Abstract
Hepatic glucokinase plays a key role in glucose metabolism as underlined by the anomalies associated with glucokinase mutations and the consequences of tissue-specific knock-out. In the liver, glucokinase transcription is absolutely dependent on the presence of insulin. The cis-elements and trans-acting factors that mediate the insulin effect are presently unknown; this is also the case for most insulin-responsive genes. We have shown previously that the hepatic expression of the transcription factor sterol regulatory element binding protein-1c (SREBP-1c) is activated by insulin. We show here in primary cultures of hepatocytes that the adenovirus-mediated transduction of a dominant negative form of SREBP-1c inhibits the insulin effect on endogenous glucokinase expression. Conversely, in the absence of insulin, the adenovirus-mediated transduction of a dominant positive form of SREBP-1c overcomes the insulin dependency of glucokinase expression. Hepatic fatty acid synthase and Spot-14 are insulin/glucose-dependent genes. For this latter class of genes, the dominant positive form of SREBP-1c obviates the necessity for the presence of insulin, whereas glucose potentiates the effect of SREBP-1c on their expression. In addition, the insulin dependency of lipid accumulation in cultured hepatocytes is overcome by the dominant positive form of SREBP-1c. We propose that SREBP-1c is a major mediator of insulin action on hepatic gene expression and a key regulator of hepatic glucose/lipid metabolism.
Sterol regulatory element binding protein-1c (SREBP-1c) belongs to a family of transcription factors involved in cholesterol and fatty acid metabolism (1). It is synthetized as a precursor form anchored in endoplasmic reticulum and nuclear membranes. After proteolytic cleavage, its mature active form migrates into the nucleus where it can bind both sterol regulatory elements (5′-TCACCCCCCAC-3′) and E-boxes (5′-CANNTG-3′) (1, 2). The factors that control the cleavage of the precursor form of SREBP-1c are presently unknown. We have shown previously in cultured rat hepatocytes that SREBP-1c expression is transcriptionally stimulated by insulin and repressed by glucagon (3). Transfection studies of promoter/reporter constructs in cell lines have suggested that this factor mediates the effect of insulin on the low-density lipoprotein receptor (4) and the fatty acid synthase genes (5). In addition, it has been hypothesized from in vivo studies in mice and in vitro studies in adipocyte cell lines (5, 6) that the nuclear abundance of SREBP-1c is enhanced by insulin. Finally, indirect evidence suggests that SREBP-1c could be a target of mitogen-activated protein (MAP) kinase, a known intermediate of insulin action (7). Insulin could then act on SREBP-1c in three different ways: increased transcription, increased mobilization to the nucleus, and activation of transcriptional activity; however, at present, there is no demonstration of an involvement of SREBP-1c in insulin action on an insulin-responsive gene in the liver.
Hepatic and β-pancreatic glucokinases play a key role in glucose metabolism and β-cell insulin secretion as underlined by the diabetes mellitus associated to glucokinase mutations or the consequences of tissue-specific knock-outs (8–11). A different promoter directs glucokinase transcription in the liver from that in pancreatic β-cells. In the liver, glucokinase transcription is absolutely dependent on the presence of insulin and is repressed by glucagon, whereas the pancreatic promoter is insensitive to these hormones (12–14). Despite numerous efforts, the cis-acting elements on the hepatic glucokinase promoter which respond to insulin and the trans-acting factors involved have never been identified. In view of the effects of insulin and glucagon at different levels on SREBP-1c nuclear activity, we have hypothesized that this transcription factor could be involved in glucokinase gene expression.
In the present study, we demonstrate in cultured rat hepatocytes by using adenovirus-mediated transduction of both dominant negative and positive forms that SREBP-1c is a major factor of insulin action on glucokinase gene expression and other hepatic insulin/glucose responsive genes.
Materials and Methods
Animals.
Animal studies were conducted according to the French national guidelines for the care and use of experimental animals. Female (200–300 g body weight) or 13-day-old suckling Wistar rats from Iffa Credo were used for isolation of hepatocytes or analysis of specific mRNA expression in whole liver. They were housed in plastic cages at a constant temperature (22°C) with light from 0700 to 1900 h for at least 1 week before the experiments. Liver sampling was performed for both suckling and adult animals at 0900 h, i.e., in the immediate postabsorptive period for adult rats. After animal anesthesia, livers were excised, frozen in liquid nitrogen, and stored at −80°C until RNA extraction. All suckling rats were in the absorptive phase as indicated by their stomach full of milk.
Preparation of Recombinant Adenovirus.
