Abstract
The spectrum of mutations induced by the naturally occurring DNA adduct pyrimido[1,2-α]purin-10(3H)-one (M1G) was determined by site-specific approaches using M13 vectors replicated in Escherichia coli. M1G was placed at position 6256 in the (−)-strand of M13MB102 by ligating the oligodeoxynucleotide 5′-GGT(M1G)TCCG-3′ into a gapped-duplex derivative of the vector. Unmodified and M1G-modified genomes containing either a cytosine or thymine at position 6256 of the (+)-strand were transformed into repair-proficient and repair-deficient E. coli strains, and base pair substitutions were quantitated by hybridization analysis. Modified genomes containing a cytosine opposite M1G resulted in roughly equal numbers of M1G→A and M1G→T mutations with few M1G→C mutations. The total mutation frequency was ≈1%, which represents a 500-fold increase in mutations compared with unmodified M13MB102. Transformation of modified genomes containing a thymine opposite M1G allowed an estimate to be made of the ability of M1G to block replication. The (−)-strand was replicated >80% of the time in the unadducted genome but only 20% of the time when M1G was present. Correction of the mutation frequency for the strand bias of replication indicated that the actual frequency of mutations induced by M1G was 18%. Experiments using E. coli with different genetic backgrounds indicated that the SOS response enhances the mutagenicity of M1G and that M1G is a substrate for repair by the nucleotide excision repair complex. These studies indicate that M1G, which is present endogenously in DNA of healthy human beings, is a strong block to replication and an efficient premutagenic lesion.
Keywords: pyrimido[1,2-α]purin-103H-one; site-specific mutagenesis; nucleotide excision repair; template utilization
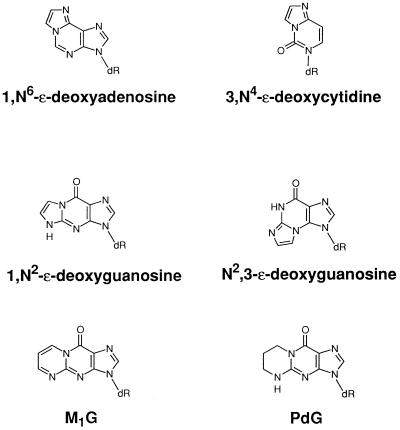
The role of endogenous DNA damage in genetic disease is an important area of contemporary research (1–3). Advances in the sensitivity and specificity of analytical methodology have enabled the detection of a number of DNA lesions in human nuclear DNA that appear to arise from metabolic sources (2). One group of endogenous adducts includes exocyclic DNA adducts such as 1,N6-ethenodeoxyadenosine, N2,3-ethenodeoxyguanosine, 3,N4-ethenodeoxycytidine, and pyrimido[1,2-α]purin-10(3H)-one (M1G) (4–9). These adducts have been detected in human tissues at levels ranging from 10 to 5400 adducts per cell (7–14). The Watson–Crick base pairing region of exocyclic adducts is obstructed, so they are anticipated to be blocks to replication and efficient premutagenic lesions. This has been demonstrated to be the case with 1,N6-ethenodeoxyadenosine and 3,N4-ethenodeoxycytidine (15–18).
M1G appears to be the most abundant endogenous exocyclic adduct in humans, and the structure of the material detected in human liver and white blood cells has been verified by mass spectrometry (14, 19). M1G is the major adduct generated by reaction of malondialdehyde (MDA) with deoxyguanosine residues in DNA. MDA is a product of lipid peroxidation and eicosanoid biosynthesis, and it is widely generated in human tissues (20). It is mutagenic to bacterial and mammalian cells and is carcinogenic in animal studies (21–25). Thus, it is important to determine the biological activity of MDA–DNA adducts, especially M1G, which is detectable in healthy human beings.
Attempts to evaluate the mutagenic potential of M1G have been hampered by its instability to the base deprotection conditions commonly used in oligonucleotide synthesis. This has prevented the synthesis of oligonucleotides containing M1G for site-specific mutagenesis experiments. Therefore, we have used 1,N2-propano-2′-deoxyguanosine (PdG) as a model for M1G and have reported that it is a strong block to replication and induces PdG→A and PdG→T mutations with approximately equal frequency (26, 27) (Fig. 1). PdG has somewhat different stereochemical and hydrogen-bonding properties from M1G, so its validity as a model for the latter is uncertain. Thus, we have made a significant effort to develop a synthetic route to incorporate M1G into oligonucleotides. We recently reported that the use of acetoxymethylbenzoyl-protecting groups provides a route for the synthesis of M1G-containing oligonucleotides because of the mild conditions used for their deprotection and removal from the solid support (K2CO3/methanol) (28). This has enabled us to construct recombinant M13 genomes containing single M1G residues for mutagenesis studies in E. coli. Double-stranded vectors were used that enabled us to evaluate the ability of M1G to block replication, induce mutations, and be repaired. We report herein that M1G is indeed a block to replication and an efficient premutagenic lesion and that its in vivo effects are attenuated by nucleotide excision repair.
