Abstract
We have identified and molecularly characterized a human protein with a Mr of 40,880 Da. After UV irradiation of HeLa cells, this protein was cross-linked to poly(A)-containing mRNA and was therefore designated mrnp 41 (for mRNA binding protein of 41 kDa). Cell fractionation and immunoblotting showed mrnp 41 in both the cytoplasm and the nucleus and particularly in the nuclear envelope. Immunofluorescence microscopy localized mrnp 41 to distinct foci in the nucleoplasm, to the nuclear rim, and to meshwork-like structures throughout the cytoplasm. The cytoplasmic meshwork staining was disrupted by prior treatment of cells with the actin filament- or microtubule-disrupting drugs cytochalasin or nocodazole, respectively, suggesting association of mrnp 41 with the cytoskeleton. Double immunofluorescence with antibodies against mrnp 41 and the cytoplasmic poly(A) binding protein showed colocalization to the cytoplasmic meshwork. Immunogold electronmicroscopy confirmed mrnp 41’s cytoplasmic and nucleoplasmic localization and revealed a striking labeling of nuclear pore complexes. Together these data suggest that mrnp 41 may function in nuclear export of mRNPs and/or in cytoplasmic transport on, or attachment to, the cytoskeleton. Consistent with a role of mrnp 41 in nuclear export are previous reports that mutations in homologs of mrnp 41 in Schizosaccharomyces pombe, designated Rae1p, or in Saccharomyces cerevisiae, designated Gle2p, result in mRNA accumulation in the nucleus although it is presently not known whether these homologs are mRNA binding proteins as well.
Keywords: cDNA sequence, UV cross-linking, immunofluorescence, immunoelectronmicroscopy
During their transcription, capping, splicing, and polyadenylation, nuclear mRNAs associate with a large number of proteins. These associations are dynamically coordinated with respect to various stages of mRNA synthesis, processing, and subsequent transport into the cytoplasm (reviewed in ref. 1). Some proteins are dissociated before nuclear mRNPs are transported across the nuclear pore complex (NPC) and remain in the nucleus (2). Whereas many proteins are dissociated shortly after nuclear mRNP transport across the NPC and are reimported into the nucleus (3), at least one of the nuclear mRNA binding proteins has been shown to remain bound to cytoplasmic mRNA (4). After transport across the NPC and displacement of nuclear binding proteins, the cytoplasmic mRNAs acquire a new set of binding proteins of which the cytoplasmic poly(A) binding protein (cPABP) is an example (5).
Using in situ hybridization with oligo(dT) probes, the bulk of cytoplasmic mRNA has been shown to be attached to cytoskeletal elements, mostly microfilaments or microtubules (6, 7). It is not known whether cytoplasmic mRNAs are directly attached to the cytoskeleton. More likely, attachment is via mRNA-associated proteins. In addition to cytoskeletal attachment, specific mRNAs can be highly sublocalized within the cytoplasm. Examples for such specific sublocalization have been described in oocytes and eggs, as well as in somatic cells. This highly specific cytoplasmic sublocalization, in all cases so far studied, is mediated by cis-acting signals in the 3′ untranslated region of these mRNAs (8). A protein that binds to a 54-nt element of the 3′ untranslated region of β-actin mRNA recently has been identified and characterized (9).
