Abstract
Among the numerous centrin isoforms identified by two-dimensional gel electrophoresis in human cells, an acidic and slow-migrating isoform is particularly enriched in a centrosome fraction. We report here that this isoform specifically reacts with antibodies raised against Saccharomyces cerevisiae Cdc31p and is present, as other centrin isoforms, in the distal lumen of centrioles. It is encoded by a new centrin gene, which we propose to name HsCEN3 (Homo sapiens centrin gene 3). This gene is more closely related to the yeast CDC31 gene, and shares less identity with algae centrin than HsCEN1 and HsCEN2. A murine CDC31-related gene was also found that shows 98% identity and 100% similarity with HsCEN3, demonstrating a higher interspecies conservation than the murine centrin gene MmCEN1 (Mus musculus centrin gene 1) with either HsCEN1, or HsCEN2. Finally, immunological data suggest that a CDC31-related gene could exist in amphibians and echinoderms as well. All together, our data suggest the existence of two divergent protein subfamilies in the current centrin family, which might be involved in distinct centrosome-associated functions. The possible implication of this new mammalian centrin gene in centrosome duplication is discussed.
Keywords: calcium-binding protein, caltractin, cell division cycle, centriole, centrosome
Centrin, also named caltractin, which is particularly enriched in centrosomes (1), is an ancient protein of the calcium-binding, EF-hand protein, or calmodulin fold, superfamily thought to have arisen within the ancestor of eukaryotes via gene duplication (2, 3). It was first discovered in the flagellar apparatus of unicellular green algae where it is responsible for the contraction of calcium-sensitive fibers that connect basal bodies to one another and to the nucleus (4). These fibers are involved in basal body localization, orientation, and segregation (5–8). Another set of calcium-contractile, centrin-containing fibers present in the transition zone between basal body and axoneme is involved in flagellum excision (9, 10). Cloning of the centrin gene from Chlamydomonas reinhardtii revealed similarity with CDC31 gene of Saccharomyces cerevisiae (11), a gene that was discovered in a genetic screen designed to isolate cell division cycle mutants (12, 13), and which is essential for spindle pole body duplication (14). Cdc31p has been localized to the bridge of the SPB, a structure that connects the two duplicated SPBs (15).
Recently, numerous centrin proteins have been described in various species from all kingdoms except prokaryotes. Centrin genes have been cloned in several protists, Giardia lamblia, a diplomonad (16), Paramecium tetraurelia (17), a ciliate, or Naegleria gruberi, an amoebo-flagellate (18), and in a land plant, Atriplex nummularia (19). Centrin genes have also been cloned in vertebrates, including the amphibian Xenopus laevis (accession no. U37538) and the mammals Mus musculus (20) and Homo sapiens (21, 22). In the latter case, two highly related genes, HsCEN1 and HsCEN2 (Homo sapiens centrin genes 1 and 2), have been cloned (we use here the nomenclature proposed in ref. 22). All centrin proteins identified so far appear associated with the centrosome.
Whereas they have been studied in some detail in green algae and in yeast, centrin functions in animal cells remain largely unknown (see refs. 23 and 24 for reviews). In a previous study (1), we showed that human centrin is present in the distal lumen of centrioles, but that a vast portion is not centrosome-associated. Injection of heterologous centrin in two-cell stage xenopus embryos led to undercleavage suggesting an essential function of animal centrin during the cell division cycle as was found in S. cerevisiae. However, centrosome duplication did not appear to be affected, as many asters of microtubules were observed in undercleaved blastomeres, which were probably organized by centrosomes (1).
In our previous study, human centrin was characterized using a mAb raised against C. reinhardtii centrin (CrCenp) that recognized several isoforms of centrin in two-dimensional electrophoresis. Here we report that one of these isoforms, which is particularly enriched in the centrosome, is specifically recognized by antibodies (Abs) raised against Cdc31p from S. cerevisiae. This acidic, slow-migrating isoform is encoded by a new gene, which we propose to name HsCEN3. In contrast to the two already known human centrin genes, HsCEN3 is more closely related to ScCDC31 (S. cerevisiae cell division cycle gene 31) than to CrCEN (C. reinhardtii centrin gene).
MATERIALS AND METHODS
Cell Culture.
The KE37 human T lymphoblastic cell line was grown in RPMI 1640 medium containing 7% fetal calf serum, and HeLa cells were maintained in DMEM supplemented with 10% fetal calf serum. Both cell lines were kept at 37°C in a humid atmosphere containing 5% CO2.
Antibodies.
Anti-CrCenp mAbs 20H5 and 11B2 were a generous gift from J. L. Salisbury (Mayo Clinic, Rochester, MN). Anti-Cdc31p polyclonal Abs were raised in rabbit and goat against a bacterially expressed glutathione S-transferase (GST)–Cdc31p fusion protein. Both Abs were affinity purified on Cdc31p (15). mAb CTR 453 recognizes a human 350-kDa pericentriolar material (PCM) antigen (25). Anti α-tubulin was purchased from Amersham.
Immunofluorescence Microscopy.
HeLa cells were grown on coverslips, washed in PBS, and fixed in methanol at −20°C for 7 min. Cells were then rinsed in PBS containing 0.1% Tween 20. Primary Abs, diluted in PBS–Tween containing 3% BSA, were added for 30 min at room temperature. Cells were then rinsed in PBS–Tween. The same procedures were used for fluorescein or rhodamine-labeled secondary Abs (Jackson ImmunoResearch). Cells were finally dehydrated in ethanol and mounted in Citifluor (City University, London).
