Abstract
Genetic mapping of traits and mutations in mammals is dependent upon linkage analysis. The resolution achieved by this method is related to the number of offspring that can be scored and position of crossovers near a gene. Higher precision mapping is obtained by expanding the collection of progeny from an appropriate cross, which in turn increases the number of potentially informative recombinants. A more efficient approach would be to increase the frequency of recombination, rather than the number of progeny. The anticancer drug cisplatin, which causes DNA strand breakage and is highly recombinogenic in some model organisms, was tested for its ability to induce germ-line recombination in mice. Males were exposed to cisplatin and mated at various times thereafter to monitor the number of crossovers inherited by offspring. We observed a striking increase on all three chromosomes examined and established a regimen that nearly doubled crossover frequency. The timing of the response indicated that the crossovers were induced at the early pachytene stage of meiosis I. The ability to increase recombination should facilitate genetic mapping and positional cloning in mice.
A prime achievement of the genome project has been the establishment of high-density genetic maps of humans and mice (1, 2). The maps, and simple sequence repeat markers that were used to construct them, have led to a revolution in genetic analysis. These reagents form a scaffold for the creation of physical maps, consisting of ordered clones spanning the entire genome. These advances have simplified and accelerated the pace at which genes or traits can be localized and positionally cloned.
With the advent of such resources, the limiting factor in positional cloning of genes has become the collection of informative families (for humans) or conducting of crosses (for mice) in which the gene of interest is segregating. Now that extensive collections of molecular markers along chromosomes are available, the bottleneck in high-resolution mapping is the ability to obtain recombination breakpoints that delimit a gene to a workably small interval. Ideally, this is well under 0.5 centimorgans. It is not uncommon for mapping crosses in mice to involve several thousand progeny to achieve such resolution. This is expensive and time consuming, especially if the phenotype is incompletely penetrant or difficult to assay or if crossovers are especially rare in the critical region.
An alternative to increasing cross size as a means to improve mapping resolution is to boost recombination frequency, either on a genomewide basis or in the vicinity of the target gene. Recombination occurs in both mitotic and meiotic cells. Although it is required for accurate chromosome segregation in meiosis, this is not the case for mitotic crossing-over, which is postulated to be a consequence of DNA repair (3). Because certain types of DNA damage induce recombinational repair, this property has been exploited in yeast, Drosophila melanogaster, and mammalian cell cultures to develop assays for screening potential mutagenic or carcinogenic agents (4–7).
The chemotherapeutic agent cis-platinum(II)diammine dichloride (cisplatin, CP) is highly recombinogenic in assays with Candida albicans, Saccharomyces cerevesiae, and somatic cells of D. melanogaster (8, 9). RecA mutants of Escherichia coli and Rad52-deficient yeast are hypersensitive to this DNA-adduct-forming drug, implying a need for efficient recombinational repair to survive its cytotoxic effects (10). Yeast deficient for the excision repair gene RAD3 are also hypersensitive, implying that both excision and recombinational repair are required to remove CP lesions. Similarly, human cells require ERCC-1 for repair of CP-induced damage (11). Presumably, the excision of interstrand CP crosslinks creates double-strand breaks, which in turn stimulate recombinational repair (11–15). Indeed, CP is known to induce double-strand breaks in Drosophila meiotic chromosomes and to disrupt synaptonemal complexes in mice (16, 17).
These combined observations raised the possibility that CP could induce meiotic recombination in mammals. We have found that CP stimulated intrachromosomal meiotic gene conversion in a transgenic mouse assay that enables the detection of such events in spermatids (refs. 18 and 19; W.H.H. and J.C.S., unpublished observations). Given the association between gene conversion and crossing-over, we hypothesized that this drug might also induce crossing-over in mice. We report that male mice treated with CP undergo meiotic crossing-over at levels up to twice that of controls. This drug regimen should be useful in genetic mapping experiments in mice whereby the number of meioses that must be screened to localize a gene can be halved.
MATERIALS AND METHODS
Mouse Breeding and CP Treatment.
