Abstract
We have developed a technique, methylation-specific PCR in situ hybridization (MSP-ISH), which allows for the methylation status of specific DNA sequences to be visualized in individual cells. We use MSP-ISH to monitor the timing and consequences of aberrant hypermethylation of the p16 tumor suppresser gene during the progression of cancers of the lung and cervix. Hypermethylation of p16 was localized only to the neoplastic cells in both in situ lesions and invasive cancers, and was associated with loss of p16 protein expression. MSP-ISH allowed us to dissect the surprising finding that p16 hypermethylation occurs in cervical carcinoma. This tumor is associated with infection of the oncogenic human papillomavirus, which expresses a protein, E7, that inactivates the retinoblastoma (Rb) protein. Thus, simultaneous Rb and p16 inactivation would not be needed to abrogate the critical cyclin D–Rb pathway. MSP-ISH reveals that p16 hypermethylation occurs heterogeneously within early cervical tumor cell populations that are separate from those expressing viral E7 transcripts. In advanced cervical cancers, the majority of cells have a hypermethylated p16, lack p16 protein, but no longer express E7. These data suggest that p16 inactivation is selected as the most effective mechanism of blocking the cyclin D–Rb pathway during the evolution of an invasive cancer from precursor lesions. These studies demonstrate that MSP-ISH is a powerful approach for studying the dynamics of aberrant methylation of critical tumor suppressor genes during tumor evolution.
During recent years, there has been a growing interest in how patterns of DNA cytosine methylation in gene promoter regions, particularly for those genes containing CpG-rich promoters or CpG islands, play a role in the regulation of gene expression. Although these islands are normally unmethylated for most genes, hypermethylation is associated with loss of expression of one copy in the normal settings of inactivation of the female X chromosome and the silenced alleles for parentally imprinted genes (1, 2). Loss of function of key genes during human tumorigenesis can occur in association with aberrant hypermethylation (3). For example, loss of expression of the tumor suppresser genes VHL in sporadic renal cancer (4), p16 in multiple tumor types (5, 6), and hMLH1 in colorectal cancer (7–9) has been correlated with hypermethylation of the corresponding promoter, and this serves as an alternative mechanism for loss of tumor suppressor gene function.
The development of sensitive PCR techniques and genomic sequencing to assess cytosine methylation status has facilitated studies of CpG island methylation (10–12). However, these procedures can be performed only with DNA extracted from total cell populations. These methods cannot easily address critical issues, such as the precise timing of DNA methylation changes in specific cell types during embryonic development and oncogenesis, or define the extent of heterogeneity for loss of expression during tumor evolution. Understanding such temporal dynamics in tumor progression would provide important insights into neoplastic development and foster new approaches to the diagnosis and management of cancer.
In the present report, we describe a modification of a previously described PCR procedure, methylation-specific PCR in situ hybridization (MSP-ISH), for the in situ detection of the methylation status of specific sequences in archival specimens. The original technique is based on treatment of DNA with sodium bisulfite, which changes unmethylated, but not methylated, cytosines to uracil (13). The DNA is then amplified by using primers specific for detecting sequence differences generated by this modification (10). In this study, MSP-ISH is used to trace the evolution of cell populations harboring hypermethylation-associated inactivation of the important tumor suppresser gene p16 during the progression from dysplasia to invasion of two common forms of human cancer, squamous cell carcinomas of the lung and cervix. p16 encodes a cyclin-dependent kinase inhibitor essential for maintenance of the retinoblastoma (Rb) protein in the hypophosphorylated active state. Our findings not only document that inactivation of p16 is an early event in progression of these cancers, but they also produce a surprising view of how competing inactivation events for the cyclin D–Rb pathway are selected during cervical carcinoma progression.
Materials and Methods
Specimen Preparation.
The specificity of the PCR in situ system was defined by using the cell lines HN12 and OH3. The HN12 cell line, derived from a squamous cell carcinoma of the head and neck region, has a hypermethylated p16 promoter region, whereas the OH3 line, derived from a small cell carcinoma of the lung, does not exhibit p16 hypermethylation (6). Near confluent cells were fixed for 1–3 days in 10% buffered formalin, then processed as previously described (14). Formalin-fixed, paraffin-embedded tissues were used to determine the incidence and cellular distribution of p16 hypermethylation within clinical samples. The human papillomavirus (HPV) type associated with samples of cervical carcinoma was determined by DNA in situ hybridization using individual probes at varying stringencies, as previously described (14).