Cellular Fractionation.
Soluble and insoluble protein fractions from KE37 cells were prepared as described (1). Centrosomes were isolated from KE 37 cells as described (26). The 15,000 × g Xenopus interphasic egg extracts were prepared according to Murray (27). Protein fraction from fertilized starfish eggs was a generous gift from André Picard (Laboratoire Arago, Banyuls, France).
Protein Analysis.
One-dimensional SDS/PAGE was performed according to Laemmli (28) using 12% polyacrylamide gels. Two-dimensional electrophoresis was done according to O’Farrell (29). Immunoblotting experiments were performed according to the protocol of Towbin et al. (30), as modified by Van Eldik and Wolchok (31). Briefly, proteins were fixed after transfer on nitrocellulose filter by incubation with 0,2% glutaraldehyde in TBS (10 mM Tris, pH 7.4/150 mM NaCl) for 15 min at room temperature. The nitrocellulose filter was washed in distilled water and saturated in TBS containing 5% nonfat dry milk during 1 h at 37°C before the incubation with Abs.
Phosphatase alkaline-conjugated secondary Abs were purchased from Promega. Biotin-conjugated secondary Abs and phosphatase alkaline-conjugated streptavidin were purchased from Amersham. Peroxydase-conjugated secondary Abs were purchased from Jackson ImmunoResearch. For anti-Cdc31p goat Ab, we used an unlabeled anti-goat secondary Ab raised in rabbit and peroxydase-coupled protein A (Zymed).
Database Search and Cloning of HsCEN3 and MmCEN3.
CDC31-related sequences were searched in dbest using the default parameters of the blastn program (GenomNet, Tokyo). Primers derived from human and mouse expressed sequence tags (ESTs) matching CDC31 were used to clone the 5′ and 3′ cDNA ends by RACE–PCR (5′ and 3′ RACE–PCR kit, GIBCO/BRL) according to the manufacturer’s instructions. Briefly, poly(A) mRNA obtained from HeLa cells, or from mouse spleen, was reverse transcribed using a gene specific primer or an oligo(dT) primer, respectively, to obtain the 5′ and 3′ ends of the genes. Then, single-stranded cDNAs corresponding to 5′ ends were elongated with a poly(C) tail and amplified by PCR using a gene-specific primer and a poly(G) primer. cDNAs corresponding to 3′ ends were directly amplified by PCR using a gene-specific primer and a poly(T) primer. Finally, PCR products were submitted to a second round of PCR using a nested gene-specific primer. cDNAs were sequenced by genomexpress (Grenoble, France). We used two nested sets of primers localized upstream of the start codon and downstream of the stop codon to obtain full-length cDNAs by reverse transcription–PCR and to check the sequence.
Recombinant Centrin Production and Purification.
Recombinant HsCen1p and HsCen2p were produced and purified as described (1). Bacterially expressed and purified CrCenp was a generous gift from J. L. Salisbury. Human purified calmodulin was obtained from Sigma.
HsCEN3 cDNA was cloned in pET3d bacterial expression vector, and the vector introduced into BL21 DE3 bacteria strain. Protein expression was induced by 10 mM isopropyl β-d-thiogalactoside for 3 h. Bacteria were pelleted by centrifugation, lysed by boiling in Laemmli buffer, and then followed by sonication.
To purify the protein from bacterial lysate, a six histidine-tag was introduced by PCR at the carboxyl terminus of the cDNA. After cloning in pET 3d vector, the recombinant protein was expressed in BL21DE3 bacteria strain and purified on nickel-chelated agarose beads according to the manufacturer’s instruction (NI2+-NTA; Qiagen, Chatsworth, CA).
RESULTS
Anti-Cdc31p Abs Recognize a 23-kDa Protein in Human Cells.
Specific anti-Cdc31p Abs were obtained by affinity-purification on recombinant Cdc31p (anti-Cdc31p Abs). These IgGs were used to test three fractions from human lymphoblasts: Triton X-100-soluble fractions (S), which contain cytosolic proteins and most of the membrane proteins, insoluble fractions (I), which contain most of the chromatin and cytoskeleton proteins, and isolated centrosome fractions (Ctr). Anti-Cdc31p Abs were found to react with a single band of 23 kDa (Fig. 1A Upper Right), which was highly enriched in the centrosome fraction in comparison to S and I.
Figure 1.