Here, we report the identification and characterization of a 41-kDa human protein and, by UV cross-linking experiments, show that it is a mRNA binding protein. We therefore designated this protein mrnp 41 (for mRNA binding protein of 41 kDa). The cDNA-deduced primary structure of mrnp 41 yielded a calculated Mr of 40,833 Da in close agreement with the estimated Mr of ≈43 kDa based on its mobility in SDS/PAGE. The mrnp 41 protein is the likely homolog of Rae1p of Schizosaccharomyces pombe (10) and of Gle2p of Saccharomyces cerevisiae (11). Although it is presently not known whether these two proteins are mRNA binding proteins, temperature-sensitive mutations in the RAE1 or GLE2 alleles have been reported to result in accumulation of poly(A)-containing mRNA in nuclei, suggesting that these proteins are involved in RNA export (10, 11). Immunoblot analysis of SDS/PAGE-resolved proteins showed mrnp 41 in both nuclear and cytosolic fractions with a striking enrichment in the nuclear envelope fraction. Immunofluorescence microscopy localized mrnp 41 to distinct foci in the nucleoplasm, to the nuclear rim, and to a cytoplasmic meshwork. Double immunofluorescence with antibodies to mrnp 41 and to cPABP (5) showed cytoplasmic colocalization of the two proteins, consistent with mrnp 41 being a mRNA binding protein. Moreover, the distinct staining pattern that we observed with anti-mrnp 41 antibodies, such as focal staining of the nucleus and a meshwork-like cytoplasmic staining pattern, was similar to the staining pattern that was previously reported after in situ hybridization with oligo(dT) probes (6, 7). Immunogold electronmicroscopy of frozen, thin sections of HeLa cells with anti-mrnp 41 antibodies showed the expected labeling in both nucleoplasm and cytoplasm and a striking labeling of NPCs. Taken together, these data suggest that mrnp 41 may function in nucleocytoplasmic transport and in directly or indirectly attaching cytoplasmic mRNPs to the cytoskeleton.
MATERIALS AND METHODS
Protein Sequencing and Sequence Analysis.
Proteins from rat liver nuclear envelopes were extracted by 2.0 M urea in buffer A [1 mM EDTA/20 mM Tris⋅HCl, pH 7.5/1 mM DTT/0.1 mM phenylmethylsulfonyl fluoride (12)], and the extract was passed directly over a Sepharose column containing immobilized wheat germ agglutinin (WGA). Proteins were eluted with 2 M urea in buffer A containing 0.5 M N-acetylglucosamine. After trichloroacetic acid precipitation, the proteins were separated by SDS/PAGE, transferred to poly(vinylidene difluoride) membrane, and stained with amido black. A 43-kDa band was prepared for peptide sequencing as described (13, 14). One sequence (YIFLRNAAEELK) matched that deduced from a partial human cDNA clone in GenBank (accession nos. R16660 and R16670).
We sequenced R16670, representing the 3′ end, and used the 5′ rapid amplification of cDNA ends system (GIBCO/BRL) to obtain the 5′ end. Three nested antisense oligonucleotide primers were used: GACATTTAAAGGTGAAGTTATCTTTGG; GAGGAGCTGAAGTGTTGGTTCCATTAGATC; and GAGGAGCTGAAGTGTTGGTTCCATTAGATC.
DNA sequencing was by the dideoxy chain termination method using Sequenase 2.0 (United States Biochemical). Similarity of protein sequences was measured by the method of Henikoff and Henikoff (15).
Expression and Purification of a mrnp 41 Fragment.
A 321-bp PCR product encoding residues 252–359 of mrnp 41 (12.2 kDa) was synthesized using nondegenerate sense (5′-AAAGATAACTTCACCTTTAAATGTCA-3′) and antisense (5′-GGCCGCATTACGCAGGAAAAT-3′) oligonucleotide primers with additional restriction sites for EcoRI (5′) and NotI (3′). The corresponding PCR product was inserted between the EcoRI and the NotI sites of the pET21-b vector (Novagen, Madison, WI) containing a tag of six histidines. The construct was transformed into Escherichia coli BL21 (DE3). The p12/6 His fusion protein was purified as described (16) and injected into mice and rabbits for antibody production. The serum was affinity-purified using the p12/6 His fusion protein bound to nitrocellulose (16).
Cell Fractionation and Immunoblot Analysis.
A rat liver homogenate was subfractionated as described (12). In brief, the homogenate was centrifuged at 800 × g for 10 min to yield a pellet of crude nuclei and a postnuclear supernatant (12). Nuclei were purified from crude nuclei and subfractionated by nuclease digestion (12) to yield a nuclease extract and nuclear envelopes. Aliquots of these fractions containing 25 μg of protein were prepared for SDS/PAGE. After electrophoretical transfer of SDS/PAGE-separated proteins to nitrocellulose membrane, the blot was incubated with affinity-purified mouse or rabbit anti-mrnp 41 antibodies and then with peroxidase-conjugated anti-mouse or anti-rabbit antibodies, respectively; the immunoreactive proteins were detected by enhanced chemiluminescence (Amersham).