DBA/2J females, more than 2 months of age, were induced into estrous by administration of pregnant mare’s serum and human chorionic gonadotropin as described (20). Because the females were more than 2 months old, they did not superovulate with the treatment. CP (10 mg/kg; Sigma) was injected intraperitoneally immediately after dissolving at 1 mg/ml in 0.9% NaCl. Control males were injected with the same volume of 0.9% NaCl. Treated and control mice were taken from the same batches of age-matched animals obtained from the production facility at The Jackson Laboratory. Each male was placed into a cage with two hormonally treated females (also age-matched between control and treated matings) at specified times after CP injection and removed the following day. Drug treatment and housing of the mice were performed with approval of the Institutional Animal Care and Use Committee according to American Association for the Accreditation of Laboratory Animal Care guidelines.
DNA Isolation and Microsatellite Marker Typing.
DNA was isolated from brains of newborn pups as described (21). Microsatellite loci were typed in a modification of a reported procedure (22). The PCR mixture contained 5 μl of brain DNA diluted 1:50 in water, 0.22 μM primers, all four dNTPs (each at 0.8 mM), 0.5 unit of Taq polymerase (Perkin–Elmer), 2.25 mM MgCl2, and 1× PCR buffer (Perkin–Elmer). Amplification was performed in a total reaction volume of 30 μl, plus a 10 μl oil overlay, in 96-well plates on a thermal cycler (MJ Research, Cambridge, MA). After denaturing at 97°C for 1 min, 45 cycles of amplification were performed as follows: 94°C for 30 sec, 55°C for 30 sec, and 72°C for 30 sec, ending with a 5-min incubation at 72°C. PCR products were separated in agarose gels (3% of a 3:1 NuSieve mixture from FMC) and stained with Sybr green (FMC). The double and triple crossovers were typed twice to ensure accuracy.
The complete allele typing data are available via the World Wide Web at: http://www.jax.org/~jcs/cisplatin.html.
Statistical Analysis.
χ2 and P values were derived from the 2 × 2 contingency test (assisted by the software fisher 2 by Kaz Matsuki). Two values were compared in each test class: recombinant and nonrecombinant. However, because there were double and triple crossovers in CP-treated animals, the recombinant class was defined as (total no. crossovers), and the nonrecombinant class was defined as (no. offspring − no. crossovers). G tests yielded nearly identical results.
RESULTS AND DISCUSSION
CP Increases Crossing-Over on Multiple Chromosomes.
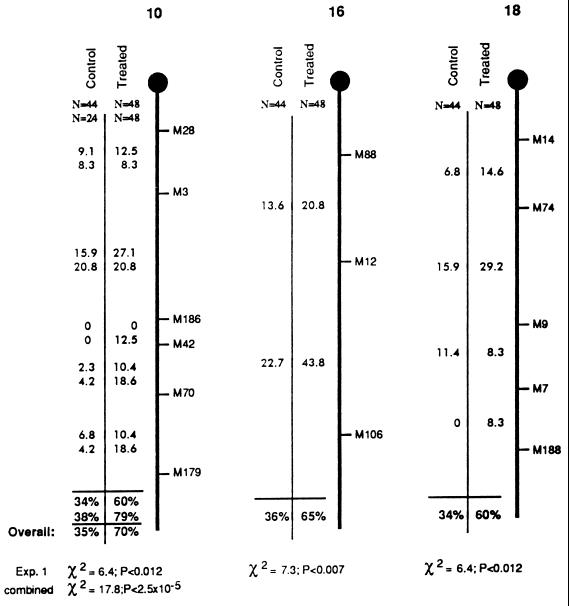
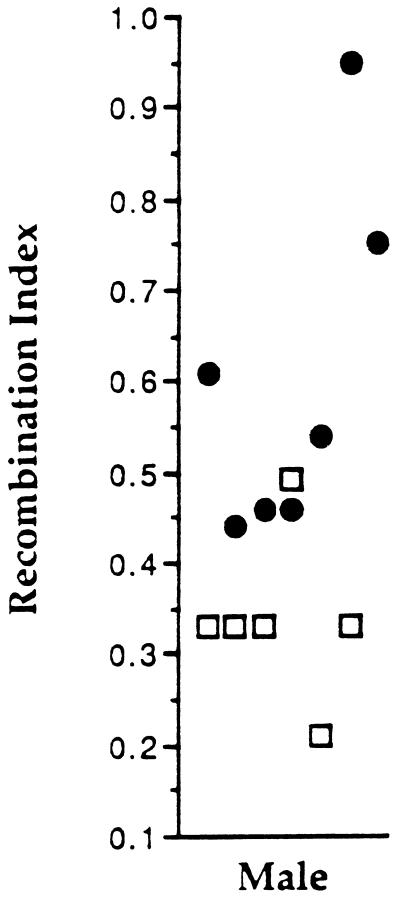
To test for possible effects of CP on crossing-over, a set of F1 hybrid males (C57BL/6J × DBA/2J) was exposed to a single CP dose of 10 mg/kg and mated 28 days later to DBA/2J females that were hormonally induced into estrous, and their offspring were scored for recombination at multiple intervals on three chromosomes. Sperm produced at this point would have been at the mid-meiosis I prophase stage of spermatogenesis at the time of treatment (ref. 23; discussed below). Compared with age-matched controls, CP-treated males exhibited no reduction in fertility; nearly all males from both groups yielded productive matings with similar litter sizes. DNA was extracted from pups of both groups and typed at microsatellite loci along three