The adenovirus vector containing the transcriptionally active amino-terminal fragment (amino acids 1–403) of SREBP-1c was constructed as in ref. 15. Briefly, the cDNA of the transcriptionally active fragment of SREBP-1c was subcloned into the shuttle vector pAd Track-CMV. The resultant plasmid was linearized by the restriction endonuclease PmeI and cotransformed with the supercoiled adenoviral vector pAd-Easy1 into Escherichia coli strain BJ5183. Recombinants were selected by kanamycin resistance and screened by restriction endonuclease digestion. Then, the recombinant adenoviral construct was cleaved with PacI and transfected into the packaging cell line 293.
The recombinant adenovirus containing the dominant negative form of SREBP-1c was constructed as described (3). The adenovirus vector containing the major late promoter with no exogenous gene (Ad null) was used as control. The adenoviral vectors were propagated in the 293 cell line, purified by cesium chloride density centrifugation, and stored as described (3).
Hepatocytes Isolation, Culture, and Treatment with Recombinant Adenovirus.
Hepatocytes were isolated from fed rats by the collagenase method (16). Cell viability was assessed by the trypan blue exclusion test and was always higher than 85%. Hepatocytes were seeded at a density of 8 × 106 cells per dish (100-mm Petri dishes) in medium M199 with Earle’s salts (GIBCO/BRL) supplemented with 100 units/ml of penicillin, 100 μg/ml of streptomycin, 0.1% (wt/vol) BSA, 2% (vol/vol) Ultroser G (GIBCO/BRL), 100 nM dexamethasone (Sigma), 1 nM insulin (Actrapid, Novo-Nordisk, Copenhagen), and 100 nM triiodothyronine (T3; Sigma). After cell attachment (4 h), the medium was replaced by a medium similar to the plating medium but free of hormones (except when indicated) Ultroser and albumin. The cells were then cultured under various conditions as described in the figure legends. For suckling rat hepatocytes, plating was done in M199 and in the presence of antibiotics exclusively.
For the experiments involving adenoviruses, hepatocytes were cultured for 16 h after cell attachment. Hepatocytes were then incubated for 120 min at 37°C in M199 either with or without adenovirus at various titers (plaque forming units, pfu). The medium was then replaced by a fresh medium as described in the figure legends.
Staining Techniques.
To detect the presence of lipid droplets, hepatocytes were fixed at the end of the culture period by 10% formaldehyde in PBS and stained with Oil Red O as described (17).
Isolation of Total RNA and Northern Blot Hybridization.
Total cellular RNAs were extracted from whole liver or cultured hepatocytes by using the guanidine thiocyanate method (18) and prepared for Northern blot hybridization as described (19). Labeling of each DNA probe with [α-32P]dCTP was performed by random priming (Rediprime labeling kit, Amersham International). Autoradiograms of Northern blots were scanned and quantified by using an image processor program. Fatty acid synthase (FAS), SREBP-1c, and Spot-14 (S14) cDNAs were as previously described (3). Rat angiotensinogen (AGE) cDNA was purchased from American Type Culture Collection. A cDNA rat glucokinase probe was prepared by reverse transcriptase-mediated PCR from fed rat liver total RNA. The PCR primers used were as follows: upper primer, 5′-ATGGCTATGGATACTACAAGGTGTG-3′, and lower primer, 5′-AGAGTGCTTAGGATGTTGTGGATCT-3′. Northern blots were hybridized with a ribosomal 18S probe to verify that equivalent amounts of total RNA were loaded in each lane.
Glucose 6-Phosphate Assay.
The intracellular concentration of glucose 6-phosphate in cultured hepatocytes was measured by spectrophotometric assay (20). Results are expressed as means ± SD of duplicate values obtained in two separate experiments.
Results
To test the involvement of SREBP-1c in hepatic glucokinase expression, we used a strategy involving the expression in cultured hepatocytes of a dominant negative form of SREBP-1c (SREBP-1c DN). SREBP-1c DN consists of the amino-terminal fragment of SREBP-1c (amino acids 1–403), which is readily directed to the nucleus although it contains an alanine mutation at amino acid 320 (21). This mutation abolishes the binding of SREBP-1c to both sterol regulatory elements and E-boxes, but still allows dimerization leading to a decreased availability of endogenous SREBP-1c. The efficiency and specificity of this dominant negative form to counteract the transcriptional activity of SREBP-1c has already been established in transient transfections in cell lines and primary cultured hepatocytes (3, 21).