PCR in Situ Hybridization (MSP-ISH).
Silane-coated slides, containing either cultured cells or sections prepared from paraffin-embedded tissue samples, were digested in pepsin (2 mg/ml in 0.1 M HCl) for 30 min, washed in sterile water for 1 min, then air dried. Three sequential sections were placed on each slide. The samples were incubated in 0.2 M NaOH for 10 min at 37°C, then placed in the 3 M sodium bisulfite solution as per the manufacturer’s recommendation (CpG Wiz p16 methylation assay; Intergen Discovery Products, Gaithersburg, MD) and incubated at 55°C for 15 h, followed by incubation in 0.3 M NaOH for 5 min.
Our PCR in situ hybridization protocol has been published (14, 15). Briefly, after a manual hot start and denaturing at 94°C for 3 min, 35 cycles were conducted at 55°C for 1.5 min and 94°C for 1 min. For each slide, one section was analyzed with the primers (each at 1 μM) specific for the unmethylated p16 sequences (10), one section with the primers specific for the methylated p16, while no primers were applied to the third tissue section. This allowed direct comparison of these variables on the same cells present in the serial sections under the same conditions. After amplification, in situ hybridization was performed using an unmethylated-specific or methylated-specific internally digoxigenin-labeled probe (1 μg/ml), corresponding to the U and M products generated by solution-phase methylation-specific PCR (MSP), diluted with Hybrisol VII (Oncor). The amplicon and probe were codenatured at 95°C for 5 min, hybridized at 37°C for 2 h, washed for 10 min in 1× SSC [0.15 M sodium chloride/0.015 M sodium citrate (pH 7)] with 2% BSA at 52°C, incubated with the anti-digoxigenin alkaline phosphatase conjugate (1:200; Roche Molecular Biochemicals), and then exposed to the chromogen, nitroblue tetrazolium and 5-bromo-4-chloro-3-indolyl phosphate (NBT/BCIP) (Enzo Diagnostics) at 37°C. The final counterstain, nuclear fast red, stains the negative cells pink in contrast to the blue signal.
Solution-Phase MSP.
MSP (10) was performed to compare results obtained with MSP-ISH. Three 4-μm sections sequential to those used for MSP-ISH were placed in a 1.5-ml sterile tube, and DNA was isolated using the Intergen EX-WAX DNA Isolation Kit. Purity of the DNA isolation was determined by amplifying 100 ng of DNA with the p16W wild-type primers (10), and samples were then evaluated for methylation state of p16 using specific methylated and unmethylated primers (10) to generate 145-bp and 154-bp PCR products, respectively. The PCR products were loaded onto nondenaturing 6% polyacrylamide gels, stained with ethidium bromide, and visualized under UV illumination.
Immunohistochemistry.
Immunohistochemistry was performed as described previously (14, 16, 17) on the two cell lines and each of the histologic sections to compare the in situ PCR-amplified DNA results with cellular steady-state p16 protein levels. Briefly, after deparaffinization, the mAb (p16 INK4/CDKN2, Sigma; or p16 INK4a, Neomarkers, Fremont, CA) was applied and incubated at 37°C for 2 h. To determine the optimal conditions for each Ab, pretreatment with antigen retrieval [microwave of tissue sections in 10 mM sodium citrate buffer (pH 6.0)], protease digestion (proteinase K at 10 μg/ml for 15 min), and no pretreatment were compared at dilutions of 1:50 to 1:250. The best results were obtained with the p16 INK4/CDKN2 Ab at a dilution of 1:100 after proteinase K pretreatment of samples. The Ab–antigen complexes were visualized with fast red, using hematoxylin as the counterstain as described by the manufacturer (BioGenex Laboratories, San Ramon, CA).
Results
Validity of the MSP-ISH Assay.