randomly chosen chromosomes—chromosomes 10, 16, and 18. The results are summarized in Fig. 1. The crossover frequency (no. crossovers/no. progeny) in offspring of CP-treated males was elevated by more than 75% on each chromosome, and all the increases were statistically significant by the χ2 test (Fig. 1). When the data for all chromosomes was combined, the recombination frequency rose from 35% per chromosome in the untreated to 62% in the CP-treated group—a highly significant increase (χ2 = 12, P < 8 × 10−6). To assess whether progeny from any individual male in the treated group contributed disproportionately to the data, a recombination index (total no. crossovers/no. pups sired) was calculated for each animal. Only one of the control males had a higher recombination index than treated animals (Fig. 2).
Figure 1.
Crossover frequencies in CP-treated and control males. The treated group refers to male mice that were injected with CP at 10 mg/kg and mated 4 weeks thereafter. The control group was sham-injected with vehicle (0.9% NaCl). Results shown are for chromosomes 10, 16, and 18, as indicated. The thick vertical line represents each chromosome, and • indicates the position of the centromere. Microsatellite loci that were typed in the crosses are indicated at the right of each chromosome, whereby the prefixes D10Mit, D16Mit, and D18Mit have been abbreviated with M. The numbers listed in the intervals between microsatellites are recombination percentages in those particular intervals. Results from two independent crosses with chromosome 10 are shown (the second cross in CP-2). The results from the first cross are presented immediately above those from the second cross. The overall recombination frequency between the most distal and proximal markers are shown under the horizontal lines near the bottom. The results of χ2 analysis in a two-way contingency test are shown at the bottom. These reflect the probability that the overall recombination frequency differs between control and treated groups. Independently summarized percentages for both experiments involving chromosome 10 are given, along with the overall combined percentage as indicated. Contingency test results are given for both the first experiment and the combined data on chromosome 10. The lengths of each chromosome as depicted do not reflect true physical or genetic size.
Figure 2.
Recombination indices for individual males. A recombination index [(total number crossovers for all chromosomes)/(number pups sired × 3)] was calculated and plotted for each of the CP-treated and control males that yielded the data in Fig. 1 (excluding CP group 2 for chromsome 10). •, CP-treated males; ○, controls.
Timing and Repeatability of CP-Induced Recombination in Meiosis.
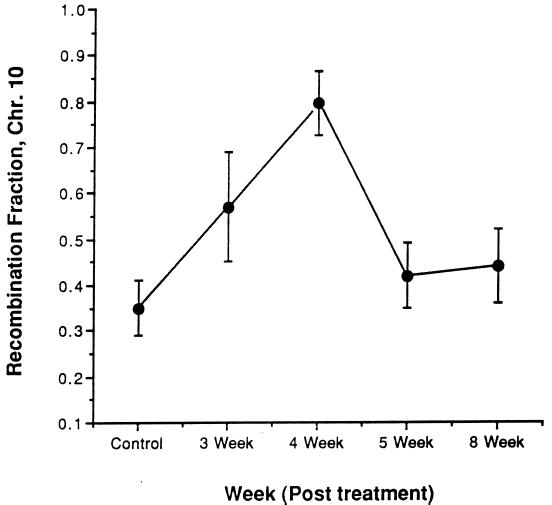
Additional experiments were performed to (i) test the repeatability of the effect, (ii) ascertain the developmental timing of the recombination induction, and (iii) determine whether the CP-mediated induction of recombination could be observed over time within individual animals. One set of males (CP-2) was injected with CP as above and mated 3, 4, and 8 weeks later. A second sham-treated control group (CON-2) was concurrently tested, and additional CP-treated mice were mated 5 weeks after exposure. Crossing-over on chromosome 10 was measured in progeny from these experiments.