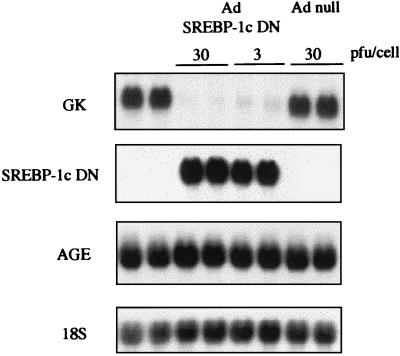
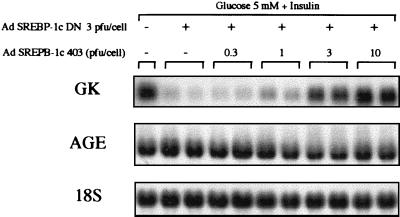
We thus infected cultured rat hepatocytes with SREBP-1c DN by using an adenoviral vector. The transgene is expressed in about 90% of plated hepatocytes (3). Using this method enables evaluation of the effects of SREBP-1c directly on endogenous gene expression rather than on promoter/reporter constructs. Rat hepatocytes cultured in the presence of insulin express glucokinase mRNA (Fig. 1). Adenovirus-mediated expression of the SREBP-1c DN totally prevents the expression of glucokinase, whereas a control gene (angiotensinogen) is not affected (Fig. 1). Infection of hepatocytes with an adenovirus that does not contain the SREBP-1c DN gene has no consequence (Fig. 1). To check that the effects of the dominant negative form of SREBP-1c are specific and not caused by the squelching of other transcription factors, we have performed a rescue experiment. An adenovirus containing the transcriptionally active 1- to 403-amino acid amino-terminal fragment of SREBP-1c (without the alanine mutation) (5) was engineered (SREBP-1c 403) and used to infect cultured hepatocytes with increasing titers (0.3 to 10 pfu per cell) together with the SREBP-1c DN (3 pfu per cell). This restores a normal glucokinase expression in a dose-dependent manner (Fig. 2). This series of experiments demonstrates that the presence of an active form of SREBP-1c is necessary in the nucleus for glucokinase expression.
Figure 1.
A dominant negative form of SREBP-1c decreases glucokinase gene expression in cultured hepatocytes of adult rats. Hepatocytes were cultured for 16 h after plating in the presence of 5 mM glucose. Cells were then incubated with 100 nM insulin and 5 mM glucose either without adenovirus or with null adenovirus (Ad null; 30 pfu per cell) or the SREBP-1c-DN adenovirus (3 or 30 pfu per cell). After 18 h, total RNAs were extracted and probed for the expression of glucokinase (GK), SREBP-1c DN and AGE genes, and 18S rRNA (18S). The Northern blot presented is representative of three experiments.
Figure 2.
A dominant positive form of SREBP-1c abolishes the effect of the dominant negative form of SREBP-1c on glucokinase gene expression in cultured hepatocytes of adult rats. Hepatocytes were cultured for 16 h after plating in the presence of 5 mM glucose. Cells were then incubated with 100 nM insulin and 5 mM glucose either without adenovirus or with the SREBP-1c DN adenovirus (3 pfu per cell) in the absence or presence of increasing titers of the SREBP-1c-403 adenovirus (0, 0.3, 1, 3, or 10 pfu per cell). After 18 h, total RNAs were extracted and analyzed for the expression of GK and AGE genes and 18S rRNA. The Northern blot presented is representative of two experiments.
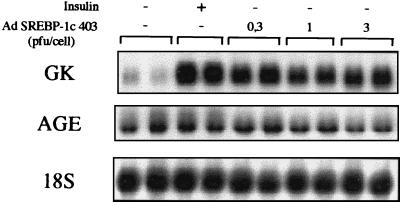
We then addressed the question of the relationship between insulin and SREBP-1c. If SREBP-1c is a major mediator of insulin action on glucokinase gene expression, it should be possible to mimic the effect of insulin by expressing SREBP-1c 403 in hepatocyte nuclei. We cultured hepatocytes in the absence or presence of insulin. In the absence of the hormone, glucokinase expression is kept low, whereas insulin induces a strong expression of glucokinase (Fig. 3). Hepatocytes infected with increasing titers of the adenovirus containing SREBP-1c 403 express glucokinase mRNA at a high level in the absence of insulin (Fig. 3). In other words, a transcriptionally active form of SREBP-1c mimics the effect of insulin on glucokinase gene expression.
Figure 3.