Immunofluorescence Microscopy.
HeLa cells were grown on coverslips until they were subconfluent. After washing briefly with ice-cold PBS, cells were fixed and permeabilized for 10 min in ice-cold methanol and blocked for 1 h with 2% BSA/0.05% Tween 20 in PBS. The fixed and permeabilized cells were incubated with affinity-purified anti-mrnp 41 antibodies [or, in the case of double immunofluorescence, also with monoclonal anti-cPABP antibodies (5) for 12 h at 4°C and washed five times for 5 min with 0.1% BSA/0.5% Tween 20 in PBS. Antibody binding was detected with tetramethylrhodamine B isothiocyanate-labeled goat anti-mouse or anti-rabbit antibodies or with Cy3-labeled polyclonal donkey anti-rabbit IgG (Jackson ImmunoResearch). Coverslips were mounted and examined as described (12). Confocal microscopy was performed with a Bio-Rad MRC 600 confocal imaging system connected to an Olympus (New Hyde Park, NY) microscope.
Immunogold Electronmicroscopy.
HeLa cells were fixed with 2% paraformaldehyde and 0.5% glutaraldehyde in 100 mM cacodylate (pH 7.4) for 30 min at 4°C. The cells were cryosectioned, and sections were incubated with affinity-purified mouse anti-mrnp 41 antibodies and then with rabbit anti-mouse IgG coupled to 10 nm of gold (Amersham). The grids were stained with uranyl acetate and viewed on a Joel 100 CX electron microscope operated at 80 kV.
UV Cross-Linking.
UV cross-linking of mRNA to proteins was done essentially as described (17). HeLa cells were grown to subconfluency on 20 dishes of 10-cm diameter. They were washed twice with PBS containing 1 mM CaCl2 and 0.5 mM MgCl2. Ten dishes were UV-irradiated with 650 × 102 mJ/mm2 for 3 min whereas 10 control dishes were not UV-irradiated. For the isolation of poly(A)-containing mRNA, cells were released from the tissue culture dishes by treatment with trypsin and homogenized in extraction buffer, and the isolated RNA was loaded onto an oligo(dT)-cellulose column as described by the manufacturer (QuickPrep mRNA purification kit, Pharmacia). The collected mRNA was precipitated with ethanol, dissolved in 200 μl of 10 mM Tris⋅HCl, pH 7.4/1 mM CaCl2/25 μg/ml chromatographically purified bovine pancreatic RNase (Sigma)/400 units micrococcal nuclease/0.5% aprotonin/1 mg/ml pepstatin A/1 mg/ml leupeptin and incubated at 37°C for 1 h. The proteins were precipitated with 3 volumes of ethanol, dissolved in sample buffer, separated by SDS/PAGE, transferred electrophoretically to nitrocellulose membrane, and probed with affinity-purified mouse anti-mrnp 41 antibodies as described above.
RESULTS
We partially sequenced an SDS/PAGE-resolved protein of 43 kDa that was present in a subfractionated 2.0 M urea extract of isolated rat liver nuclear envelopes (see Materials and Methods). A partial amino acid sequence of a tryptic peptide of the 43-kDa protein matched the translated sequence of a partial human cDNA clone in GenBank (accession nos. R16660 and R16670). We sequenced clone R16670 and used the 5′ rapid amplification of cDNA ends system to obtain the complete cDNA sequence and the deduced amino acid sequence (Fig. 1A). The calculated Mr of the protein is 40,833 kDa, close to the 43 kDa estimated from its mobility on SDS/PAGE. Because we found this protein to be directly associated with poly(A)-containing mRNA (see below), we designated it mrnp 41. There are three β transducin/WD40 motifs in mrnp 41. These motifs occur in several families of proteins and were proposed to function in protein–protein interactions (18). The protein does not contain any of the known RNA binding motifs (19). It remains to be shown whether mrnp 41 is a WGA-binding protein. Extracted nuclear envelope proteins were not denatured by SDS before WGA–Sepharose chromatography (see Materials and Methods), so it is possible that mrnp 41 did not directly bind to WGA but bound indirectly, through interaction with a WGA-binding protein.