The results confirmed the sharp increase in recombination induced by CP. The CON-2 group displayed a recombination rate nearly identical to the original controls (34% vs. 38%), and the CP-2 group mated after 4 weeks showed a frequency of 79% (Fig. 1), representing a more than 2-fold increase over the controls. Interestingly, this was significantly higher (P < 0.045) than the original cohort (60%). The only difference between the two experiments was that CP-2 males were also mated 1 week before the 4-week sampling point. One possible explanation is that because sperm survive for up to 2 weeks after exit from the testis (24), the ejaculates from males of the first group would have contained a mixture of sperm descended from precursors at different developmental stages. The possibility was considered that the mating of CP-2 males 3 weeks after exposure may have enabled them to clear sperm whose precursors were postmeiotic at the time of CP exposure, thereby effectively enriching the ejaculate for sperm that were derived from CP-exposed pachytene spermatocytes. However, this effect was not observed for chromosomes 16 and 18. Recombination between the proximal and distal markers in CP-2 was not statisically different from the CP-1 values on these chromosomes (chromosome 16, 65% vs. 57%, P > 0.47; chromosome 18, 60% vs. 50%, P > 0.3. Data are available on the World Wide Web at http://www.jax.org/~jcs/cisplatin.html). When the data from all three chromosomes are combined, the recombination fraction in CP-1 was 0.618 (89 of 144) and that of CP-2 was 0.62 (89 of 143). Thus, the overall level of induced crossovers appeared to be highly repeatable. It must be considered, however, that cryptic double crossovers might not have been detected on chromosomes 16 and 18 in CP-2.
The combined data reveal a peak of CP-induced recombination on chromosome 10 in gametes from males exposed 4 weeks before mating (Fig. 3). Importantly, this effect was consistent not only between cohorts of treated animals but also within the group (CP-2) that was sequentially mated. The crossover rate in CP-2 rose from 57% at 3 weeks (see below) to 79% at 4 weeks before declining at 8 weeks to a level (44%) that was not significantly different than the combined frequency (35%) of the two control groups (P < 0.37; Table 1). Spermatozoa present 8 weeks after treatment would have been at the spermatogonium stage when exposed, so it appears that CP did not have a marked effect upon mitotic recombination in stem cells. The inductive effect of CP disappeared within 7 days after the peak at 4 weeks after treatment; males mated 5 weeks after exposure did not differ significantly (P < 0.45) from the combined controls (Table 1). These results suggest that the spermatogenic cell types subject to induction of recombination are those that require no more than 4–5 weeks to develop into mature spermatozoa in the ejaculate.
Figure 3.
Time course effects of CP on recombination. Data are the crossover values (mean ± SEM) for chromosome 10. Data reported for controls are combined from two experiments. All other data points, except for week 5, were collected from the same males mated at different times after exposure.
Table 1.
Time course of recombination induction in CP-treated mice
| Interval | No. of crossovers (%)
|
|||
|---|---|---|---|---|
| 3 week (n = 23) | 4 week (n = 48) | 5 week (n = 48) | 8 week (n = 41) | |
| M28–M3 | 2 (8.7) | 4 (8.3) | 3 (6.3) | 4 (9.8) |
| M3–M186 | 8 (34.8)* | 10 (20.8) | 8 (16.7) | 8 (19.5) |
| M186–M42 | 0 | 6 (12.5) | 0 | 0 |
| M42–M70 | 2 (8.7) | 9 (18.8) | 3 (6.3) | 3 (7.3) |
| M70–M179 | 1 (4.4) | 9 (18.8) | 6 (12.5) | 3 (7.3) |
| Total | 13 (57) | 38 (79) | 20 (41.6) | 18 (44) |
Time course of recombination induction in CP-treated mice. Six males were treated with CP and mated 3, 4, and 8 weeks later. An independent set of males that were mated 5 weeks after CP exposure is also shown. The number of recombinants in chromosome 10 intervals are listed. M is an abbreviation for D10Mit. Recombination percentages (shown in parentheses) equal (total no. crossovers) × 100/n.