Figure 1.
Structures of exocyclic adducts.
MATERIALS AND METHODS
Materials.
KspI was purchased from Boehringer Mannheim. BssHII and T4 DNA ligase were from New England Biolabs. Formamide was from Aldrich Chem (Metuchen, NJ). GELase was from Epicentre Technologies (Madison, WI). T4 polynucleotide kinase was from Amersham. Tris⋅HCl, EDTA, Mops, calf thymus DNA, and lauryl sulfate were from Sigma. X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) and isopropyl β-d-thiogalactoside (IPTG) were from Gold Biotechnology (St. Louis). Nitrocellulose transfer membranes (BA85 82.5 mm diameter) were from Schleicher & Schuell. Ultrafree probind filters (0.45 μm) were from Millipore. [γ-32P]ATP (3000 Ci/mmol) was from DuPont/NEN. The M1G-adducted 8-mer oligonucleotide 5′-GGT(X)TCCG-3′ (X = M1G) was synthesized, purified, and characterized as described (28). After purification, the M1G oligonucleotide was checked by a 20% denaturing polyacrylamide gel to ensure that the 8-mer ran a single band. The M1G 8-mer was determined to be 99.7% pure. Oligonucleotides used as hybridization probes were prepared using an Applied Biosystems automated DNA synthesizer in the Vanderbilt University Molecular Toxicology Molecular Genetics Core and purified using a SurePure oligonucleotide purification kit from Amersham.
Bacterial Strains and DNA Isolation.
All strains used in this study are derivatives of the Escherichia coli strain AB1157 (thr-1, ara-14, leuB6, Δ(gpt-proA)62, lacY1, tsx-33, supE44, galK2, λ-, rac-, hisG4, rfbD1, mgl-51, rpsL31, kdgK51, xyl-5, mtl-1, argE3, and thi-1) except for E. coli strain NR10148. The strains used in this study were LM102 [AB1157; (F′, traD36, proAB, lacIQZΔM15)], LM103 [AB1157uvrA6; (F′, traD36, proAB, lacIQZΔM15)], LM113 [AB1157umuC122; (F′, traD36, proAB, lacIQZΔM15)], LM115 [BH190; (F′, traD36, proAB, lacIQZΔM15)], and NR10148 (ara, thi, Δprolac, zcf-117::Tn10, F′prolac(F′128–27), Δ(uvrB-bio)). LM102, LM103, LM113, and LM115 were constructed as described (22). BH190 (as AB1157 but fpg-1::Knr, uvrA::Tn10) and NR10148 were generous gifts from J. Laval (Institut Gustave–Roussy) and R. Schaaper (National Institutes of Health), respectively.
Single-stranded M13MB102 containing either a cytosine or thymine at site 6256 of the (+)-strand were isolated as described (29). Double-stranded M13MB102 DNAs were harvested using Qiagen columns (Chatsworth, CA). The strain JM105 was used as the host bacterium for the production of M13MB102 and for the indicator plates.
Construction of Site-Specifically Modified M13MB102 Genomes.
Construction of gapped-duplex M13MB102 DNA and the ligation of adducted or unadducted 8-mers were as described previously with a few modifications (26). In brief, double-stranded M13MB102 DNA was linearized with KspI and BssHII then dialyzed with a 12-fold excess of single-stranded M13MB102 DNA in decreasing concentrations of formamide. The resultant gapped-duplex DNA was isolated by a 0.8% low melting point agarose gel run in 40 mM Tris⋅acetate/1 mM EDTA buffer (pH 8). The gapped-duplex band was excised, and the DNA was recovered using the enzyme GELase, according to the procedure provided by Epicentre Technologies. From this point on, the use of Tris buffers was avoided because of observations that Tris can ring-open the M1G adduct in basic and frozen conditions (30). M1G- and G-containing 8-mers (100 pmol) were phosphorylated before ligation using ATP (50 μM) and T4 polynucleotide kinase in 50 mM Mops buffer (pH 7.2). For the ligations, gapped-duplex DNA was added to each of the phosphorylated G- and M1G-containing 8-mers along with 400 units of T4 DNA ligase and ATP (1 mM). The ligation reaction proceeded for 4 hrs at 16°C in 50 mM Mops buffer. The reaction mixtures were then brought up to a volume of 100 μl with water, and the DNA was purified by spinning through modified poly(vinylidene difluoride) membranes from Millipore. The ligation products were then resolved on a 40-mM Mops, 0.8% low melting point agarose gel. Doubly ligated DNA was excised from the gel and recovered using GELase except the supplied 50X Bis⋅Tris⋅NaCl buffer was not used. No additional buffer was added. The amount of enzyme used for digestion was increased from 1 unit of enzyme/600 mg of gel slice to 1 unit/300 mg. Formation of G:T- and M1G:T-mismatched M13MB102 DNA was the same as above except during the formamide dialysis a 12-fold excess of single-stranded M13MB102 was used that contained a T at position 6256.