Specificity of anti-Cdc31p Abs in human cells. (A Upper) Western blot analysis of low-speed Triton X-100 soluble (S) and insoluble (I) protein fractions and of a centrosome-enriched fraction (Ctr) obtained from human lymphoblastic KE37 cells, using the 20H5 mAb (Left) and the rabbit anti-Cdc31p Ab (Right). Both halves correspond to the same nitrocellulose filter. The Ctr lane has been split in two. (Lower) Same analysis as in A using another monoclonal anti-CrCenp Ab (11B2 mAb) and goat polyclonal anti-Cdc31p Abs. Note that the 23-kDa band recognized by anti-Cdc31p Abs corresponds to a minor band in both 20H5 mAb and mAb 11B2 signals (arrows). Proteins corresponding to 5 × 105 (Upper) and 2 × 105 (Lower) cells were loaded on each soluble and insoluble lanes. Centrosome lanes correspond to 5 × 107 and 3 × 107 centrosomes, respectively. (B) Western blot analysis of the Ctr fraction by two-dimensional electrophoresis. The same blot was first incubated with 20H5 mAb, and the signal was detected using a biotin-conjugated anti-mouse secondary Ab and alkaline phosphatase-conjugated streptavidin (Left). The blot was then incubated with the rabbit anti-Cdc31p Ab and revealed by a peroxydase-coupled anti-rabbit secondary Ab (Right). The blot is oriented with the acidic pIs on the left. Note that a single spot is identified by the anti-Cdc31p Ab, which corresponds to the isoform having the higher molecular weight and the more acidic pI in the 20H5 mAb pattern (arrows). The other spots previously stained by 20H5 mAb are seen in negative, enabling one to position accurately one pattern with respect to the other. The same result is obtained if anti-Cdc31p Abs are used first (data not shown). Proteins from 108 centrosomes were loaded. (C) Specificity of a monoclonal anti-CrCen Ab (20H5 mAb, Left) and of rabbit polyclonal anti-Cdc31p Abs (Right) on purified Cdc31p (lane 1), HsCen1p (lane 2), HsCen2p (lane 3), CrCenp (lane 4), and purified human calmodulin (lane 5). Protein (10 ng) was loaded on each lane. (D) Specificity of a monoclonal anti-CrCen Ab (20H5 mAb, Left) and of rabbit polyclonal anti-Cdc31p Abs (Right) on bacterially produced HsCen1p (lane 1), HsCen2p (lane 2), HsCen3p (lane 3), and histidine tagged-HsCen3p (lane 4). Similar amount of purified HsCen1p, HsCen2p, and of HsCen3p from a bacterial lysate (as estimated by Coomassie blue staining) were loaded on each lane, whereas a 10-fold amount of purified histidine-tagged purified HsCen3p was loaded. Arrowheads point to HsCen3p. Positions of molecular weights (31, 21, and 14 kDa) are indicated on the left-hand side of the blots.
The anti-Cdc31p signal was compared with that obtained with a mAb 20H5 raised against CrCenp that reacts with human centrin (1). The 23-kDa anti-Cdc31p signal did not correspond to the major signal obtained with 20H5 mAb. Instead, it correlated with a minor 20H5 mAb signal, which was only observed in the Ctr fraction, as demonstrated in Fig. 1A Upper. Similar results were obtained using the anti-CrCenp 11B2 mAb and affinity-purified goat anti-Cdc31p Abs (Fig. 1A Lower).
The 23-kDa Protein Recognized by Anti-Cdc31p Abs Corresponds to an Acidic, Slow-Migrating Isoform of Centrin.
To ascertain that the 23-kDa band stained with anti-CrCenp Abs and anti-Cdc31p Abs corresponded to the same protein, we used two-dimensional electrophoresis separation of centrosomal proteins. Anti-Cdc31p and anti-CrCenp Abs were tested on the same blot to precisely compare both staining patterns (Fig. 1B). The blot was first incubated with 20H5 mAb. A complex centrin pattern was obtained as described (1). The blot was then incubated with the rabbit anti-Cdc31p Ab. A single spot was detected, corresponding to the centrin isoform having the higher molecular weight and the more acidic pI. This demonstrated that the 23-kDa band obtained with the anti-Cdc31p Abs and with the anti-CrCenp Abs corresponded to the same isoform of centrin.
Anti-Cdc31p Abs Do Not Cross-React with HsCEN1 and HsCEN2 Gene Products.
We next compared the specificity of anti-Cdc31p Abs to that of the anti-CrCenp 20H5 mAb on recombinant proteins CrCen, ScCdc31, HsCen1 and HsCen2 and on human calmodulin. The 20H5 mAb was found to react with CrCenp and with human centrin but not with ScCdc31p (Fig. 1C). In contrast, anti-Cdc31p Abs only recognized ScCdc31p. As expected, neither Ab recognized human calmodulin (lane 5), showing the strict specificity of both Abs for members of the centrin family. All together, these data suggest that the acidic, slow migrating isoform of centrin specifically recognized by anti-Cdc31p Abs might be encoded by an as yet uncharacterized centrin gene, more closely related to the yeast CDC31 gene.
Human and Mouse Cells Contain an Additional Centrin Gene.
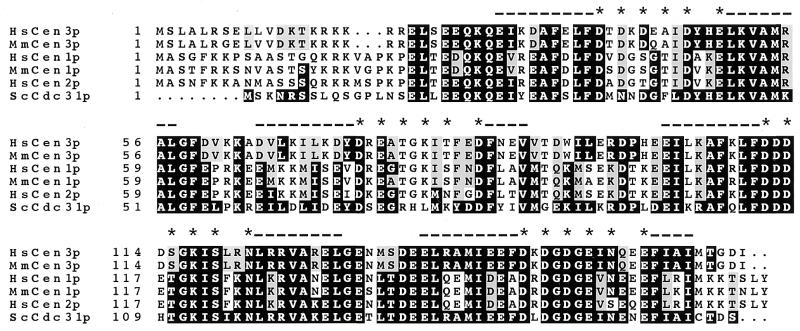
We were interested in determining whether a third human centrin gene, closer to ScCDC31 than HsCEN1 and HsCEN2, exists. Whereas all initial attempts to clone such a gene from human cDNA libraries using PCR, hybridization or Ab screening proved unsuccessful, a search for ScCDC31-related sequences in databases of ESTs has revealed human, mouse and rat ESTs showing high conservation with ScCDC31 gene (accession nos. AA031510, AA033392, and H33906). As these clones represented incomplete cDNAs, we cloned the corresponding human and murine full-length cDNAs by reverse transcription–PCR. Two successive rounds of PCR using nested primers were necessary to obtain 500 ng of product, suggesting a low abundance of the mRNAs. Human and murine cDNAs, which we named HsCEN3 and MmCEN3 (Mus musculus centrin gene 3), respectively (accession nos. Y12473 and Y12474), encode identical proteins except for 3 amino acid residues out of 167 (Fig. 2). These proteins have a calculated molecular weight of 19.5 kDa and a pI of 4.6. HsCen3p shares 54% identity and 75% similarity with both HsCen1p and HsCen2p. It is closer to ScCdc31p than HsCen1p and HsCen2p (Table 1). Conversely, HsCen1p and HsCen2p are closer to CrCenp than HsCen3p.