Figure 1.
DNA-deduced amino acid sequence of human mrnp 41 and alignment with homologs. (A) Three repeats of Try-Asp (WD) motifs are boxed; the amino acid sequence retrieved from peptide sequencing is underlined. (B) Alignment of amino acid sequences of mnrp 41 and putative homologs in S. pombe (Rae1p), S. cerevisiae (Gle2p), C. elegans, and A. thaliana. Alignment was by the program lasergene navigator (DNAstar, Madison, WI).
Homology searches in the databanks revealed (Fig. 1B) that mrnp 41 is 45% identical to the S. pombe protein Rae1p (10), 39% identical to the Saccharomyces cerevisiae protein, Gle2p (11) and 50% identical to yet uncharacterized proteins of Caenorhabditis elegans and Arabidopsis thaliana, respectively (see Discussion).
We expressed a 12-kDa C-terminal fragment (lacking the WD motifs) in E. coli and raised polyclonal antibodies against the purified protein in mice and rabbits. Affinity-purified rabbit antibodies reacted with a 43-kDa band of SDS/PAGE-resolved proteins of a rat liver homogenate (Fig. 2, lane 1). Upon fractionation of the homogenate, the antibody-reactive 43-kDa band was present both in purified nuclei (lane 3) and a postnuclear supernatant (lane 2). After subfractionation of purified nuclei, the antibody-reactive 43-kDa band was present in the nuclease extract (lane 5) and was enriched in nuclear envelopes (lane 4). Identical data were obtained with affinity-purified mouse anti-mrnp 41 antibodies (not shown). These data indicate that mrnp 41 is both a cytoplasmic and a nuclear protein that is enriched in the nuclear envelope fraction.
Figure 2.
Cell fractionation and Immunoblot with anti-mrnp 41 antibodies. A rat liver homogenate (lane 1) was fractionated by differential centrifugation into crude nuclei and a postnuclear supernatant (lane 2). Purified nuclei (lane 3) were fractionated by nuclease digestion yielding a nuclease extract (lane 5) and a nuclear envelope fraction (lane 4). Aliquots containing 25 μg of protein were subjected to SDS/PAGE. Proteins were transferred to nitrocellulose and immunoblotted using affinity-purified rabbit antibodies against mrnp 41.
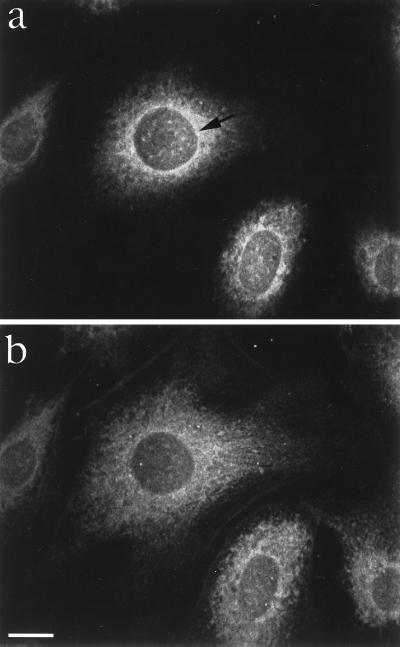
We used the affinity-purified antibodies for immunofluorescence analysis by confocal laser microscopy using methanol-permeabilized and fixed HeLa cells. When focusing on a plane close to the coverslip, we observed a fine meshwork-like cytoplasmic staining pattern (Fig. 3b). When focusing near the equatorial plane of the nucleus, the fine meshwork pattern merged into a more coarse one and intensified at the nuclear rim (Fig. 3a). In addition, there was focal intranuclear staining (Fig. 3). These data indicate that mrnp 41 is both a nuclear and a cytoplasmic protein with pronounced enrichment around the nuclear rim and a striking meshwork-like distribution in the cytoplasm.
Figure 3.