Transformation of E. coli Cells and Determination of Mutation Frequency.
Cells were SOS-induced and transformed by electroporation as described (27). In brief, bacteria in logarithmic-growth phase were SOS-induced with UV light before making them competent for transformation. The UV dose was determined by irradiating cells at increasing times from 0 to 3 min and then plating dilutions of the irradiated cells on Luria–Bertani plates. The optimal UV dose corresponded to roughly a 10% survival rate of the cells compared with no exposure. For transformation, 3 μl of DNA sample (≈25 ng/μl) was added to 20 μl of cells. The cell/DNA mixture was placed into a chilled GIBCO/BRL microelectroporation cuvette, and the electroporations were performed at 1.5 kV/cm using a GIBCO/BRL Cell-Porator E. coli electroporation system. After electroporation, 1 ml of SOC medium (20 g/liter bacto-tryptone/5 g/liter bacto-yeast extract/20 mM glucose/2.5 mM kCl/10 mM MgCl2/9 mM NaCl) (31) was added, the bacteria were plated on LB plates in the presence of competent bacteria and IPTG, and the bacteria were allowed to grow overnight.
To determine mutation frequencies, phages were eluted from the primary transformation plates, diluted, and then replated with JM105 on X-Gal/IPTG indicator plates to give roughly 300 plaques per plate (32). The plaques on the secondary plates were then lifted using nitrocellulose membranes and probed for base pair substitution mutations at position 6256 by differential hybridization with 13-mer probes (26). Membranes from 12 modified phage plates and 12 unmodified phage plates were split evenly into four dishes. Each dish contained one of the four probes. There was only one lift per plate, not four identical lifts with one membrane being placed into each dish, so the summation of mutations detected along with G hybridizations sometimes did not add up to exactly 100%. The specificity of the probes for a 1-base change at position 6256 has been shown (26, 27).
Frameshift mutations induced by M1G were detected by phenotypic screening with X-Gal/IPTG during the secondary plating. The adduct site in M13MB102 is upstream of the lacZα coding region, so mutations that cause a shift in the reading frame are detected as colorless plaques against a background of blue plaques. The frameshift mutation frequency was determined by counting the number of colorless mutant plaques as a proportion of the total plaque population (32).
RESULTS
Site-specific, M1G-, and G-containing M13MB102 genomes were constructed by the gapped-duplex method (26). In brief, RF M13MB102 was linearized by KspI/BssHII followed by formamide dialysis with a 12-fold excess of single-stranded DNA containing either a C or T at position 6256. The resultant product isolated after agarose gel electrophoresis was a gapped duplex in which the (+)-strand contained either a C or T at site 6256 and the (−)-strand contained an 8-base gap. An 8-base oligonucleotide containing either M1G or G was ligated into this gap. Both the M1G-containing and G-containing 8-mers were purified by two successive rounds of HPLC. The M1G 8-mer was determined to be 99.7% pure by denaturing PAGE. The fully ligated M1G- or G-M13MB102 containing C or T in the (+)-strand was purified by electrophoresis in ethidium bromide-containing agarose gels. Phosphorylation and ligation of the oligonucleotides as well as isolation of the recombinant genomes was conducted in the presence of 50 mM Mops buffer instead of the usual Tris buffer to avoid decomposition of the M1G adduct (30). Incubating the M1G 8-mer for up to 8 h at 20°C in a 50 mM Mops/10 mM MgCl2/5 mM DTT buffer resulted in no degradation of M1G as determined by UV spectroscopy (data not shown). The substitution of Mops for Tris did not inhibit any of the reactions used for genome construction.
The isolated M1G-adducted M13MB102 DNA containing a C or a T at position 6256 of the (+)-strand [(M1G:C)-M13MB102 or (M1G:T)-M13MB102] was then electroporated into SOS-induced E. coli cells. The transformed cells were plated to produce a lawn of plaques. The plaques were eluted and an aliquot of the stock was replated to yield roughly 300 plaques per plate. Plaque DNA from this secondary plating was lifted with nitrocellulose membranes and probed by differential hybridization with radiolabeled probes specific for each type of base substitution. Frameshift mutations were detected by phenotypic screening with X-Gal/IPTG during the secondary plating. Any frameshifts induced by M1G would result in clear plaques instead of wild-type blue plaques. M1G did not increase the frequency of frameshift mutations in comparison to unadducted genomes in any of the strains tested. Control experiments were performed with unmodified M13MB102 DNA containing a C or a T at position 6256 of the (+)-strand [(G:C)- or (G:T)-M13MB102] DNA.
Mutagenicity of M1G in a Wild-Type Repair Background.