Most of these recombinants were contributed by a single male (see text).
Inspection of crossover distributions revealed some unusual patterns. It is possible that the stimulation of recombination by CP was nonuniform along the length of the chromosomes. For example, although there was little difference between control and treated in proximal chromosome 10 between D10Mit28 and D10Mit186, distal recombination frequencies were markedly higher (Fig. 1). Typing of larger data sets with additional markers should reveal whether CP has preferred sites of action and where such sites may exist in the genome. Another unusual observation was made with regards to the offspring of CP-2 males mated 3 weeks after exposure. This group had a higher crossover frequency on chromosome 10 relative to controls (Table 1; the small sample size renders the results not highly significant, P < 0.073). However, the majority of crossovers (8 of 13) occurred in the D10Mit3–D10Mit186 interval, and 5 of these arose in a single male’s litter of 7. This could not be explained by a “jackpot” event in stem cells, because offspring from this animal assayed at later time points (4 and 8 weeks) did not show elevated recombination in the same interval. It is unclear whether this curiosity was related to CP treatment.
The timing of recombination events in mammalian meiosis is poorly characterized, in contrast to yeast. Crossing-over is presumed to occur sometime during the long (7 day) pachytene phase, but it is not clear just when these events are initiated. Because the CP-induced crossing-over occurred during a defined window of time, this information may be useful in elucidating this issue. On the basis of the time course of spermatogenesis in the mouse established by classical anatomical and mutagenesis studies, CP-induced recombination events, presumably a consequence of causing double-strand breaks, can be traced to primary spermatocytes that have progressed about 2 days into pachynema (23, 25, 26).
In yeast, double-strand breaks are detectable at the same time as lateral axes of the synaptonemal complex form (3). They disappear by the end of the zygotene and early pachytene stages, when the synaptonemal complex axes align to form the tripartite structure. Thus in yeast, initial events of recombination occur well before exchange in pachynema. Our results suggest that CP initiates recombination later than spontaneous double-strand break formation and disappearance in yeast meiosis. However, because the rate of spermatogenesis can vary between strains of mice and the time required for passage from the testis to the ejaculate (estimated to be 7.5–8 days) is not concretely established, our conclusion that CP induces recombination in early pachynema must be considered an estimate.
It has been reported that preleptotene/leptotene spermatocytes are much more sensitive to CP-induced chromosome aberrations than are spermatocytes at the zygotene or pachytene stages (ref. 27; preleptotene spermatocytes require about 4 days to progress to pachynema), but it is not clear that these clastogenic events are the same that would stimulate crossing-over. Recent evidence from mice suggests that by the early to midpachytene stage, events have progressed to a point where they can be resolved rapidly into chiasmata (28). This suggests that spontaneous initiation of recombination in spermatocytes occurs in the early pachytene stage or sooner. This observation is generally consistent with our results. It is possible that CP can be exploited as a tool to further refine the window of time during which recombination is initiated during meiosis in mice.
Recombination and Interference.
Crossing-over is essential to ensure accurate disjunction of homologous chromosomes during meiosis. In most organisms, the number of crossovers per meiosis is relatively constant. Recombinational interference ensures that each chromosome pair has at least one crossover. Otherwise, larger chromosomes would have multiple crossovers, and smaller ones would often have none (29, 30). The results presented herein indicate that induction of recombination by CP did not occur by elimination of interference. A total of 67 crossovers on chromosome 10 were observed in the two 4-week treatment groups. This included one triple and four double crossovers (there were none in the controls). In the absence of interference, a Poisson distribution of the 67 crossovers among the 96 offspring predicts significantly more nonrecombinants and double recombinants than were observed (Table 2). The overrepresentation of single recombinants implies that interference (or another mechanism) operates to distribute crossovers evenly between chromosomes, avoiding nonrecombinants that would lead to possible aneuploidy.
Table 2.