Figure 2.
Predicted protein sequences for HsCen3 and MmCen3. The two new sequences are compared with ScCdc31p and to the known centrin proteins from both human and mouse origin. Alignement was performed using the pileup program (Infobiogen, Villejuif, France). Comparison to ScCdc31p was performed using the boxshade program (Institut Suisse de Recherches Experimentales sur le Cancer, Lausanne, Switzerland). Amino acids identical to ScCdc31p and present in at least half of the sequences are boxed in black. Conservative changes are boxed in grey. The position of the four EF-hand domains is indicated by dashes (position of the helix) and stars (conserved positions of the loops or fixation pockets). The pileup program from Infobiogen aligned the new sequences with the known human and murine centrin sequences by creating a gap of 3 amino acids between the NH2 terminus and the conserved central part of the sequence. This suggests that a deletion or an addition in the amino terminus appeared when these genes diverged. The default parameters of the programs were used in all cases. The GenBank accession numbers for HsCen3 and MmCen3 are Y12473 and Y12474, respectively.
Table 1.
Sequence comparison of mammalian centrin proteins with Cdc31p from S. cerevisiae and with C. reinhardtii centrin
| HsCen1 | HsCen2 | MmCen3 | MmCen1 | ScCdc31 | CrCen | |
|---|---|---|---|---|---|---|
| HsCen3 | 54/75 | 54/75 | 98/100 | 55/76 | 59/76 | 51/73 |
| HsCen1 | 100 | 84/92 | 54/75 | 91/94 | 54/68 | 68/85 |
| HsCen2 | 100 | 54/75 | 82/92 | 56/66 | 71/85 | |
| MmCen3 | 100 | 54/74 | 58/76 | 51/73 | ||
| MmCen1 | 100 | 53/68 | 65/83 | |||
| ScCdc31 | 100 | 50/66 |
Percentages of identity/similarity have been calculated using the besfit program (Infobiogen).
Noteworthy, special features characteristic of the yeast Cdc31p are present in HsCen3p, such as a tyrosine in the first and the second EF-hand domains, a proline in the central region between the second and the third EF-hand domains, and a stretch of 12 amino acids (from residues 136 to 147) in the fourth and most conserved EF-hand domain. The amino-terminal region is divergent: it is five residues longer than that of the yeast Cdc31p and three residues shorter than those of HsCen1p and HsCen2p, but highly positively charged as in classical centrin (see ref. 18). Functional calmodulin homologs from human and yeast (32) show a conservation similar to the conservation between ScCdc31p and HsCen3p/MmCen3p (60% identity compared with 59%).
HsCEN3 Encodes a Protein with an Apparent Molecular Weight of 23 kDa That Reacts with Anti-Cdc31p Abs.
Recombinant HsCen3p and 6His-tagged HsCen3p were produced in bacteria and assessed for reactivity with anti-Cdc31p Abs and anti-CrCenp mAb. The two proteins of apparent molecular weights of 23 kDa and 25 kDa, respectively, were recognized by both Abs (Fig. 1D). These Ab-reactivity and migration properties of HsCen3p demonstrate that HsCen3p corresponds to the acidic, slow migrating isoform of centrin previously described.