A dominant positive form of SREBP-1c induces glucokinase gene expression in cultured hepatocytes of adult rats in the absence of insulin. Hepatocytes were cultured for 16 h after plating in the presence of 5 mM glucose. Cells were then incubated with 5 mM glucose either without adenovirus and with or without 100 nM insulin, or with increasing titers of the SREBP1c-403 adenovirus (0, 0.3, 1, or 3 pfu per cell). After 18 h, total RNAs were extracted and analyzed for the expression of GK and AGE genes and 18S rRNA. The Northern blot presented is representative of three experiments.
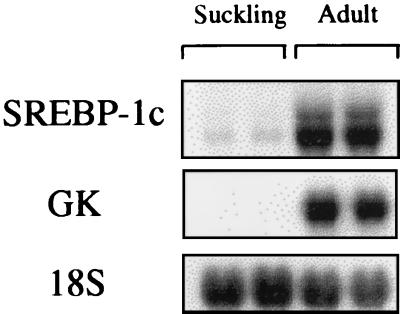
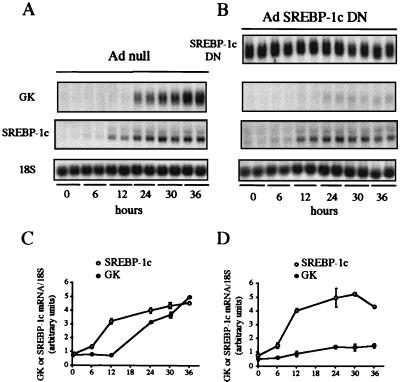
To back up the finding that SREBP-1c is involved in insulin action on glucokinase expression, we took advantage of a physiological model in which transcription of the glucokinase gene occurs for the first time in rat liver, the suckling/weaning transition (22). During the suckling period, which lasts about 2 weeks in rats, insulin concentrations are low and glucagon concentrations are high because of the ingestion of milk, which is a high-fat, low-carbohydrate diet, and the glucokinase gene is not transcribed (23). Glucokinase is expressed for the first time after transition from a milk diet to a high-carbohydrate diet after weaning, which is concomitant with an increase in insulin level. Glucokinase expression in suckling rat liver can also be induced earlier in vivo by giving a carbohydrate diet or in vitro by adding insulin to cultured hepatocytes (20, 24). If our hypothesis relating glucokinase expression to SREBP-1c is true, the expression of SREBP-1c should be absent from the suckling rat liver because of the low insulin and high glucagon concentrations. Its appearance should precede the induction of glucokinase in the presence of insulin in cultured suckling rat hepatocytes. The expression of SREBP-1c is indeed low in the liver of suckling rats as is glucokinase expression (Fig. 4) when compared with fed adult rats. When insulin is added to cultured suckling rat hepatocytes, it induces the appearance of SREBP-1c mRNA between 6 and 12 h (Fig. 5 A and C). Glucokinase mRNA is not detectable until a high expression of SREBP-1c is apparent, i.e., between 12 and 24 h after insulin addition (Fig. 5 A and C). To functionally relate the appearance of SREBP-1c and glucokinase expression, the same experiment was repeated in the presence of SREBP-1c DN. Under these conditions, insulin induces the normal appearance of endogenous SREBP-1c mRNA, but the appearance of glucokinase mRNA is markedly blunted (Fig. 5 B and D). The combined evidence presented demonstrates that SREBP-1c is a major mediator of the effect of insulin on glucokinase gene expression.
Figure 4.
Comparison of the expression of glucokinase and SREBP-1c genes in the liver of suckling and fed adult rats. Total RNAs were extracted and analyzed for the expression of GK and SREBP-1c genes and 18S rRNA. Each lane represents data obtained from the liver of one animal. This Northern blot is representative of data obtained with six different animals in each group.
Figure 5.
Insulin-induced expression of SREBP-1c precedes the appearance of glucokinase mRNA. A dominant negative form of SREBP-1c abolishes the effect of insulin on glucokinase gene expression in cultured hepatocytes of suckling rats. (A and C) Cells were incubated with 5 mM glucose with the Ad null (3 pfu/cell) or (B and D) the SREBP-1c-DN adenovirus (3 pfu per cell). After 24 h, cells were incubated with 100 nM insulin and 5 mM glucose for 0, 6, 12, 24, 30, or 36 h. Total RNAs were extracted and analyzed for the concentration of GK, endogenous SREBP-1c and SREBP-1c DN mRNAs, and 18S rRNA. The Northern blot presented is representative of two experiments. (C and D) Graphs show the concentrations of GK and endogenous SREPB-1c mRNA, expressed as a ratio to the corresponding 18S signal and as the means ± SD of duplicate values obtained in two separate experiments.