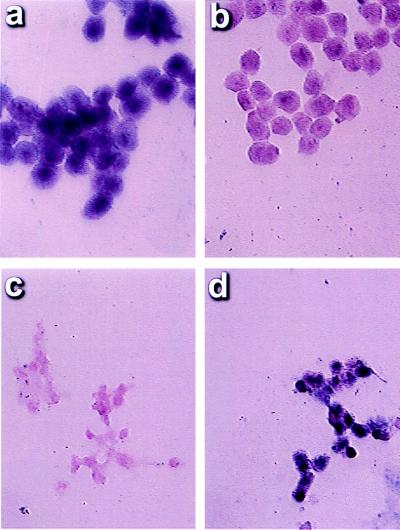
The cell lines HN12 (hypermethylated at p16) and OH3 (unmethylated at p16) were used to determine the specificity of MSP-ISH for discerning the methylation status of the p16 promoter. MSP-ISH using the methylation-specific p16 primers in conjunction with the digoxigenin-labeled p16 probe detected an intense signal in the nuclei of all HN12 cells (Fig. 1a). No signal was seen on the cells on the same glass slide if the unmethylated primers and probe were used (Fig. 1b). Conversely, no signal was evident in the OH3 cells using the methylated primer/probe set, whereas a strong signal was present in all cells when the unmethylated primer/probe set was used (Fig. 1 c and d). Signal was absent with the methylated or unmethylated primer/probe set for all samples when sodium bisulfite treatment was omitted (data not shown).
Figure 1.
Specificity of MSP-ISH in cancer cell lines. The cancer cell line HN12 has a p16 promoter that is inactivated because of hypermethylation. MSP-ISH after sodium bisulfite treatment demonstrated a signal corresponding to methylated cytosines in the cells (a); no signal was evident with the primer/probe set specific for unmethylated CpG islands (b). Conversely, the cell line OH3 does not contain a methylated p16 promoter. These cells were negative after MSP-ISH assay for the methylated promoter (c) and positive when the primer/probe set for unmethylated p16 was used (d).
Previous studies indicate that OH3 cells express and HN12 cells do not express p16 at the mRNA level (6). Immunohistochemistry for p16 protein was performed to correlate this expression status further with the MSP-ISH results. OH3 cells stained strongly for the protein, whereas no staining was detected in HN12 cells (data not shown), corroborating the accuracy of the MSP-ISH technique to specifically identify the nuclei of cells carrying methylated p16 alleles, which do not express the p16 protein.
Lung Carcinoma and Its Precursors.
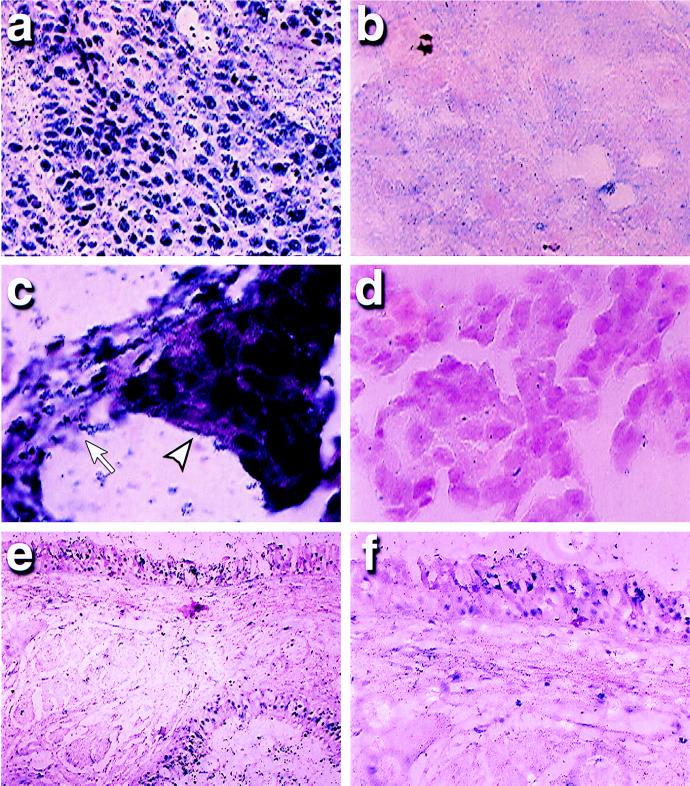
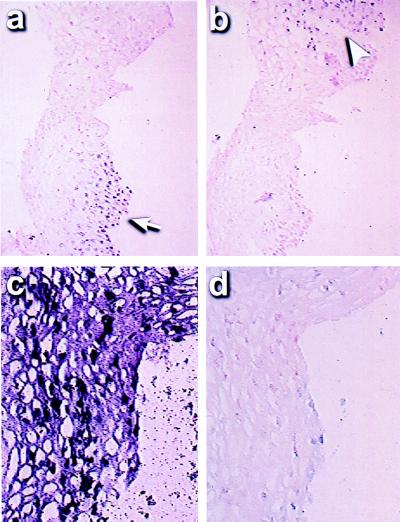
Four squamous cell carcinomas of the lung (Fig. 2) were analyzed. One had an extensive area of carcinoma in situ (CIS), in addition to the invasive lung cancer. Solution MSP analysis of extracted DNA had demonstrated distinct signals for both methylated and unmethylated p16 sequences (18). MSP-ISH revealed that the signal for the methylated alleles was associated only with invasive cells in all four of these tumors (Fig. 2a). The percentage of invasive cancer cells that were positive after in situ PCR amplification with the methylated primer and probe set was at least 90% in three of four cancers, whereas unmethylated p16 sequences were seen in <1–3% of these cells. Each of these tumors demonstrated no signal with the primers for unmethylated sequences in the cancer cells (Fig. 2b). In the other cancer case, only 20% of the tumor cells had hypermethylated p16 sequences. However, no cancer cell gave a signal with the primers for unmethylated sequences, perhaps reflecting methylation of only some of the CpG sites in this region in most of the tumor cells. In this regard, six CpG sites are incorporated under the two PCR primers, and most or all of these have to be either methylated or unmethylated for initial annealing of the primers to occur. In addition to the squamous cell carcinomas of the lung, we also studied a lung adenocarcinoma that had only unmethylated alleles by MSP analysis of extracted DNA. Cells in this tumor showed a signal with only the unmethylated primer set after MSP-ISH (Fig. 2 c and d). Importantly, in all of the above samples, normal tissue adjacent to the tumors, consisting primarily of the alveolar capillaries and associated pneumocytes, demonstrated nuclei with only unmethylated p16 after in situ amplification (example, Fig. 2 c and d).