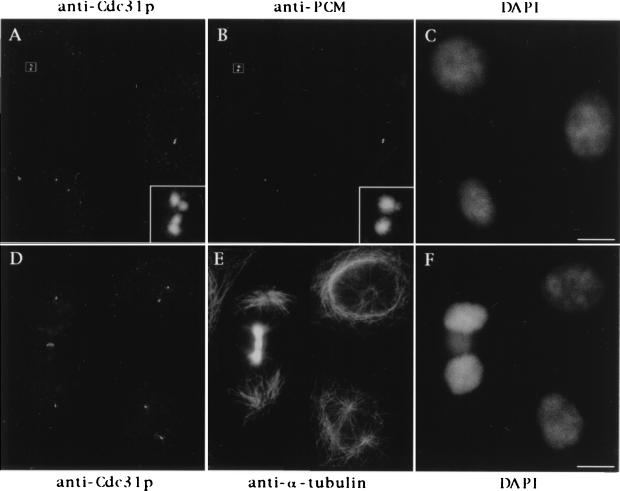
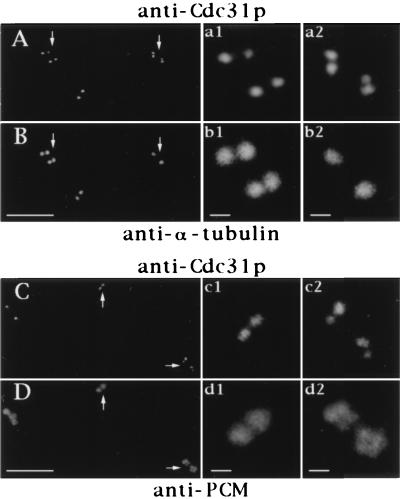
We wondered whether HsCen3p could display a distribution distinct from that of the other centrin isoforms previously described in human cells (1). This question was addressed using the HsCen3p-specific anti-Cdc31p Abs. We found that the staining obtained with the rabbit anti-Cdc31p Abs was quite similar to general centrin staining obtained previously with 20H5 mAb (1): in HeLa cells, one or two pairs of dots corresponding to the centrioles were observed (Fig. 3). On centrosomes isolated from human lymphoblastic KE37 cells, anti-Cdc31p Ab staining was more discrete than tubulin decoration of centrioles, showing sometimes two pairs of very closely associated dots whereas corresponding tubulin staining showed only two dots (Fig. 4). This suggested that the growing procentrioles could be stained by anti-Cdc31p Abs and could be discriminated from parental centriole staining earlier than with anti-tubulin Abs. The presence of HsCen3p in the distal lumen of centrioles and in growing centriole buds was further demonstrated at the electron microscopy level (data not shown).
Figure 3.
Cellular distribution of HsCen3p. Cells were triple stained with rabbit anti-Cdc31p Abs (A), an anti-PCM mAb (CTR 453, B), and 4′,6-diamidino-2-phenylindole (DAPI) (C), or with rabbit anti-Cdc31p Abs (D), an anti-α-tubulin mAb (E), and DAPI (F). A 6-fold magnification of the duplicated centrosome framed in A and B is shown on the bottom right of A and B. Note that the two distinct pairs of closely associated dots stained with anti-Cdc31p Abs correspond to one pair of duplicated centrioles. Note that the midbody joining the two sister-cells shown in D–F is slightly stained with the anti-Cdc31p Ab. (Bars = 10 μm.)
Figure 4.
Localization of HsCen3p in isolated centrosomes. Centrosomes isolated from KE 37 cells were double-labeled with rabbit anti-Cdc31p Abs (A) and an anti-α-tubulin mAb (B) or with rabbit anti-Cdc31p Abs (C) and an anti-PCM mAb (CTR453, D). Four-fold magnification of centrosomes pointed out by arrows are shown on the right side of the figure (a1–b1, a2–b2). Given the distances between the centers of the dots obtained with anti-Cdc31p Abs, the pair figure a1–b1 may show two G1 centrosomes with disoriented centrioles, whereas the pair a2–b2 may represent a duplicating centrosome containing not fully grown daughter centrioles. Note that in this case, centrioles and their corresponding procentrioles are not resolved by the tubulin staining whereas they are clearly resolved by the anti-Cdc31p Abs staining. In C and D, the c1–d1 magnified pair pictures correspond to a centrosome that has not yet initiated centriole duplication according to centrin staining whereas duplication is largely engaged in the centrosome shown on c2–d2 pair pictures. (Bars = 5 μm in A–D and 0.5 μm in high magnification windows.)
Anti-Cdc31p Abs React with a Slow-Migrating Isoform of Centrin in Amphibians and Echinoderms.
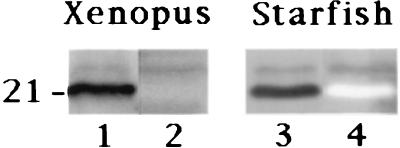
We wondered whether Cdc31-related proteins were also present in other animal species. We first looked at Xenopus XR cells, and observed that anti-Cdc31p Abs specifically decorated centrosomes (data not shown). When used on protein fractions from low-speed interphasic Xenopus egg extracts, 20H5 mAb reacted with a major band at 21 kDa and a minor one at ≈23 kDa, whereas anti-Cdc31p Abs reacted with the minor, slow-migrating band only, as in human cells (Fig. 5).
Figure 5.
Analysis of centrin in Xenopus and starfish eggs by Western blot analysis. Total proteins from low speed Xenopus egg extracts (Left) and from starfish fertilized eggs (Right) were submitted to SDS/PAGE on a 12% polyacrylamide gel. For Xenopus, two different lanes were incubated with 20H5 mAb (lane 1) and anti-Cdc31p Abs (lane 2), using biotin-conjugated anti-mouse and anti-rabbit secondary Abs, respectively. For starfish, the same lane was incubated first with 20H5 mAb (lane 3), and the detection performed as described, and second with rabbit anti-Cdc31p Abs (lane 4), using peroxydase-conjugated anti-rabbit secondary Abs. The slow-migrating band in the 20H5 pattern is specifically recognized by anti-Cdc31p Abs in Xenopus and starfish.
We next investigated for the presence of centrin in echinoderms by Western blot analysis. Anti-CrCenp 20H5 mAb and anti-Cdc31p Abs were successively probed on a protein fraction from fertilized starfish eggs. The 20H5 mAb was found to react with two bands, a major one at 20 kDa and a minor one at ≈22 kDa (Fig. 5). Anti-Cdc31p Abs reacted with the slow-migrating minor band only (Fig. 5), in a manner quite reminiscent of the pattern seen in human cells and Xenopus egg extracts. Similar results were obtained in sea urchin egg extracts (data not shown). Altogether, these data suggest that a ScCDC31-related gene may also exist in amphibians and echinoderms.