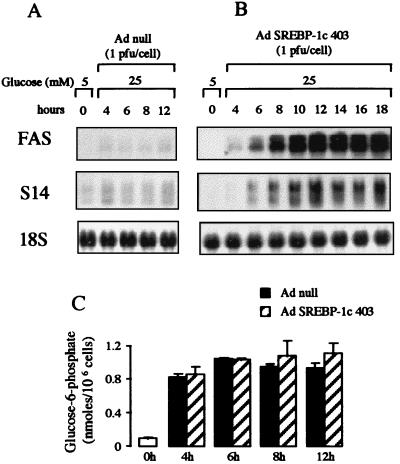
Hepatic genes involved in fatty acid synthesis from glucose (lipogenesis), such as FAS and S14, require both high insulin and high glucose concentrations for their activation in the liver (25, 26). In addition, glucose must be metabolized via glucokinase (25). Until now, the prevailing idea has been that insulin was necessary only to induce glucokinase expression and activity; however, it is also conceivable that a direct effect of SREBP-1c on lipogenesis-related gene promoters is part of the effect of insulin as suggested by experiments on the FAS gene promoter in adipocytes (5). To address this question, a kinetic experiment was performed in which the induction of the expression of these genes by SREBP-1c 403 and the concentration of glucose 6-phosphate, the product of the glucokinase reaction, were measured. As can be seen from Fig. 6A , cells cultured in the presence of 25 mM glucose alone and the null adenovirus have a low FAS and S14 gene expression. In the presence of 25 mM glucose but without insulin, infection of the cells with the adenovirus containing SREBP-1c 403 induces a time-dependent increase in FAS and S14 gene expression which is already visible 4 h after the infection (Fig. 6B). Addition of 25 mM glucose in the culture medium clearly increases glucose 6-phosphate concentration (Fig. 6C), showing that glucokinase is active in these hepatocytes; however, the increase in glucose 6-phosphate (Fig. 6C) is similar in the absence or presence of SREBP-1c 403. This result demonstrates that under these experimental conditions (see Discussion), the effect of SREBP-1c on lipogenic gene expression is independent of an increase in the glucokinase phosphorylating capacity; thus, there is a direct SREBP-1c-mediated effect of insulin on FAS and S14 promoters.
Figure 6.
The effect of the dominant positive form of SREBP-1c in the presence of a high glucose concentration on FAS and S14 gene expression and the determination of the glucose 6-phosphate concentrations in cultured hepatocytes of adult rats. After plating, hepatocytes were cultured for 16 h in the presence of 5 mM glucose. Cells were then incubated with 25 mM glucose in the absence of insulin either with the Ad null (A) or with SREBP-1c 403 (B). After 4, 6, 8, 10, 12, 14, 16, and 18 h, total RNA was extracted and analyzed for the expression of FAS and S14 genes and 18S rRNA and (C) the concentrations of glucose 6-phosphate were determined. The Northern blot presented is representative of three experiments. Glucose 6-phosphate concentrations are the means ± SD from values obtained from two separate experiments.
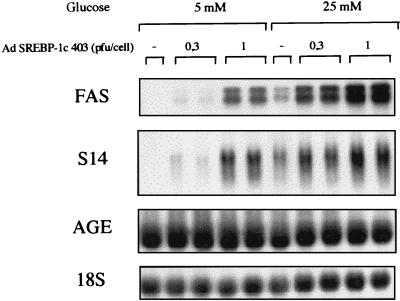
Because the expression of these genes is insulin/glucose dependent, we questioned next the role of glucose in this context. Cultured hepatocytes were infected with increasing amounts of the dominant positive form of SREBP-1c in the absence of insulin, and in the presence of low (5 mM) or high (25 mM) glucose. Glucose (25 mM) in the absence of insulin induces only a small increase in FAS and S14 gene expression (Fig. 7). In the presence of 5 mM glucose, SREBP-1c 403 already demonstrates a significant inducing effect on FAS and S14 gene expression. A high (25 mM) glucose concentration potentiates the effect of increasing amounts of SREBP-1c 403 on the expression of these genes (Fig. 7).
Figure 7.
Comparison of the effects of the dominant positive form of SREBP-1c in the presence of a low or high glucose concentration on lipogenic gene expression in cultured hepatocytes of adult rats. After plating, hepatocytes were cultured for 16 h in the presence of 5 mM glucose. Cells were then incubated for 16 h with 5 or 25 mM glucose either with or without the SREBP-1c 403 adenovirus (0.3 and 1 pfu per cell). Total RNAs were extracted and analyzed for the expression of FAS, S14, and AGE genes and 18S rRNA. The Northern blot presented is representative of three experiments.