Both (M1G:C)- and (G:C)-M13MB102 were transformed into the strain LM102, which is wild-type for DNA repair. The presence of the M1G adduct resulted in predominately M1G→A and M1G→T mutations with very few M1G→C mutations (Table 1). The percentages of mutations measured were 0.35 ± 0.09, 0.4 ± 0.2, and 0.12 ± 0.06% for M1G→A, M1G→T, and M1G→C mutations, respectively. Adding these precentages together yields an overall mutation frequency for M1G of 0.9 ± 0.4%. No G→A, G→T, or G→C mutation was detected within the limit of sensitivity of the assay (≈0.1%) when (G:C)-M13MB102 DNA was used.
Table 1.
Percentages of base pair substitutions detected in progeny of dG- and M1G-adducted M13MB102 containing either a C or a T at position 6256 of the (+)-strand transformed into wild-type LM102
| Strain | M1G→A | M1G→A | M1G→T | M1G→C | Total mutation frequency |
|---|---|---|---|---|---|
| LM102* | |||||
| M1G:C | 99.2 ± 0.4 | 0.35 ± 0.09 | 0.4 ± 0.2 | 0.12 ± 0.06 | 0.9 |
| dG:C | 100 | <0.09 | <0.08 | <0.09 | |
| LM102† | |||||
| M1G:T | 19 ± 3 | 82 ± 3‡ | 2 ± 1 | 0.3 ± 0.2 | 4.3 |
| dG:T | 88 ± 4 | 13 ± 3 | <0.07 | <0.07 |
Results are expressed as mutation frequencies ± SD × 102 and were obtained by screening 1000 plaques per experiment for each probe. Values are the average of six independent DNA constructions, transformations, and hybridizations.
Values are the average of three independent DNA constructions, transformations, and hybridizations.
The values for the replication of the (+)-strand contain a small contribution from mutagenic replication (M1G→A) of the (−)-strand. The value is assumed to be equivalent to the frequency of M1G→T induced by replication of the (−)-strand.
The amount of template utilization of the adducted and unadducted strands was determined with an M13MB102 genome that contains a thymine opposite the M1G adduct at position 6256 of the (+)-strand [(M1G:T)-M13MB102]. Repair of the adduct or replication of the (+)-strand lead to hybridizations with the A-containing probe in phages that arose as progeny of the initial replication event. The resultant increase in A hybridizations, as compared with A mutations detected using a (M1G:C)-M13MB102 genome, indicated the amount of utilization of the (+)-strand as template. Replication of the (−)-strand was predominant in a (G:T)-M13MB102 genome; 88% of the phage population that was probed had a guanine at position 6256 (Table 1). Adenine was detected at position 6256 in 13% of the plaques probed. No G→T or G→C mutation was detected within the limit of sensitivity of the assay. Transformation of the M1G:T genome into wild-type LM102 resulted in a dramatic increase to 82% of the phage population containing an A at position 6256, indicating that there was a high percentage of replication using the (+)-strand as template (Table 1). Subtracting 2% to account for A mutations induced by replication past the M1G adduct on the (−)-strand resulted in a value of 80% for use of the (+)-strand as a template when M1G was present in the M13MB102 genome. This latter correction was necessary because the frequencies of M1G→A and M1G→T mutations were equal when C was opposite M1G in the genome. Use of the (−)-strand as a template when M1G was present in double-stranded M13MB102 genomes was estimated by summing the percentages of M1G→G (19%), M1G→T (2%), and M1G→C (0.3%) events and adding an equivalent number of M1G→A events as M1G→T events (2%). Thus, the total number of replication events that occurred in which the M1G-containing strand was the template was 23.3%. Of these, 19% proceeded with incorporation of dCMP opposite M1G, which would be considered “accurate” replication. Thus, the total percentage of mutations to T, A, or C induced by M1G in SOS-induced wild-type LM102 was (1–19/23.3) × 100 or 18%. This is significantly higher than the value estimated by calculating percentage mutations based on the total number of plaques probed (0.9%).
Mutagenesis in a umuC− Background.
LM113 cells are deficient in the SOS mutagenesis pathway because they carry a Tn5 insertion in the umuC gene. This results in the expression of a truncated and nonfunctional UmuC protein. The UmuC protein participates with UmuD′ and RecA to facilitate bypass of DNA lesions that are blocks to replication (33, 34). SOS-dependent bypass occurs with a higher error frequency than normal replication and is responsible for the mutations induced by many chemicals in E. coli. Transformation of (M1G:C)-M13MB102 genomes into LM113 (umuC−) resulted in a significant decrease in M1G→A, M1G→T, and M1G→C mutations, indicating that a functioning SOS pathway significantly enhances the mutagenicity of M1G in E. coli (Table 2).
Table 2.