Localization of mrnp 41 in HeLa cells by confocal immunofluorescence microscopy. HeLa cells were grown to subconfluency on a coverslip, permeabilized and fixed with methanol, and then probed with affinity-purified mouse anti-mrnp 41 antibodies (see Materials and Methods). An optical section corresponding to the equatorial plane of the nucleus shows staining of intranuclear foci, of the nuclear rim (see arrow head), and of a coarse meshwork (a). An optical section close to coverslip shows in addition a fine meshwork-like staining in the cytoplasmic periphery (b). (Bar = 10 μm.)
The cytoplasmic staining pattern is similar to that reported previously for cytoplasmic mRNAs localized by in situ hybridization with oligo(dT) probes (6, 7). This staining pattern was shown to reflect attachment of mRNAs to cytoskeletal element because it was dramatically altered when cells were treated with cytoskeleton-disrupting agents, such as colcemid or cytochalasin D (6). Indeed, when HeLa cells were preincubated with cytochalasin D or colcemid before immunofluorescence, as described by Taneja et al. (6), the meshwork-like staining in the cytoplasm collapsed into coarse aggregates (not shown).
To test whether mrnp 41 is a mRNA binding protein, we carried out UV cross-linking. Plates of HeLa cells were UV-irradiated, with control plates receiving no UV irradiation. Poly(A)-containing mRNAs were extracted and purified on oligo(dT) cellulose. Under the protein denaturing conditions used, only proteins covalently cross-linked to the mRNA were purified. The purified mRNAs were digested extensively with RNase, and the digest was analyzed by SDS/PAGE followed by blotting of the SDS/PAGE-separated proteins with anti-mrnp 41 antibodies. An immunoreactive band at 43 kDa was found to be associated with mRNAs only from UV-irradiated cells and not with mRNAs from non-UV-irradiated control cells (Fig. 4). These data indicate that mrnp 41 is a mRNA binding protein.
Figure 4.
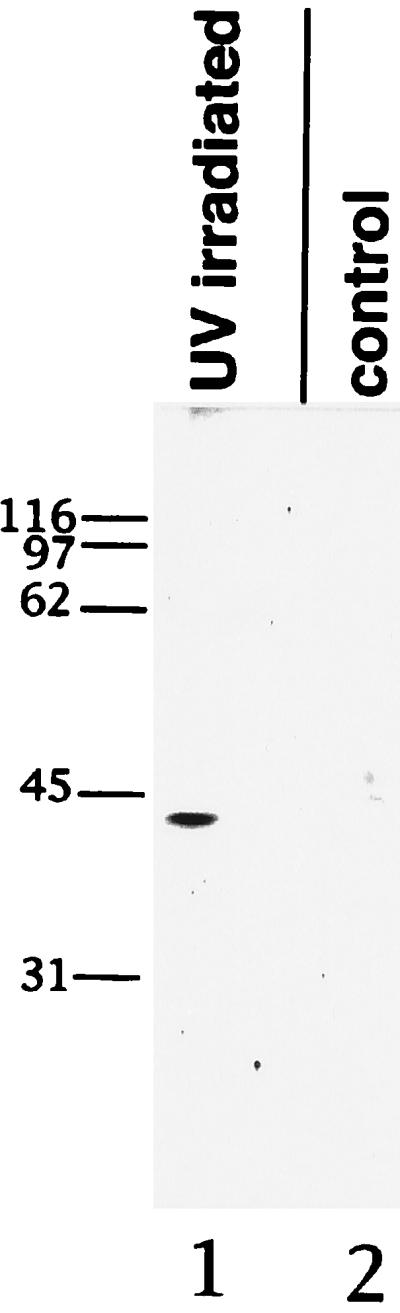
UV cross-linking of mrnp 41 to poly(A)-containing mRNA. Poly(A)-containing mRNA was isolated from HeLa cells that were either UV-irradiated (UV irradiated) or not UV-irradiated (control). The poly(A)-containing mRNA was digested with ribonucleases, the digest separated by SDS/PAGE, and the resolved proteins probed with affinity-purified mouse anti-mrnp 41 antibodies. Numbers to the left indicate Mr of marker proteins.