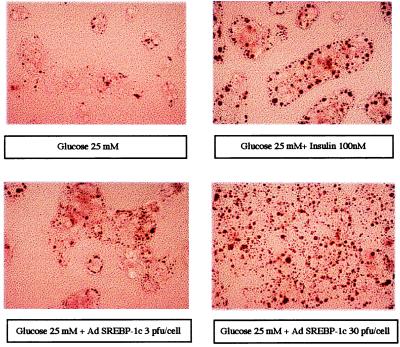
All the proteins encoded by the genes described above (glucokinase, FAS, and S14) are involved in the fatty acid synthesis from glucose. To underline the overall insulin-mimicking effect of SREBP-1c on these pathways, we visualized their end products, namely triglycerides, in hepatocytes cultured under various conditions. Glucose (25 mM) alone (Fig. 8) does not induce lipid accumulation in cultured hepatocytes whereas in the presence of both glucose and insulin, lipid droplets are visible. As expected, in the presence of 25 mM glucose alone, SREBP-1c 403 mimics the effect of insulin on lipid accumulation (Fig. 8).
Figure 8.
The effect of a dominant positive form of SREBP-1c on lipid accumulation in cultured hepatocytes of adult rats. Hepatocytes were cultured for 16 h after plating in the presence of 5 mM glucose. Cells were then incubated with 25 mM glucose either without adenovirus and with or without 100 nM insulin, or with the SREBP1c-403 adenovirus (3 or 30 pfu per cell). After 24 h, hepatocytes were stained for the presence of lipid droplets. Microscopic views of hepatocytes at a magnification of ×400 are shown.
Discussion
The liver has a central role in glucose homeostasis. It can produce glucose from glycogen stores or through the gluconeogenic pathway when glucose is not provided by the diet. It is also the first organ to handle glucose arising from intestinal absorption through the portal vein for glycogen storage. When glucose is ingested in large quantities, it is used by the liver for the synthesis of fatty acids that will be exported as triglycerides embedded in lipoproteins. Insulin is a major regulator of hepatic glucose metabolism and controls the expression of specific genes. Despite the importance of insulin in hepatic gene regulation, little is known about hepatic transcription factors which mediate insulin action (27). This was also true for the glucokinase gene, which encodes the first enzyme involved in glucose utilization in the liver and is entirely dependent on insulin for its transcription (12–14).
We demonstrate that SREBP-1c is a major factor for insulin-induced glucokinase gene expression because (i) abolishment of endogenous SREBP-1c activity leads to a marked decrease in insulin-induced glucokinase expression, and (ii) the active mature form of SREBP-1c is able to mimic the insulin effect. We cannot infer from the present experiments what the mechanisms are by which insulin stimulates transcriptional activity of SREBP-1c; however, it is conceivable that insulin acts at different levels. We have already established that insulin stimulates SREBP-1c expression in the liver (3). Insulin could also enhance the nuclear abundance of SREBP-1c (5, 6), possibly by activating a specific protease. The fact that the mature form of SREBP-1c does not require the presence of insulin to activate glucokinase expression (Fig. 2) suggests that SREBP-1c does not undergo an additional activation such as phosphorylation (7); however, we cannot rule out that adenovirus-mediated expression of the mature form of SREBP-1c leads to anomalously high SREBP-1c content in the nucleus, thus overriding the necessity for an additional posttranslational activation by insulin of the nuclear form of SREBP-1c.
Transcription of the glucokinase gene is initiated in the liver at a liver-specific promoter downstream from the pancreatic β-cell promoter (28). Until now, it has been impossible to delineate an insulin response element on the hepatic glucokinase promoter either by classical transfection experiments or by using transgenic animals (29, 30). The finding that SREBP-1c is a transcription factor that mediates the effect of insulin should help to identify this insulin response element on the hepatic glucokinase promoter.
Genes belonging to the lipogenic pathway such as FAS and S14 have a more complex regulation than the glucokinase gene because their expression depends on both a high insulin and glucose concentration and glucose metabolization by glucokinase (25). We suggested in a previous paper that the insulin dependency of this class of genes in the liver could also be related to a SREBP-1c-mediated effect of insulin on their promoter independent of a stimulation of glucokinase expression (3). We now demonstrate that this is indeed the case. In postabsorptive animals or under our experimental conditions (hepatocytes isolated from fed rats cultured for a short period of time), the long half-life (30 h) of the glucokinase protein (28) abolishes the necessity for additional glucokinase synthesis. Under these conditions, the major functional effect of insulin for lipogenesis-related genes would be the direct SREBP-1c-mediated activation of their promoter; however, in vivo, under conditions in which glucokinase activity is extremely low, such as long-term starvation, activation of lipogenesis-related genes would require both a direct SREBP-1c-mediated insulin action on their promoter as well as an indirect action through the activation of glucokinase transcription.