Figure 2.
p16 methylation in lung cancers and precancerous lesions by MSP-ISH. An invasive squamous cell carcinoma demonstrates a strong signal after MSP-ISH with the methylated primer/probe set in cancer cells (a), but is negative with the unmethylated primer/probe set (b). However, in an adenocarcinoma, both the cancer cells and normal cells of the alveolus (arrowhead, cancer cells; arrow, normal cells) demonstrated a signal after MSP-ISH with the unmethylated primer set for the p16 promoter (c), but were negative with the methylated primer/probe (d). In a dysplastic lesion, a positive signal was seen only in the dysplastic cells lining the airways and in adjoining subjacent glands (top and bottom right regions of e) and was negative in the normal stromal cells between these preneoplastic cells. On higher magnification, this lesion is found to contain positive methylation signal in only a portion of the dysplastic cells (f).
Four cases of carcinoma in situ of the lung (including the one case with an associated invasive component), a precursor lesion to invasive squamous cell cancer, were evaluated for p16 methylation. In each case, cells constituting the in situ lesions comprised a very small population (<1%) of the total cells in the biopsy. Solution MSP analysis of extracted DNA revealed that two of these early lesions contained both unmethylated and methylated p16 alleles, whereas only unmethylated alleles were detected in the other two in situ lesions. Interestingly, all four precursor lesions, like the advanced squamous cell tumor shown in Fig. 2a, demonstrated a signal after in situ amplification and hybridization with the methylated primer/probe set in >90% of the dysplastic cells (data not shown). This result may reflect the great sensitivity of MSP-ISH vs. solution MSP for detection of very few transformed cells within a sample. In contrast, with the unmethylated primer and probe, no signal was seen in the transformed cells in two cases, whereas the other two showed rare positive cells (<1% of the total CIS cells). One case also contained an area of mild dysplasia, a lesion less advanced than CIS. Methylation of p16 was confined to the dysplastic cells (Fig. 2e) and was noted in only approximately 40% of these dysplastic cells (Fig. 2f), suggesting a progression of methylation between the dysplastic and CIS lesions.
Importantly, comparison of the present data with previous studies of p16 protein expression in the above tumors and precursor lesions showed a 100% concordance between protein expression and methylation status (18). p16 protein was previously not detected in the neoplastic cells, which, in the current study, contain a positive nuclear methylation signal in the squamous precursor or invasive tumors examined. However, p16 protein was readily detected in virtually all cells from the lung adenocarcinoma, which contained only cells with a signal for unmethylated p16 gene sequences and in most normal cells surrounding either the established squamous cells tumors or the precursor lesions.