Percentages of base pair substitutions detected in progeny of dG- and M1G-adducted M13MB102 transformed into NER- and SOS mutagenesis-deficient E. coli
| Strain | M1G→G | M1G→A | M1G→T | M1G→C | Total mutation frequency |
|---|---|---|---|---|---|
| LM113 (umuC−)* | |||||
| M1G | 99.9 | 0.03 | <0.04 | <0.04 | 0.03 |
| dG | 100 | <0.05 | <0.05 | <0.07 | |
| LM103 (uvrA−)† | |||||
| M1G | 98 ± 1 | 1.0 ± 0.8 | 1.3 ± 0.4 | 0.3 ± 0.2 | 2.6 |
| dG | 100 | <0.10 | 0.10 | <0.11 | |
| LM115 (fpg−, uvrA−)‡ | |||||
| M1G | 97.42 | 0.57 | 1.44 | 0.34 | 2.35 |
| dG | 100 | 0.09 | <0.12 | 0.12 | |
| NR10148 (uvrB−)‡ | |||||
| M1G | 98.24 | 0.73 | <0.30 | 0.70 | 1.43 |
| dG | 100 | <0.09 | <0.06 | 0.10 |
Values presented are the highest result from two independent experiments.
Results are expressed as mutation frequenceis ± SD × 102 and were obtained by screening 1000 plaques per experiment for each probe. Values are the average of six independent DNA constructions, transformations, and hybridizations.
Values are from a single transformation experiment.
Mutagenesis in a Nucleotide Excision Repair-Deficient Background.
Experiments were performed to determine if nucleotide excision repair plays a role in the removal of M1G. LM103 contains a uvrA6 mutation, resulting in a nonfunctional UvrA protein, which is essential for nucleotide excision repair (NER). If NER is involved in the removal of M1G, a deficiency in this repair pathway should result in an increase in the overall mutation frequency compared with repair-proficient bacteria. Transformation of (M1G:C)-M13MB102 into LM103 (uvrA6) resulted in M1G→A, M1G→T, and M1G→C mutations of 1.0 ± 0.8, 1.3 ± 0.4, and 0.3 ± 0.2%, respectively, (2.6% total) (Table 2). This is a 2.9-fold increase in the overall mutation frequency compared with repair-proficient, wild-type LM102, suggesting that NER is involved in the removal of the M1G adduct (Tables 1 and 2). (M1G:C)-M13MB102 also was transformed into two other strains of bacteria [LM115 (uvrA::Tn10) and NR10148 (uvrB)] that carry different mutations in the NER pathway than the uvrA6 carried in LM103 (Table 2). There were 1.6- and 2.6-fold increases in the overall percentage mutations for NR10148 (uvrB) and LM115 (uvrA::Tn10), respectively. These results further support the role of NER in the repair of the M1G adduct. Also, because there was a similar overall mutation frequency between the double mutant strain LM115 (fpg-1::Knr, uvrA::Tn10) and the LM103 (uvrA6) strain, the FaPy–DNA glycosylase does not appear to play a role in the removal of the M1G adduct.
Further studies with LM103 (uvrA6) were performed using (G:T)- and (M1G:T)-M13MB102 genomes. For nonadducted genomes, the (−)-strand was used as template 94% of the time with replication using the (+)-strand as template 6% of the time (Table 3). There was no detection of G→T or G→C mutations within the level of sensitivity of the assay. Upon transformation of M1G:T genomes, M1G→T, M1G→C, and M1G→G events were detected concomitant with a decrease in the number of M1G→A hybridizations (Table 3). Summing the M1G→T (4%) and M1G→C (0.6%) mutation frequencies and adding an equal frequency of M1G→A mutations as M1G→T (4%) results in an overall mutation frequency of 8.6%. This is a 2-fold increase in overall mutation frequency compared with similar experiments in wild-type LM102, indicating that, without a functional NER system, the half-life of M1G is longer, thus increasing the potential for mutations to occur (Tables 1 and 3). There was also a comparable increase in the relative frequency of (−)-strand use measured between wild-type LM102 and LM103 (uvrA6) (1.8-fold). This result is consistent with a role for NER in the removal of M1G. Removal of M1G in wild-type LM102 by NER would lead to increased incorporation of A residues during repair synthesis because of the presence of T at position 6256 on the (+)-strand. Although the percentage of replication of M1G was higher in the NER-deficient cells, the mutation frequency was similar to that observed in wild-type LM102 when corrected for template use. Eighteen percent of the (−)-strand replication events led to mutations in wild-type LM102, and 20% led to mutations in LM103 (uvrA6).
Table 3.
Replication of M13MB102 genomes containing dG:T or M1G:T at position 6256 in NER-deficient LM103
| Strain | M1G→G | M1G→A | M1G→T | M1G→C | Total mutation frequency |
|---|---|---|---|---|---|
| LM103 (uvrA−) | |||||
| M1G:T | 34 ± 4 | 64 ± 4* | 4 ± 1 | 0.6 ± 0.2 | 8.6 |
| dG:T | 94 ± 2 | 6 ± 2 | <0.08 | <0.09 |
Values represent the average of three separate constructions, transformations, and hybridizations.