The role of glucose in the system is only partially unraveled. Our present data suggest that glucose increases the efficiency of nuclear SREBP-1c. It is noteworthy that in the absence of insulin, a small but consistent effect of a high glucose concentration is observed on FAS and S14 expression. This could be related to an effect of glucose on a residual nuclear SREBP-1c activity. The glucose signal could either potentiate the transcriptional capacity of SREBP-1c or alleviate the action of a repressor of SREBP action specific for this class of genes. The fact that glucokinase gene expression is insensitive to glucose would favor the latter possibility. Further work is necessary to discriminate between these hypotheses.
In summary, this and previous work (3, 31) reveal that SREBP-1c is a master gene for the regulation of lipid and glucose metabolism in the liver. It could then be a gene involved in metabolic dysfunctions such as non-insulin-dependent diabetes mellitus, obesity, and hepatic insulin resistance syndromes. Finally, the present study opens new perspectives for the unraveling of the cellular mechanisms involved in the insulin cascade leading to the regulation of gene expression.
Acknowledgments
We thank Drs. B. Spiegelman and J. B. Kim (Boston) and Dr. R. Plannels (Marseille, France) for the kind gift of cDNAs; Drs. P. Lemarchand (Paris), B. Vogelstein and T. C. He (Baltimore), and S. Hauguel-De Mouzon (Meudon, France) for their help in the production of adenoviruses; Dr. J. Boilot (Paris) for her help with the suckling rat experiments; Dr. J. Girard (Meudon, France) for helpful discussions; and Dr. A. Woods (London) and I. Dugail (Paris) for a critical review of the manuscript. We thank J. Benmamar for taking care of the animals. This work was supported by European Commission FAIR Program Grant CT 97-3011. P.F. is supported by the Centre National de la Recherche Scientifique. M.F. is a recipient of a grant from the Ministère de l’Enseignement Supérieur et de la Recherche.
Abbreviations
- Ad
adenovirus
- FAS
fatty acid synthase
- SREBP-1c
sterol regulatory element binding protein-1c
- SREBP-1c DN
dominant negative form of SREBP-1c
- SREBP-1c-403
dominant positive form of SREBP-1c
- S14
Spot 14
- pfu
plaque-forming unit
- GK
glucokinase
- AGE
angiotensinogen
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Brown M S, Goldstein J L. Cell. 1997;89:331–340. doi: 10.1016/s0092-8674(00)80213-5. [DOI] [PubMed] [Google Scholar]
- 2.Kim J B, Spotts G D, Halvorsen Y D, Shih H M, Ellenberger T, Towle H C, Spiegelman B M. Mol Cell Biol. 1995;15:2582–2588. doi: 10.1128/mcb.15.5.2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Foretz M, Pacot C, Dugail I, Lemarchand P, Guichard C, Le Liepvre X, Berthelier-Lubrano C, Spiegelman B M, Kim J B, Ferré P, Foufelle F. Mol Cell Biol. 1999;19:3760–3768. doi: 10.1128/mcb.19.5.3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Streicher R, Kotzka J, Muller-Wieland D, Siemeister G, Munck M, Avci H, Krone W. J Biol Chem. 1996;271:7128–7133. doi: 10.1074/jbc.271.12.7128. [DOI] [PubMed] [Google Scholar]
- 5.Kim J B, Sarraf P, Wright M, Yao K M, Mueller E, Solanes G, Lowell B B, Spiegelman B M. J Clin Invest. 1998;101:1–9. doi: 10.1172/JCI1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shimano H, Horton J D, Shimomura I, Hammer R E, Brown M S, Goldstein J L. J Clin Invest. 1997;99:846–854. doi: 10.1172/JCI119248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kotzka J, Muller-Wieland D, Koponen A, Njamen D, Kremer L, Roth G, Munck M, Knebel B, Krone W. Biochem Biophys Res Commun. 1998;249:375–379. doi: 10.1006/bbrc.1998.9161. [DOI] [PubMed] [Google Scholar]