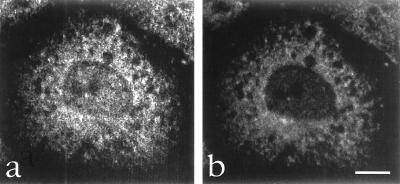
To confirm that the observed meshwork-like staining pattern in the cytoplasm represents staining of cytoplasmic mRNAs, we performed double immunofluorescence using antibodies against a bona fide cytoplasmic mRNA binding protein, cPABP, and anti-mrnp 41 antibodies. The cPABP is an example of a mRNA binding protein that associates with mRNA only after its transport from the nucleus into the cytoplasm (5). As expected, there was no nuclear staining with anti-cPABP antibodies, in contrast to the focal nuclear staining with anti-mrnp 41 antibodies (Fig. 5, compare a and b). Most importantly, there was colocalization to a cytoplasmic meshwork-like pattern of cPABP and mrnp 41 (Fig. 5). We conclude that the cytoplasmic meshwork-like staining pattern elicited by anti-mrnp 41 antibodies indeed reflects localization of cytoplasmic mRNPs.
Figure 5.
Double immunofluorescence confocal microscopy using mouse mAb against cPABP and affinity-purified rabbit antibodies against mrnp 41. Methanol-permeabilized and fixed HeLa cells were incubated with affinity-purified rabbit anti-mrnp 41 antibodies (a) and with monoclonal anti-cPAPB antibodies (b), and the bound antibodies were visualized with fluorescently labeled secondary antibodies. (Bar = 10 μm.)
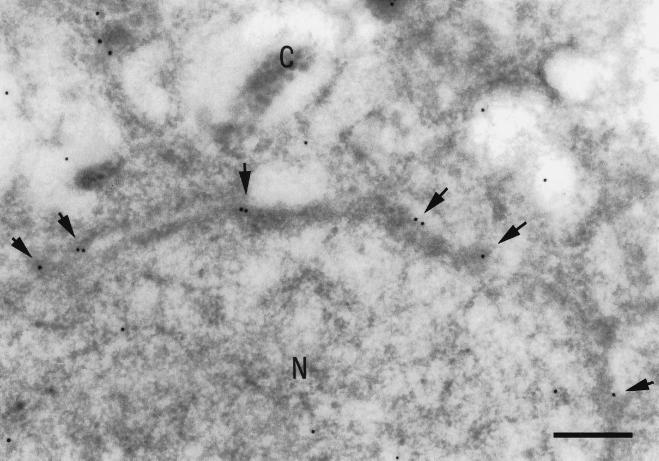
To determine whether the observed enrichment of mrnp 41 in the nuclear envelope fraction as well as the intense nuclear rim staining in immunofluorescence is due to an accumulation of mrnp 41 at NPCs, we carried out Immunogold labeling of frozen thin sections of HeLa cells (Fig. 6). Besides the expected sporadic gold labeling of the nucleoplasm and the cytoplasm, there was a striking labeling at the level of the nuclear pore complexes (see Discussion).
Figure 6.
Immunogold electron microscopy of HeLa cells using anti-mrnp 41 antibodies. Frozen ultrathin sections of HeLa cells were incubated with affinity-purified mouse anti-mrnp 41 antibodies and binding was detected with 10 nm of gold-coupled secondary antibodies. N, nucleoplasm; C, cytoplasm. Arrows point to gold particles at nuclear pore complexes: (Bar = 300 nm.)