The values for the replication of the (+)-strand contain a small contribution from mutagenic replication (M1G→A) of the (−)-strand. The value is assumed to be equivalent to the frequency of M1G→T induced by the replication of the (−)-strand.
DISCUSSION
The present study investigated both the mutagenicity and repair of an M1G adduct site specifically positioned in M13MB102 and transformed into both wild-type and repair-deficient E. coli. M1G was mutagenic in repair-proficient bacteria and resulted in roughly equal numbers of M1G→A and M1G→T mutations with few M1G→C mutations; the total mutation frequency was 9 × 10−3 or 0.9%. Previous experiments in our laboratory have demonstrated that the “background” frequency of G→T transversions in recombinant, unmodified M13MB102 genomes is <0.002% (26). Thus, the frequency of mutations induced by M1G (≈1%) was at least 500-fold greater than the spontaneous mutation frequency. However, the use of duplex M13 genomes for site-specific mutagenesis experiments masks the true mutagenic potency of adducts that block replication. When corrections were made for the strand utilization of replication, the increase in mutations induced by M1G was 9000-fold higher than control genomes.
The mutation spectrum of M1G is quite similar to that of PdG when the two adducts are compared in an identical sequence context (26, 27). PdG induces equal numbers of transitions to A and transversions to T but the total mutation frequency is ≈2-fold higher than observed with M1G (27). Neither M1G nor PdG induced frameshift mutations in the sequence used in the present study, but PdG does induce frameshifts in a sequence containing reiterated CGs (M. Hashim, N. Schnetz–Boutaud, and L.J.M., unpublished results). The similarities in potency and mutation spectra validate our previous use of PdG as a model for M1G. This is further substantiated by comparison of the effects of M1G and PdG on template utilization and by the sensitivity of both adducts to NER. Electroporation of M1G-containing vectors into E. coli strains that are deficient in genes coding for components of Uvr(A)BC increases the apparent mutation frequency 3-fold. An increase of approximately the same magnitude in the utilization of the M1G-containing strand as template also was seen in excision repair-deficient strains, so the actual mutation frequency (mutations/replication event) remains the same.
Several exocyclic adducts, including 1,N6-ethenodeoxyadenosine and 3,N4-ethenodeoxycytidine, have been evaluated as premutagenic lesions by site-specific approaches (15–18, 35). Most of them appear to be mutagenic in E. coli, as would be anticipated from their structure. It is difficult to make a quantitative comparison of the mutagenic potency of these adducts because different vectors, sequence contexts, bacterial strains, and repair backgrounds were used in the various assays. Moriya et al. have determined the mutagenicity of PdG and 3,N4-ethenodeoxycytidine in E. coli and simian kidney (COS) cells using single-stranded vectors that are not subject to repair (35). PdG was a strong block to replication and was highly mutagenic in E. coli but was less mutagenic in mammalian cells. For example, the frequency of mutations induced by PdG decreased from 1 in E. coli to 0.1 in COS cells. We found that the mutation frequency of PdG carried on duplex vectors was ≈0.3 in E. coli, which is comparable to the frequency of mutations induced by M1G in the present study. The differences in mutagenicity between our experiments and those of Moriya et al. could be attributable to differences in replication of single-stranded and double-stranded genomes or the sequence context in which the adduct is contained in the vector. For example, we found dramatic differences in the ability of PdG to induce base pair substitutions during in vivo or in vitro replication depending on the identity of the base 5′ to it (36). Based on these considerations, it seems likely that M1G is mutagenic in mammalian cells, but perhaps at a lower frequency than in E. coli.
The relative increases in mutagenicity of M1G and PdG observed in uvrA− strains implies that the two adducts are repaired by Uvr(A)BC with similar efficiencies. We have recently found that PdG is a relatively poor substrate for repair by the E. coli nucleotide excinuclease in vitro but is an excellent substrate for repair by the mammalian enzyme complex (37). This implies that M1G also is a good substrate for removal by the mammalian enzyme. The steady-state levels of DNA adducts detected in human tissues repesent a balance between formation and repair. If M1G is as good a substrate for the mammalian nucleotide excinuclease as PdG, significantly higher levels of M1G may be formed in tissues but lowered by efficient repair.