- 8.Froguel P, Vaxillaire M, Sun F, Velho G, Zouali H, Butel M O, Lesage S, Vionnet N, Clement K, Fougerousse F, et al. Nature (London) 1992;356:162–164. doi: 10.1038/356162a0. [DOI] [PubMed] [Google Scholar]
- 9.Froguel P, Zouali H, Vionnet N, Velho G, Vaxillaire M, Sun F, Lesage S, Stoffel M, Takeda J, Passa P, et al. N Engl J Med. 1993;328:697–702. doi: 10.1056/NEJM199303113281005. [DOI] [PubMed] [Google Scholar]
- 10.Velho G, Petersen K F, Perseghin G, Hwang J H, Rothman D L, Pueyo M E, Cline G W, Froguel P, Shulman G I. J Clin Invest. 1996;98:1755–1761. doi: 10.1172/JCI118974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Postic C, Shiota M, Niswender K D, Jetton T L, Chen Y, Moates J M, Shelton K D, Lindner J, Cherrington A D, Magnuson M A. J Biol Chem. 1999;274:305–315. doi: 10.1074/jbc.274.1.305. [DOI] [PubMed] [Google Scholar]
- 12.Iynedjian P B, Gjinovci A, Renold A E. J Biol Chem. 1988;263:740–744. [PubMed] [Google Scholar]
- 13.Iynedjian P B, Jotterand D, Nouspikel T, Asfari M, Pilot P R. J Biol Chem. 1989;264:21824–21829. [PubMed] [Google Scholar]
- 14.Magnuson M A, Andreone T L, Printz R L, Koch S, Granner D K. Proc Natl Acad Sci USA. 1989;86:4838–4842. doi: 10.1073/pnas.86.13.4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He T C, Zhou S, da Costa L T, Yu J, Kinzler K W, Vogelstein B. Proc Natl Acad Sci USA. 1998;95:2509–2514. doi: 10.1073/pnas.95.5.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berry M N, Friend D S. J Cell Biol. 1969;43:506–520. doi: 10.1083/jcb.43.3.506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Green H, Kehinde O. Cell. 1975;5:19–26. doi: 10.1016/0092-8674(75)90087-2. [DOI] [PubMed] [Google Scholar]
- 18.Chomczynski P, Sacchi N. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 19.Coupé C, Perdereau D, Ferré P, Hitier Y, Narkewicz M, Girard J. Am J Physiol. 1990;258:E126–E133. doi: 10.1152/ajpendo.1990.258.1.E126. [DOI] [PubMed] [Google Scholar]
- 20.Bossard P, Decaux J F, Juanes M, Girard J. Eur J Biochem. 1994;223:371–380. doi: 10.1111/j.1432-1033.1994.tb19003.x. [DOI] [PubMed] [Google Scholar]
- 21.Kim J B, Spiegelman B M. Genes Dev. 1996;10:1096–1107. doi: 10.1101/gad.10.9.1096. [DOI] [PubMed] [Google Scholar]
- 22.Iynedjian P B, Ucla C, Mach B. J Biol Chem. 1987;262:6032–6038. [PubMed] [Google Scholar]
- 23.Girard J, Ferré P, Pégorier J P, Duée P H. Physiol Rev. 1992;72:507–562. doi: 10.1152/physrev.1992.72.2.507. [DOI] [PubMed] [Google Scholar]
- 24.Bossard P, Parsa R, Decaux J F, Iynedjian P, Girard J. Eur J Biochem. 1993;215:883–892. doi: 10.1111/j.1432-1033.1993.tb18106.x. [DOI] [PubMed] [Google Scholar]
- 25.Girard J, Ferré P, Foufelle F. Annu Rev Nutr. 1997;17:325–352. doi: 10.1146/annurev.nutr.17.1.325. [DOI] [PubMed] [Google Scholar]
- 26.Towle H C, Kaytor E N, Shih H M. Annu Rev Nutr. 1997;17:405–433. doi: 10.1146/annurev.nutr.17.1.405. [DOI] [PubMed] [Google Scholar]
- 27.O’Brien R M, Noisin E L, Granner D K. Biochem J. 1994;303:737–742. doi: 10.1042/bj3030737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iynedjian P B. Biochem J. 1993;293:1–13. doi: 10.1042/bj2930001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iynedjian P B. Biochem J. 1998;333:705–712. doi: 10.1042/bj3330705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Niswender K D, Postic C, Jetton T L, Bennett B D, Piston D W, Efrat S, Magnuson M A. J Biol Chem. 1997;272:22564–22569. doi: 10.1074/jbc.272.36.22564. [DOI] [PubMed] [Google Scholar]
- 31.Shimomura I, Shimano H, Korn B S, Bashmakov Y, Horton J D. J Biol Chem. 1998;273:35299–35306. doi: 10.1074/jbc.273.52.35299. [DOI] [PubMed] [Google Scholar]