Cervical Carcinoma and Its Precursors.
The methylation status of p16 has not been studied in cervical carcinoma, a tumor that also arises via the squamous cell pathway. A priori, it might be expected that inactivation of p16 would not be found in cervical carcinomas because these tumors arise in association with HPV infection. This virus expresses the E7 protein, which inactivates the Rb protein (19), critical for cell-cycle control. This pathway is inactivated in multiple tumor types, either directly by mutations in Rb or by inactivation of p16. Simultaneous inactivation of both Rb and p16, including loss of p16 function with promoter hypermethylation, is virtually never found in a single tumor because lesions in either gene are sufficient to disrupt the cyclin D–Rb pathway (6).
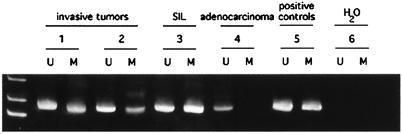
Surprisingly, solution MSP detected both methylated and unmethylated p16 gene alleles in four of eight low-grade noninvasive squamous intraepithelial lesions (SILs), a precursor lesion to invasive cervical carcinomas, and in four of four invasive squamous cell cancers of the cervix (Fig. 3). All four invasive lesions contained HPV (three contained HPV 16 and one HPV 18). Five of the SILs contained HPV types known to be associated with cervical cancer (HPVs 51 in three cases, HPV 52 in one case, and HPV 16-related in one case), whereas the other three had so called “benign” HPV types that are not associated with cancer (HPVs 2, 6, and 11). Two of the low-grade lesions that contained methylated copies of p16 had nononcogenic HPV types. In contrast to the squamous cell lesions, only unmethylated p16 was detected in three of three adenocarcinomas of the cervix (Fig. 3), two of which contained HPV 18 and one that contained HPV 16.
Figure 3.
Methylation-specific PCR of p16 in invasive cervical carcinomas and squamous intraepithelial lesions. Both the unmethylated (U) and methylated (M) PCR products are shown for each sample. Invasive squamous cell cancers of the cervix (samples 1 and 2) contain both unmethylated and methylated alleles, as does a low-grade squamous intraepithelial lesion (SIL; sample 3). An adenocarcinoma of the cervix displays only unmethylated alleles (sample 4). Also shown are positive controls for the U and M PCRs (sample 5) and water controls (sample 6).
To determine the distribution of cells containing the hypermethylated copies of p16 in the cervical neoplasias, tissues were examined by the MSP-ISH technique. Each of these tissues contained foci of residual normal squamous and glandular epithelium and stromal cells, which did not demonstrate a signal with the methylated primer/probe set, but did show a signal with the unmethylated primer/probe set (data not shown). Likewise, each of the three well differentiated adenocarcinomas had unmethylated p16 promoter sequences in >90% of the tumor cells. Rare tumor cells (<1% of the invasive component) showed a signal with the methylated primers/probe system in two of the three adenocarcinomas (data not shown).
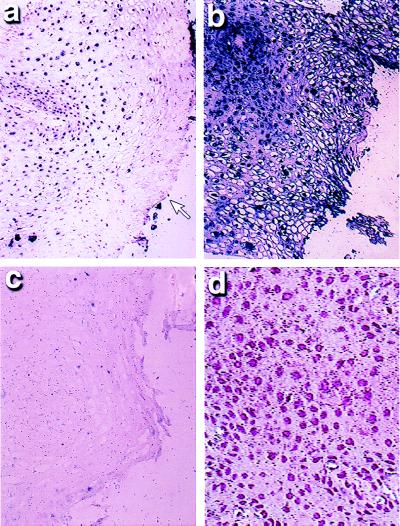
A striking difference was found between the invasive and preinvasive stages of the cervical lesions for distribution of cells with hypermethylated copies of p16. In the four low-grade SILs that contained hypermethylated p16 copies by solution MSP, the cells positive for this change aggregated into small groups and constituted only 5–30% of the dysplastic cells in each lesion (Fig. 4a). Each of the early-stage invasive cancers also had associated extensive areas of high-grade SIL, and, in these areas and in contrast to the low-grade lesions, most cells demonstrated hypermethylation of p16 (Fig. 4b), whereas no signal was seen with the unmethylated primer/probe set (Fig. 4c). In each of four cases of invasive cancer, >75% of the tumor cells showed p16 promoter hypermethylation.