DISCUSSION
Our data indicate that a hitherto uncharacterized protein of isolated nuclear envelopes is a mRNA binding protein. This protein was therefore termed “mrnp 41,” based on its cDNA-deduced Mr of 40,880 Da. Cell fractionation and immunoblotting of SDS/PAGE-separated proteins of a cytosolic, a nucleoplasmic, and a nuclear envelope fraction of rat liver revealed that mrnp 41 occurs in all three fraction but that it is enriched in the nuclear envelope fraction. An enrichment at the nuclear envelope also was suggested by pronounced immunofluorescence staining of the nuclear rim, in addition to that in the cytoplasm and the nucleoplasm. This nuclear rim staining was further resolved by Immunogold electron microscopy of frozen thin sections and was shown to result from a striking location of mrnp 41 in NPCs. An accumulation of mRNPs at NPCs has been reported previously and is suggested to represent a rate-limiting step of mRNP transport across the NPC (20). Electron microscope tomography has shown that the large mRNPs of the Chironomus tentans salivary gland nuclei traverse the center of NPCs in a rod-like configuration with the 5′ end of the mRNA in the lead (21). The observed conversion of a globular mRNP into a rod-like structure at the nucleoplasmic side of NPCs (22) may require the coordinated, transport factor-mediated docking of several mRNA-associated proteins to multiple docking sites that are provided by peptide repeat-containing nucleoporins on the nucleoplasmic and cytoplasmic sides of the NPC. These coordinated docking reactions may represent rate limiting steps in mRNP transport across the NPC and therefore might explain the accumulation of mRNPs and mrnp 41 at the NPC. It should be noted that the procedure for the preparation of the rat liver nuclear envelope fraction used here included incubation with pancreatic RNase and micrococcal nuclease, which most likely fragment the mRNAs in mRNPs. Although we do not know whether RNase treatment resulted in some release of mrnp 41 from the isolated nuclear envelopes, the striking enrichment of mrnp 41 in this fraction suggests that its association with the envelope was largely resistant to RNase treatment. The region of mrnp 41 that binds to mRNAs and a possible cis-acting element in mRNAs remain to be identified. The protein does not contain any of the known RNA binding motifs (19).
Homology searches in the databanks revealed (Fig. 2b) that mrnp 41 is 45% identical to the S. pombe protein, Rae1p (10) and 39% identical to the S. cerevisiae protein, Gle2p (11). Temperature-sensitive alleles of RAE1 and GLE2 have been shown to result in accumulation of poly(A)-containing RNA in the nucleus, suggesting that Rae1p and Gle2p are involved in RNA export. It is not yet known whether Rae1p and Gle2p are mRNA binding proteins. Gle2p has been proposed to be a structural component of the NPC or a nuclear transport factor (11). If Gle2p and Rae1p were mRNA binding proteins as well, it is conceivable that the reported RNA export defect is caused by the mutant protein’s failure to effect transport factor-mediated docking of mRNP at the NPC.
Most interesting is the observed dual nuclear and cytoplasmic location of mrnp 41. Such a dual location has been established so far only for one other mRNA binding protein, hrp36 of Chironomus tentans (4). The hrp36 that is similar to the mammalian hnRNP A1 was found associated not only with nuclear mRNA but also with mRNA assembled into polyribosomes (4). Most striking is the cytoplasmic location of mrnp 41 in a cytoplasmic meshwork that overlaps with the location of cPABP (Fig. 5). A similar cytoplasmic meshwork-like staining pattern has been seen after in situ hybridization with oligo(dT) probes and has been shown to be due to association of most cellular mRNAs with cytoskeletal elements, primarily microfilaments and microtubules (6, 7). Hence, it is possible that mrnp 41, besides functioning in nuclear export, also plays a role, directly or indirectly, in motor-driven movement of mRNPs on cytoskeletal elements or in their attachment to them.
Acknowledgments
We thank Dr. Michael Matunis for suggesting the UV cross-linking experiment, for helpful discussions, and for critically reading this manuscript and Dr. Gideon Dreyfuss for generously supplying us with mAb to cPABP. Protein sequence analysis was provided by The Rockefeller University Protein/DNA Technology Center, which is supported in part by the National Institutes of Health shared instrumentation grants and by funds provided by the U.S. Army and Navy for purchase of equipment.
ABBREVIATIONS
- mrnp 41
mRNA-associated protein of 41 kDa
- mRNP
messenger ribonucleoprotein
- cPABP
cytoplasmic poly(A) binding protein
- NPC
nuclear pore complex
- WGA
wheat germ agglutinin
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. U85943).