The steady-state levels of M1G present in human liver, white blood cells, and pancreas range from 6/108 to 9/107 bases (refs. 10–12 and J. P. Plastaras, K. Anderson, F. Kadlubar, I. A. Blair, and L.J.M , unpublished results). The levels in liver are the highest reported for an endogenously occurring exocyclic adduct and are ≈3- to 4-fold lower than the levels of the endogenous DNA oxidation product, 8-oxodeoxyguanosine (10, 38, 39). The levels of M1G in white cells are comparable to the levels of another endogenous adduct, 7-methyldeoxyguanosine (40). M1G is detectable in human genomic DNA and we have found that it is an efficient premutagenic lesion, so M1G should be considered an important potential source of endogenous mutations. This should be most important in cells making MDA as a result of lipid peroxidation or eicosanoid biosynthesis. The latter point is of particular interest because of recent findings that the inducible form of cyclooxygenase (cyclooxygenase-2), which makes MDA, is expressed in human colonic epithelial cells as an early event in colon carcinogenesis (41, 42).
Acknowledgments
We are grateful to M. Hashim for his assistance in the Mops buffer studies and for helpful discussions. This work was supported by research (CA47479), training (CA09582), and center grants (ES00267 and CA68485) from the National Institutes of Health.
ABBREVIATIONS
- MDA
malondialdehyde
- M1G
pyrimido[1,2-α]purin-10(3H)-one
- PdG
1,N2-propano-2′-deoxyguanosine
- NER
nucleotide excision repair
- (G:C)- or (G:T)-M13MB102
unadducted M13MB102 DNA containing a C or T at position 6256 of the (+)-strand
- (M1G:C)- or (M1G:T)-M13MB102
M1G-adducted M13MB102 DNA containing a C or T at position 6256 of the (+)-strand
- X-Gal
5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside
- IPTG
isopropyl β-d-thiogalactoside
References
- 1.Ames B N, Gold L S. Mutat Res. 1991;250:3–16. doi: 10.1016/0027-5107(91)90157-j. [DOI] [PubMed] [Google Scholar]
- 2.Marnett L J, Burcham P C. Chem Res Toxicol. 1993;6:771–785. doi: 10.1021/tx00036a005. [DOI] [PubMed] [Google Scholar]
- 3.Lindahl T. Nature (London) 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 4.Singer B, Bartsch H. The Role of Cyclic Nucleic Acid Adducts in Carcinogenesis and Mutagenesis. Lyon: International Agency for Research on Cancer; 1986. [Google Scholar]
- 5.Barbin A, El Ghissassi F, Nair J, Bartsch H. Proc Am Assoc Cancer Res. 1993;34:136. (abstr.). [Google Scholar]
- 6.Marnett L J. In: DNA Adducts: Identification and Biological Significance: IARC Scientific Publication No. 125. Hemminki K, Dipple A, Shuker D E G, Kadlubar F F, Segerbäck D, Bartsch H, editors. Lyon: International Agency for Research on Cancer; 1994. pp. 151–163. [Google Scholar]
- 7.Swenberg J A, La D K, Scheller N A, Wu K Y. Toxicol Lett. 1995;82:751–756. doi: 10.1016/0378-4274(95)03593-1. [DOI] [PubMed] [Google Scholar]
- 8.Mitro K L, Scheller N A, Ranasinghe A, Swenberg J A. Proc Am Assoc Cancer R. 1995;36:142. (abstr. ). [Google Scholar]
- 9.Nair J, Carmichael P L, Fernando R C, Phillips D H, Bartsch H. Proc Am Assoc Cancer Res. 1996;37:117. (abstr.). [Google Scholar]
- 10.Chaudhary A K, Nokubo M, Reddy G R, Yeola S N, Morrow J D, Blair I A, Marnett L J. Science. 1994;265:1580–1582. doi: 10.1126/science.8079172. [DOI] [PubMed] [Google Scholar]
- 11.Nair J, Barbin A, Guichard Y, Bartsch H. Carcinogenesis. 1995;16:613–617. doi: 10.1093/carcin/16.3.613. [DOI] [PubMed] [Google Scholar]
- 12.Vaca C E, Fang J-L, Mutanen M, Valstar L. Carcinogenesis. 1995;16:1847–1851. doi: 10.1093/carcin/16.8.1847. [DOI] [PubMed] [Google Scholar]
- 13.Fang J-L, Vaca C E, Valsta L M, Mutanen M. Carcinogenesis. 1996;17:1035–1040. doi: 10.1093/carcin/17.5.1035. [DOI] [PubMed] [Google Scholar]