DISCUSSION
Biochemical analysis of centrin in human cells has unraveled an unexpected complex picture: as many as 10 isoforms can be identified by two-dimensional gel electrophoresis of proteins from human lymphoblastic cells (1). Furthermore, cell fractionation has revealed two important features: first, in marked contrast with what could be inferred from immunofluorescence experiments, most of centrin is not centrosome-associated. Second, certain centrin isoforms are enriched in centrosomes (1). The most enriched isoform displays a slightly higher apparent molecular weight than the major centrin isoforms (23 kDa instead of 20 kDa) and a more acidic pI. Centrin profile was not observed to shift in SDS/PAGE depending on free calcium concentration (data not shown).
The 23-kDa isoform was the only one to react with two independent Abs raised against ScCdc31p (Fig. 1A). These two polyclonal anti-Cdc31p were produced against unmodified, bacterially produced Cdc31p and did not recognize bacterially produced HsCen1 or HsCen2 protein (Fig. 1C). Thus we hypothesized that the 23 kDa was encoded by an as yet uncharacterized gene sharing more similarity with the ScCDC31 gene than HsCEN1 or HsCEN2. The existence of human, mouse, and rat ESTs showing high conservation with ScCDC31 supported our hypothesis and allowed us to obtain the full sequence of a new human and a new murine centrin gene, which we name HsCEN3 and MmCEN3. The human and the mouse proteins are almost identical with two conservative changes in the NH2 terminus and a change of a glutamine for a glutamic acid in the first Ca2+-binding domain. A rat EST predicts a sequence identical to that of HsCen3p between residues 20 and 75. HsCen3p and MmCen3p demonstrate a higher interspecies conservation than HsCen1p and MmCen1p (see Table 1). The conservation between HsCen2p and MmCen2p cannot be estimated as MmCEN2 is only known so far as an EST.
Several features which distinguish ScCdc31p from all other centrin sequences are found in HsCen3p and MmCen3p (see Fig. 2). The central region, for example, is divergent from that of HsCen1 and -2, and contains a proline as in ScCdc31p. The fourth EF-hand domain is clearly divergent among the two groups of sequences. Furthermore this EF-hand domain is predicted to be the only functional one in HsCen3p (using the scanprosite program located at expasy). This is to be expected since this EF-hand is nearly identical to the fourth EF-hand of ScCdc31p which has been shown to bind Ca2+ (33). Finally, the COOH terminus of HsCen3 and MmCen3 is short, as in ScCdc31p, lacking the final LY motif typical of classical centrin sequences (18). Why the protein migrates with an apparent molecular weight higher than the other centrin proteins is not clear.
The comparison of mammalian centrin proteins with ScCdc31p suggests the existence of two distinct groups of sequences, one containing the three already known mammalian centrin sequences, and the other, closer to ScCdc31 sequence, containing the two new mammalian centrin sequences (Fig. 2 and Table 1). The existence of these two groups is even more obvious when mammalian centrin sequences and ScCdc31p are all compared with CrCenp (Table 1): HsCen3p, MmCen3p, and ScCdc31p show only 50–51% identity (66–73% similarity) to CrCenp whereas HsCen1p, HsCen2p, and MmCen1p show between 65 and 71% identity (83–85% similarity). These data strongly suggest the existence of two divergent subfamilies.
The possibility that Cdc31p could define a subfamily distinct from that of centrin has already been pointed out (18, 34). The recent identification of a new centrin sequence in Giardia intestina (accession number U59300) that is divergent from the published centrin sequence of Giardia lamblia (16) further supports this view. Compared with CrCenp, G. lamblia centrin is 57% identical (74% similar) whereas G. intestina centrin is 67% identical (87% similar).
The existence of two distinct subfamilies could help understanding why centrin has been involved in contractile functions in green algae, whereas Cdc31p functions in spindle pole body duplication in S. cerevisiae. Centrin from C. reinhardtii and S. dubia cannot complement CDC31 mutations in yeast (35, 36), suggesting that algae centrin and Cdc31p are not functional homologs. We thus propose that two divergent subfamilies of centrin proteins have evolved possibly due to their implication into distinct centrosome-associated functions. One family, to which CrCenp belongs, would form calcium-dependent contractile structures associated with centrosomes, whereas the other family, to which ScCdc31p belongs, would be involved in centrosome biogenesis.
From our observation that anti-Cdc31p Abs specifically recognize a slow-migrating isoform of centrin in xenopus and starfish as in human cells, and from the existence of two divergent genes in Giardia, we would conjecture that all eukaryotes possess the two types of genes. However the complete sequence of the S. cerevisiae genome does not contain any other centrin gene than CDC31; but this organism also has no flagellar apparatus or centrosome-associated contractile structures. Therefore, in certain organisms, one of the two genes could be lost secondarily due to the loss of function associated with it. In this respect, it would be interesting to know more on the centrin genes in plants, as they have no centrosome.