References
- 1.Daneholt B. Cell. 1997;88:585–588. doi: 10.1016/s0092-8674(00)81900-5. [DOI] [PubMed] [Google Scholar]
- 2.Piñol-Roma S, Dreyfuss G. Trends Cell Biol. 1993;3:151–155. doi: 10.1016/0962-8924(93)90135-n. [DOI] [PubMed] [Google Scholar]
- 3.Piñol-Roma S, Dreyfuss G. Nature (London) 1992;355:730–735. doi: 10.1038/355730a0. [DOI] [PubMed] [Google Scholar]
- 4.Visa N, Alzhanova-Ericsson A T, Sun X, Kiseleva E, Björkroth B, Wurtz T, Daneholt B. Cell. 1996;84:253–264. doi: 10.1016/s0092-8674(00)80980-0. [DOI] [PubMed] [Google Scholar]
- 5.Görlach M, Burd C C, Dreyfuss G. Exp Cell Res. 1994;211:400–407. doi: 10.1006/excr.1994.1104. [DOI] [PubMed] [Google Scholar]
- 6.Taneja K L, Lifshitz L M, Fay F S, Singer R H. J Cell Biol. 1992;119:1245–1260. doi: 10.1083/jcb.119.5.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bassell G J, Singer R H, Kosik K S. Neuron. 1994;12:571–582. doi: 10.1016/0896-6273(94)90213-5. [DOI] [PubMed] [Google Scholar]
- 8.St. Johnston D. Cell. 1995;81:161–170. doi: 10.1016/0092-8674(95)90324-0. [DOI] [PubMed] [Google Scholar]
- 9.Ross A F, Oleynikov Y, Kislaukis E H, Taneja K L, Singer R H. Mol Cell Biol. 1997;17:2158–2165. doi: 10.1128/mcb.17.4.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brown J A, Bharathi A, Ghosh A, Whalen W, Fitzgerald E, Dhar R. J Biol Chem. 1995;270:7411–7419. doi: 10.1074/jbc.270.13.7411. [DOI] [PubMed] [Google Scholar]
- 11.Murphy R, Watkins J L, Wente S R. Mol Biol Cell. 1996;7:1921–1937. doi: 10.1091/mbc.7.12.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Radu A, Blobel G, Wozniak R W. J Cell Biol. 1993;121:1–9. doi: 10.1083/jcb.121.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fernandez J, DeMott M, Atherton D, Mische S M. Anal Biochem. 1992;201:255–264. doi: 10.1016/0003-2697(92)90336-6. [DOI] [PubMed] [Google Scholar]
- 14.Kraemer D, Strambio-de-Castillia C, Blobel G, Rout M P. J Biol Chem. 1995;270:19017–19021. doi: 10.1074/jbc.270.32.19017. [DOI] [PubMed] [Google Scholar]
- 15.Henikoff S, Henikoff J G. Proc Natl Acad Sci USA. 1992;89:10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kraemer D, Wozniak R W, Blobel G, Radu A. Proc Natl Acad Sci USA. 1994;91:1519–1523. doi: 10.1073/pnas.91.4.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pinol-Roma S, Adam S A, Choi Y D, Dreyfuss G. Methods Enzymol. 1989;180:410–418. doi: 10.1016/0076-6879(89)80114-4. [DOI] [PubMed] [Google Scholar]
- 18.Neer E J, Schmidt C J, Nambudripad R, Smith T F. Nature (London) 1994;371:297–300. doi: 10.1038/371297a0. [DOI] [PubMed] [Google Scholar]
- 19.Burd C G, Dreyfuss G. Science (Washington, DC) 1994;265:615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- 20.Visa N, Izaurralde E, Ferreira J, Daneholt B, Mattaj I W. J Cell Biol. 1996;133:5–14. doi: 10.1083/jcb.133.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mehlin H, Daneholt B, Skoglund U. Cell. 1992;69:605–613. doi: 10.1016/0092-8674(92)90224-z. [DOI] [PubMed] [Google Scholar]
- 22.Mehlin H, Daneholt B, Skoglund U. J Cell Biol. 1995;129:1205–1216. doi: 10.1083/jcb.129.5.1205. [DOI] [PMC free article] [PubMed] [Google Scholar]