Distribution of crossovers in offspring of CP-treated males
| XO/Chr | No. of crossovers
|
|||
|---|---|---|---|---|
| Obs. | Exp. | χ2 | P | |
| 0 | 35 | 47.8 | 3.41 | >0.065 |
| 1 | 56 | 33.4 | 15.3 | <0.0001 |
| 2 | 4 | 11.7 | 5.1 | <0.024 |
| 3 | 1 | 2.7 | 1.1 | >0.3 |
Distribution of crossovers in offspring of CP-treated males. Data are a composite of the two groups of CP-treated males for chromosome 10 (Fig. 1). XO/Chr is the number of crossovers on chromosome 10 in any single offspring. The next two columns are the observed (Obs.) and expected (Exp.) numbers of animals with the corresponding number of crossovers. Expected values are calculated by using the Poisson distribution.
It remains to be seen whether interference represents an absolute barrier to an upper level of recombination or whether more effective drug treatments can increase crossing-over to an extent limited only by chromosomal stability and meiotic progression. A physical mechanism of interference is implied by the existence of ZIP1, a yeast synaptonemal complex protein whose absence abolishes interference (29). This suggests that interference may put an upper level to the number of recombination events on a single chromosome, unless there is experimental modification of synaptonemal complex components.
Utility in Genetic Analyses.
The ability to increase crossover frequency can be of considerable practical benefit to mouse genetics. It can markedly decrease time, cost, and effort in conducting large crosses or in creating mapping reagents such as advanced intercross lines (31). It should be kept in mind that because CP is known to cause chromosome aberrations during at certain stages of spermatogenesis (27), there is a possible risk of creating mutations in the progeny of treated males. However, we are unaware of any specific locus test data for CP.
CP more than doubled crossing-over (on chromsome 10) in these experiments, but it is possible that other protocols could be developed that further increase the frequency. In the present work, a single drug was tested at a single dosage, in one genotype of mice, at 7-day sampling intervals. Furthermore, the effect of CP on females was not investigated, so the current benefit is restricted to recombinational mapping in males. Adjusting the treatment parameters or using different strains and both sexes of mice (perhaps containing mutations in certain DNA repair genes) might coax crossover frequencies to even higher levels.
Acknowledgments
We thank Eva Eicher, Rosemary Elliot, Wayne Frankel, and Mary Ann Handel for critical reviews and advice on the manuscript. This work was supported by grants from the National Institutes of Health (GM45415), American Cancer Society (CN-118), and National Science Foundation (Presidential Young Investigator Award) to J.C.S. W.H.H. and M.E.L. are recipients of postdoctoral fellowship (ES05743) and Physician Scientist (ES00251) awards, respectively, from National Institute on Environmental Health Sciences.
ABBREVIATION
- CP
cisplatin
References
- 1.Dietrich W F, Miller J, Steen R, Merchant M A, Damron Boles D, Husain Z, Dredge R, Daly M J, Ingalls K A, O’Connor T J, Evans C A, DeAngelis M M, Levinson D M, Kruglyak L, Goodman N, Copeland N G, Jenkins N A, Hawkins T L, Stein L, Page D C, Lander E S. Nature (London) 1996;380:149–152. doi: 10.1038/380149a0. [DOI] [PubMed] [Google Scholar]
- 2.Dib C, Faure S, Fizames C, Samson D, Drouot N, Vignal A, Millasseau P, Marc S, Hazan J, Seboun E, Lathrop M, Gyapay G, Morissette J, Weissenbach J. Nature (London) 1996;380:152–154. doi: 10.1038/380152a0. [DOI] [PubMed] [Google Scholar]