Do green algae also possess two centrin genes? In the case of C. reinhardtii, the existence of a unique centrin gene has been assumed as only two or three isoforms of centrin, corresponding to unmodified, phosphorylated or hyperphosphorylated products of centrin gene were detected (4, 37). However, the immunological reactivity of ScCdc31p-like proteins with anti-CrCenp Abs is low. This could explain why a ScCdc31p-related protein has not been detected in C. reinhardtii, especially if it is not as abundant as CrCenp. It is noteworthy that in the vfl2 mutant of C. reinhardtii, the mutated protein cannot participate in the formation of the nucleus–basal body connector (5, 8). Yet, centrin Abs decorate basal bodies. This could mean that the mutated protein is still able to associate with basal bodies. Alternatively, it could correspond to the product of a CDC31-related gene, specifically concentrated in basal bodies, where it would participate in the duplication of the flagellar apparatus.
Higher constraint on the CEN3 gene could suggest that the function of its product requires interactions with numerous components, precluding the possibility of compensatory mutations to occur. It could reflect for example a tightly conserved function in the centrosome duplication pathway. If the CEN3 gene is demonstrated to be really involved in centrosome duplication, it should be rewarding to identify interacting components. This might provide a first step toward understanding the remarkable lack of species barrier of the centrosome-induced parthenogenetic development of Xenopus (for a review, see 38).
Cellular localization of HsCen3p could, in addition to gene conservation, represent a criterium for its specific involvement in centrosome duplication. HsCen3p likely represents the most abundant centrosomal centrin isoform: a 10-fold increase in the amount of recombinant HsCen3p was necessary to obtain a mAb20H5 signal comparable to that of HsCen1p and HsCen2p (Fig. 1B compared with Fig. 1D). Its localization in distal lumen of centrioles is similar to that of other centrin isoforms. However, a thorough ultrastructural analysis will be necessary to accurately determine the mutual localization of the three centrin isoforms during centrosome biogenesis.
A comprehensive view on centrin functions has remained difficult until now. Characterization of a new centrin gene in mammals that encodes a protein closer to ScCdc31p than the known mammalian centrin genes may radically change the situation. Functional studies, currently undertaken both in yeast and in animal cells, should allow us to determine whether HsCEN3 is the functional homolog of ScCDC31, and whether it participates in centrosome duplication. A thorough analysis of the phylogenetic evolution of these genes should also tell when the Cdc31p-like subfamily and the CrCenp-like subfamily have diverged. This might be important for understanding the evolution of the centrosome.
Acknowledgments
We thank J. L. Salisbury for providing us with anti-C. reinhardtii centrin Abs and CrCenp, A. Picard for starfish proteins, S. Holmes for valuable help in identifying a protein reacting with anti-Cdc31p Abs in Xenopus extract, C. Celati and A. M. Tassin for purification of bacterially expressed His-tagged HsCenp3, and Yves Blouquit for purification of bacterially expressed HsCen1p and HsCen2p. We thank Daniel Meur for the art work. We also thank Hervé Philippe for valuable discussion on the phylogeny of calmodulin and centrin, and E. Bailly, V. Doye, S. Holmes, T. Küntziger and A. M. Tassin for critical reading of the manuscript. The work was supported by Centre National de la Recherche Scientifique, by an European Economic Community Grant HCP CHRX CT 94-0642 to M.B. and E.S., and by Deutsche Forschungsgemeinschaft Grant Schi295/2-2 to E.S. A.P. received fellowships from Ministère de l’Enseignement Supérieur et de la Recherche and Fondation pour la Recherche Médicale. S.M. received fellowships from Ministère de l’Enseignement Supérieur et de la Recherche and from Association pour la recherche sur le cancer.
ABBREVIATIONS
- Ab
antibody
- ScCDC31
Saccharomyces cerevisiae cell division cycle gene 31
- CrCEN
Chlamydomonas reinhardtii centrin gene
- EST
expressed sequence tags
- HsCEN1
-2, -3, Homo sapiens centrin genes 1, 2, and 3
- MmCEN1 and -3
Mus musculus centrin genes 1 and 3
- PCM
pericentriolar material
Footnotes
References
- 1.Paoletti A, Moudjou M, Paintrand M, Salisbury J L, Bornens M. J Cell Sci. 1996;109:3089–3102. doi: 10.1242/jcs.109.13.3089. [DOI] [PubMed] [Google Scholar]
- 2.Nakayama S, Moncrief N D, Kretzinger R H. J Mol Evol. 1992;34:416–448. doi: 10.1007/BF00162998. [DOI] [PubMed] [Google Scholar]
- 3.Bhattacharya D, Steinkotter J, Melkonian M. Plant Mol Biol. 1993;23:1243–1254. doi: 10.1007/BF00042357. [DOI] [PubMed] [Google Scholar]