- 14.Rouzer C A, Chaudhary A K, Nokubo M, Ferguson D M, Reddy G R, Blair I A, Marnett L J. Chem Res Toxicol. 1997;10:181–188. doi: 10.1021/tx9601216. [DOI] [PubMed] [Google Scholar]
- 15.Basu A K, Wood M L, Niedernhofer L J, Ramos L A, Essigmann J M. Biochemistry. 1993;32:12793–12801. doi: 10.1021/bi00210a031. [DOI] [PubMed] [Google Scholar]
- 16.Palejwala V A, Simha D, Humayun M Z. Biochemistry. 1991;30:8736–8743. doi: 10.1021/bi00100a004. [DOI] [PubMed] [Google Scholar]
- 17.Palejwala V A, Rzepka R W, Simha D, Humayun M Z. Biochemistry. 1993;32:4105–4111. doi: 10.1021/bi00066a036. [DOI] [PubMed] [Google Scholar]
- 18.Pandya G A, Moriya M. Biochemistry. 1996;35:11487–11492. doi: 10.1021/bi960170h. [DOI] [PubMed] [Google Scholar]
- 19.Chaudhary A K, Nokubo M, Oglesby T D, Marnett L J, Blair I A. J Mass Spectrom. 1995;30:1157–1166. [Google Scholar]
- 20.Janero D R. Free Radical Biol Med. 1990;9:515–540. doi: 10.1016/0891-5849(90)90131-2. [DOI] [PubMed] [Google Scholar]
- 21.Mukai F H, Goldstein B D. Science. 1976;191:868–869. doi: 10.1126/science.766187. [DOI] [PubMed] [Google Scholar]
- 22.Benamira M, Johnson K, Chaudhary A, Bruner K, Tibbetts C, Marnett L J. Carcinogenesis. 1995;16:93–99. doi: 10.1093/carcin/16.1.93. [DOI] [PubMed] [Google Scholar]
- 23.Basu A K, Marnett L J. Carcinogenesis. 1983;4:331–333. doi: 10.1093/carcin/4.3.331. [DOI] [PubMed] [Google Scholar]
- 24.Yau T M. Mech Aging Dev. 1979;11:137–144. doi: 10.1016/0047-6374(79)90031-9. [DOI] [PubMed] [Google Scholar]
- 25.Spalding J W. NTP Tech Rep. 1988;331:5–13. [Google Scholar]
- 26.Burcham P C, Marnett L J. J Biol Chem. 1994;269:28844–28850. [PubMed] [Google Scholar]
- 27.Fink S P, Reddy G R, Marnett L J. Chem Res Toxicol. 1996;9:277–283. doi: 10.1021/tx950060w. [DOI] [PubMed] [Google Scholar]
- 28.Reddy G R, Marnett L J. J Am Chem Soc. 1995;117:5007–5008. [Google Scholar]
- 29.Benamira M, Marnett L J. Chem Res Toxicol. 1993;6:317–327. doi: 10.1021/tx00033a011. [DOI] [PubMed] [Google Scholar]
- 30.Niedernhofer L J, Riley M, Schnetz-Boutaud N, Sanduwaran G, Chaudhary A K, Reddy G R, Marnett L J. Chem Res Toxicol. 1997;10:556–561. doi: 10.1021/tx960191c. [DOI] [PubMed] [Google Scholar]
- 31.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 32.Benamira M, Singh U S, Marnett L J. J Biol Chem. 1992;267:22392–22400. [PubMed] [Google Scholar]
- 33.Echols H, Goodman M F. Annu Rev Biochem. 1991;60:477–511. doi: 10.1146/annurev.bi.60.070191.002401. [DOI] [PubMed] [Google Scholar]
- 34.Rajagopalan M, Lu C, Woodgate R, O’Donnell M, Goodman M F, Echols H. Proc Natl Acad Sci USA. 1992;89:10777–10781. doi: 10.1073/pnas.89.22.10777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moriya M, Zhang W, Johnson F, Grollman A P. Proc Natl Acad Sci USA. 1994;91:11899–11903. doi: 10.1073/pnas.91.25.11899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hashim M F, Marnett L J. J Biol Chem. 1996;271:9160–9165. doi: 10.1074/jbc.271.15.9160. [DOI] [PubMed] [Google Scholar]
- 37.Johnson K A, Fink S P, Marnett L J. J Biol Chem. 1997;272:11434–11438. doi: 10.1074/jbc.272.17.11434. [DOI] [PubMed] [Google Scholar]
- 38.Shimoda R, Nagashima M, Sakamoto M, Yamaguchi N, Hirohashi S, Yokota J, Kasai H. Cancer Res. 1994;54:3171–3172. [PubMed] [Google Scholar]
- 39.Asami S, Hirano T, Yamaguchi R, Tomioka Y, Itoh H, Kasai H. Cancer Res. 1996;56:2546–2549. [PubMed] [Google Scholar]
- 40.Mustonen R, Hemminki K. Carcinogenesis. 1992;13:1951–1955. doi: 10.1093/carcin/13.11.1951. [DOI] [PubMed] [Google Scholar]
- 41.Eberhart C E, Coffey R J, Radhika A, Giardiello F M, Ferrenbach S, DuBois R N. Gastroenterology. 1994;107:1183–1188. doi: 10.1016/0016-5085(94)90246-1. [DOI] [PubMed] [Google Scholar]
- 42.Kargman S L, O’Neill G P, Vickers P J, Evans J F, Mancini J A, Jothy S. Cancer Res. 1995;55:2556–2559. [PubMed] [Google Scholar]