- 3.Padmore R, Cao L, Kleckner N. Cell. 1991;66:1239–1256. doi: 10.1016/0092-8674(91)90046-2. [DOI] [PubMed] [Google Scholar]
- 4.Schiestl R, Gietz R, Mehta R, Hastings P. Carcinogenesis. 1989;10:1445–1455. doi: 10.1093/carcin/10.8.1445. [DOI] [PubMed] [Google Scholar]
- 5.Schiestl R. Nature (London) 1989;337:285–288. doi: 10.1038/337285a0. [DOI] [PubMed] [Google Scholar]
- 6.Graf U, Wurgler F, Katz A, Frei H, Juon C, Hall C, Kale P. Environ Mutagen. 1984;6:153–188. doi: 10.1002/em.2860060206. [DOI] [PubMed] [Google Scholar]
- 7.Wang Y, Maher V, Liskay R, McCormick J. Mol Cell Biol. 1988;8:196–202. doi: 10.1128/mcb.8.1.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Katz A J. Environ Mol Mutagen. 1987;10:197–203. doi: 10.1002/em.2850100210. [DOI] [PubMed] [Google Scholar]
- 9.Vogel E W, Zijlstra J A. Mutat Res. 1987;182:243–264. doi: 10.1016/0165-1161(87)90010-0. [DOI] [PubMed] [Google Scholar]
- 10.Hannan M A, Zimmer S G, Hazle J. Mutat Res. 1984;127:23–30. doi: 10.1016/0027-5107(84)90136-2. [DOI] [PubMed] [Google Scholar]
- 11.Larminat F, Bohr V. Nucleic Acids Res. 1994;22:3005–3010. doi: 10.1093/nar/22.15.3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sancar A. Annu Rev Genet. 1995;29:69–106. doi: 10.1146/annurev.ge.29.120195.000441. [DOI] [PubMed] [Google Scholar]
- 13.Szostak J, Orr-Weaver T, Rothstein R, Stahl F. Cell. 1983;33:25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- 14.Sun H, Treco D, Schultes N P, Szostak J W. Nature (London) 1989;338:87–90. doi: 10.1038/338087a0. [DOI] [PubMed] [Google Scholar]
- 15.Wu T C, Lichten M. Science. 1994;263:515–518. doi: 10.1126/science.8290959. [DOI] [PubMed] [Google Scholar]
- 16.Brodberg R K, Lyman R F, Woodruff R C. Environ Mutagen. 1983;5:285–297. doi: 10.1002/em.2860050307. [DOI] [PubMed] [Google Scholar]
- 17.Allen J W, Poorman P A, Backer L C, Gibson J B, Westbrook Collins B, Moses M J. Cell Biol Toxicol. 1988;4:487–494. doi: 10.1007/BF00117776. [DOI] [PubMed] [Google Scholar]
- 18.Murti J R, Bumbulis M, Schimenti J. Mol Cell Biol. 1992;12:2545–2552. doi: 10.1128/mcb.12.6.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murti J R, Schimenti K J, Schimenti J C. Mutat Res. 1994;307:583–595. doi: 10.1016/0027-5107(94)90268-2. [DOI] [PubMed] [Google Scholar]
- 20.Hogan B, Constantini F, Lacy E. Manipulating the Mouse Embryo. Cold Spring Harbor, NY: Cold Spring Harbor Lab. Press; 1986. [Google Scholar]
- 21.Schimenti J. Mouse Genome. 1991;89:573. [Google Scholar]
- 22.Taylor B A, Phillips S J. Mamm Genome. 1995;6:493–498. doi: 10.1007/BF00356164. [DOI] [PubMed] [Google Scholar]
- 23.Oakberg, E. F. (1960) J. Dairy Sci. 43 (Suppl.), 54–67.
- 24.DeLeon P A, Boice M L. Hum Genet. 1982;62:70–77. doi: 10.1007/BF00295606. [DOI] [PubMed] [Google Scholar]
- 25.Oakberg E F. Am J Anat. 1956;99:391. doi: 10.1002/aja.1000990303. [DOI] [PubMed] [Google Scholar]
- 26.Sirlin J L, Edwards R G. Nature (London) 1957;179:725–727. doi: 10.1038/179725a0. [DOI] [PubMed] [Google Scholar]
- 27.Adler I-D, Tarras A. Mutat Res. 1990;243:173–178. doi: 10.1016/0165-7992(90)90087-z. [DOI] [PubMed] [Google Scholar]
- 28.Wiltshire T, Park C, Caldwell K A, Handel M A. Dev Biol. 1995;169:557–567. doi: 10.1006/dbio.1995.1169. [DOI] [PubMed] [Google Scholar]
- 29.Sym M, Roeder G S. Cell. 1994;79:283–292. doi: 10.1016/0092-8674(94)90197-x. [DOI] [PubMed] [Google Scholar]
- 30.Kaback D B, Guacci V, Barber D, Mahon J W. Science. 1992;256:228–32. doi: 10.1126/science.1566070. [DOI] [PubMed] [Google Scholar]
- 31.Darvasi A, Soller M. Genetics. 1995;141:1199–207. doi: 10.1093/genetics/141.3.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]