- 4.Salisbury J L, Baron A, Surek B, Melkonian M. J Cell Biol. 1984;99:962–970. doi: 10.1083/jcb.99.3.962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wright R L, Salisbury J, Jarvik J W. J Cell Biol. 1985;101:1903–1912. doi: 10.1083/jcb.101.5.1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mc Fadden G I, Schulze D, Surek B, Salisbury J L, Melkonian M. J Cell Biol. 1987;105:903–912. doi: 10.1083/jcb.105.2.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wright R L, Adler S A, Spanier J G, Jarvik J W. Cell Motil Cytoskeleton. 1989;14:516–526. doi: 10.1002/cm.970140409. [DOI] [PubMed] [Google Scholar]
- 8.Taillon B E, Adler S A, Suhan J P, Jarvik J W. J Cell Biol. 1992;119:1613–1624. doi: 10.1083/jcb.119.6.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sanders M A, Salisbury J L. J Cell Biol. 1989;108:1751–1760. doi: 10.1083/jcb.108.5.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sanders M A, Salisbury J L. J Cell Biol. 1994;124:795–805. doi: 10.1083/jcb.124.5.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang B, Mengersen A, Lee V D. J Cell Biol. 1988;107:133–140. doi: 10.1083/jcb.107.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Byers B. In: The Molecular Biology of the Yeast Saccharomyces: Life Cycle and Inheritance. Strathern J N, Jones E W, Broach J R, editors. Plainview, NY: Cold Sping Harbor Lab. Press; 1981. pp. 59–96. [Google Scholar]
- 13.Schild D, Ananthaswany H N, Mortimer R K. Genetics. 1981;97:551–562. doi: 10.1093/genetics/97.3-4.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baum P, Furlong C, Byers B. Proc Natl Acad Sci USA. 1986;83:5512–5516. doi: 10.1073/pnas.83.15.5512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Spang A, Courtney I, Fackler U, Matzner M, Schiebel E. J Cell Biol. 1993;123:405–416. doi: 10.1083/jcb.123.2.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meng T-C, Aley S B, Svard S G, Smith M W, Huang B, Kim J, Gillin F D. Mol Biochem Parasitol. 1996;79:103–108. doi: 10.1016/0166-6851(96)02636-9. [DOI] [PubMed] [Google Scholar]
- 17.Madeddu L, Klotz C, Le Caer J P, Beisson J. Eur J Biochem. 1996;238:121–128. doi: 10.1111/j.1432-1033.1996.0121q.x. [DOI] [PubMed] [Google Scholar]
- 18.Levy Y Y, Lai E Y, Remillard S P, Heinzelman M B, Fulton C. Cell Motil Cytoskeleton. 1996;33:298–323. doi: 10.1002/(SICI)1097-0169(1996)33:4<298::AID-CM6>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 19.Zhu J-K, Bressan R A, Hasegawa P M. Plant Physiol. 1992;99:1734–1735. doi: 10.1104/pp.99.4.1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ogawa K, Shimizu T. Biochim Biophys Acta. 1993;1216:126–128. doi: 10.1016/0167-4781(93)90048-i. [DOI] [PubMed] [Google Scholar]
- 21.Lee V D, Huang B. Proc Natl Acad Sci USA. 1993;90:11039–11043. doi: 10.1073/pnas.90.23.11039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Errabolu R, Sanders M A, Salisbury J L. J Cell Sci. 1994;107:9–16. doi: 10.1242/jcs.107.1.9. [DOI] [PubMed] [Google Scholar]
- 23.Salisbury J L. Curr Opin Cell Biol. 1995;7:39–45. doi: 10.1016/0955-0674(95)80043-3. [DOI] [PubMed] [Google Scholar]
- 24.Schiebel E, Bornens M. Trends Cell Biol. 1995;5:197–201. doi: 10.1016/s0962-8924(00)88999-0. [DOI] [PubMed] [Google Scholar]
- 25.Bailly E, Dorée M, Nurse P, Bornens M. EMBO J. 1989;8:3985–3995. doi: 10.1002/j.1460-2075.1989.tb08581.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moudjou M, Bornens M. In: Cell Biology: A Laboratory Handbook. Celis J E, editor. San Diego: Academic; 1994. pp. 595–604. [Google Scholar]
- 27.Murray A W. Methods Cell Biol. 1991;36:581–605. [PubMed] [Google Scholar]
- 28.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 29.O’Farrell P H. J Biol Chem. 1975;250:4007–4012. [PMC free article] [PubMed] [Google Scholar]
- 30.Towbin H, Staehelin T, Gordon J. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Van Eldik L J, Wolchok S R. Biophys Biochem Res Commun. 1984;124:752–759. doi: 10.1016/0006-291x(84)91022-2. [DOI] [PubMed] [Google Scholar]
- 32.Davis T N, Thorner J. Proc Natl Acad Sci USA. 1989;86:7909–7913. doi: 10.1073/pnas.86.20.7909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Geier B M, Wiech H, Schiebel E. J Biol Chem. 1996;271:28366–28374. doi: 10.1074/jbc.271.45.28366. [DOI] [PubMed] [Google Scholar]
- 34.Moncrief N D, Kretzinger R H, Goodman M. J Mol Evol. 1990;30:522–562. doi: 10.1007/BF02101108. [DOI] [PubMed] [Google Scholar]
- 35.Lee V D, Stapleton M, Huang B. J Cell Biol. 1989;109:118a. (abstr.). [Google Scholar]
- 36.Wiech H, Geier B M, Paschke T, Spang A, Grein K, Steinkotter J, Melkonian M, Schiebel E. J Biol Chem. 1996;271:22453–22461. doi: 10.1074/jbc.271.37.22453. [DOI] [PubMed] [Google Scholar]
- 37.Martindale V E, Salisbury J L. J Cell Sci. 1990;96:395–402. doi: 10.1242/jcs.96.3.395. [DOI] [PubMed] [Google Scholar]
- 38.Tournier F, Bornens M. In: Microtubules. Hyams J S, Llyod C W, editors. New York: Wiley; 1994. pp. 303–324. [Google Scholar]