Biochemistry. 1995;34:13125–13132. doi: 10.1021/bi00040a025. [DOI] [PubMed] [Google Scholar]
- 27.Meyers S, Lenny N, Hiebert S W. Mol Cell Biol. 1995;15:1974–1982. doi: 10.1128/mcb.15.4.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jimenez G, Pinchin S M, Ish-Horowicz D. EMBO J. 1996;15:7088–7098. [PMC free article] [PubMed] [Google Scholar]
- 29.Zaiman A L, Lenz J. J Virol. 1996;70:5618–5629. doi: 10.1128/jvi.70.8.5618-5629.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun W, Graves B J, Speck N A. J Virol. 1995;69:4941–4949. doi: 10.1128/jvi.69.8.4941-4949.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hernandez-Munain C, Krangel M S. Mol Cell Biol. 1994;14:473–483. doi: 10.1128/mcb.14.1.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takahashi A, Satake M, Yamaguchi-Iwai Y, Bae S, Lu J, Maruyama M, Zhang Y W, Oka H, Arai N, Arai K, Ito Y. Blood. 1995;86:607–616. [PubMed] [Google Scholar]
- 33.Redondo J M, Pfohl J L, Hernandez-Munain C, Wang S, Speck N A, Krangel M S. Mol Cell Biol. 1992;12:4817–4823. doi: 10.1128/mcb.12.11.4817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cameron S, Taylor D S, Te Pas E C, Speck N A, Mathey-Pervot B. Blood. 1994;83:2851–2859. [PubMed] [Google Scholar]
- 35.Evan G I, Wyllie A H, Gilbert C S, Littlewood T D, Land H, Brooks M, Waters C M, Penn L Z, Hancock D C. Cell. 1992;69:119–128. doi: 10.1016/0092-8674(92)90123-t. [DOI] [PubMed] [Google Scholar]
- 36.Boyd R L, Hugo P. Immunol Today. 1991;12:71–78. doi: 10.1016/0167-5699(91)90161-L. [DOI] [PubMed] [Google Scholar]
- 37.Fulton R, Forrest D, McFarlane R, Onions D, Neil J C. Nature (London) 1987;326:190–194. doi: 10.1038/326190a0. [DOI] [PubMed] [Google Scholar]
- 38.Kurokawa M, Tanaka T, Tanaka K, Ogawa S, Mitani K, Yazaki Y, Hirai H. Oncogene. 1996;12:883–892. [PubMed] [Google Scholar]
- 39.Tanaka T, Tanaka K, Ogawa S, Kurokawa M, Mitani K, Nishida J, Shibata Y, Yazaki Y, Hirai H. EMBO J. 1995;14:341–350. doi: 10.1002/j.1460-2075.1995.tb07008.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tanaka T, Mitani K, Kurokawa M, Ogawa S, Tanaka K, Nishida J, Yazaki Y, Shibata Y, Hirai H. Mol Cell Biol. 1995;15:2383–2392. doi: 10.1128/mcb.15.5.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hiebert S W, Sun W, Davis J N, Golub T, Shurtleff S, Buijs A, Downing J R, Grosveld G, Roussel M F, Gilliland D G, Lenny N, Meyers S. Mol Cell Biol. 1996;16:1349–1355. doi: 10.1128/mcb.16.4.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ghozi M, Bernstein Y, Negreanu V, Levanon D, Groner Y. Proc Natl Acad Sci USA. 1996;93:1935–1940. doi: 10.1073/pnas.93.5.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]