Figure 4.
Correlation of histology and p16 methylation status in low-grade SILs of the cervix. This cervical SIL showed areas of both low-grade and high-grade SIL on routine histology. In the region of low-grade SIL, some of the cells demonstrated a strong signal after MSP-ISH specific for hypermethylated p16, whereas those toward the surface (open arrow) did not (a). However, in an adjoining area where high-grade SIL was evident, all of the dysplastic cells were positive for p16 hypermethylation (b), but had no signal seen in the corresponding serial section for unmethylated p16 (c). Immunohistochemical analysis for p16 showed a signal in the normal squamous cells, which are identified by their well ordered appearance and uniform nuclei (d); a strong signal was seen in these cells after MSP-ISH for the unmethylated DNA sequences only.
The relationship between cells with hypermethylated copies of p16 and inactivation of the gene was apparent in the cervical carcinomas as in the lung tumors. There was strong concordance between the MSP-ISH data and immunohistochemistry. p16 protein was absent from the invasive carcinomas and from the pockets of cells in the low-grade SILs that contained the MSP-ISH signal for methylated p16 (data not shown). p16 protein was detected in the normal squamous epithelium (Fig. 4d), as well as in the stromal cells and in the malignant cells of the adenocarcinomas that did not contain p16 hypermethylation.
As noted previously, the high frequency of p16 inactivation in cervical carcinoma is surprising, given the association of this tumor with infection by a virus capable of inactivating the Rb gene by means of the E7 protein. However, when we explored the distribution of cells containing hypermethylated p16 and those expressing viral E7 and E6, a protein capable of inactivating p53 (20), hypermethylated p16 sequences were confined to cells distinct from those containing viral E7 or E6 RNA in each of five cases of low-grade SIL (Fig. 5 c and d). In advanced lesions, relatively few cells expressed E6 or E7 (approximately 30%) (16, 17), and most contained hypermethylated p16, just as found in the cancers and low-grade SILs with oncogenic HPV types.
Figure 5.
Inactivation of different tumor suppressor genes occurs in distinct cell groups in low-grade SIL of the cervix. (a) An area of a low-grade SIL that contains HPV 18, where expression of the E6 ORF is evident in some but not all dysplastic cells (arrow). Note that a different group of cells in the serial section contained the E7 transcript (b; arrowhead). Hypermethylated p16 was not seen in this section (data not shown). (c) Another area of the same low-grade lesion, where many cells contained hypermethylated p16 DNA as determined by MSP-ISH. No E6 (d) or E7 (data not shown) expression was evident in these cells as determined by reverse transcriptase in situ PCR.
Discussion
Transcriptional silencing of p16, in association with hypermethylation of the CpG island located in the proximal promoter region, has been associated with virtually every form of human cancer and appears to be particularly common in squamous cell tumors (18). Recent studies of a rat model for lung carcinogenesis (18) and of precursor lesions to human esophageal tumors (21) and squamous cell lung cancers (18) have suggested that hypermethylation of p16 may be an early event in tumorigenesis. However, with the techniques employed in these previous studies, it has been difficult to assess the precise timing of these methylation events in tumor progression, the cellular origins of the methylated alleles, and the precise cellular consequences of the methylation for gene expression. The MSP-ISH technique has allowed us to address these key points and illustrate how hypermethylation and concomitant inactivation of p16 expression is a common and early event in the evolution of squamous cell neoplasia at two disparate sites—the lung and the cervix.
The MSP-ISH study of squamous cell lung cancers verifies not only that p16 can be abnormally methylated in the precursor lesions of these cancers but also that the neoplastic cells are the sole source for the involved alleles. Furthermore, abnormal methylation of p16 appears to be a clonal event involving most cells in the CIS lung lesions and, importantly, is accompanied by complete absence of p16 protein in the involved cells. Thus, whether one or two alleles of p16 are present in these early lesions, inactivation of all gene copies in these precursor lesions is apparent and may be pivotal for the evolution of the invasive cancer. Methylation of both alleles of p16, in contrast to deletion of one allele and methylation of the retained allele, occurs in other tumor types, for example, colon cancer (5). The observation that methylation of p16 in dysplastic lesions is heterogeneous and appears to progress in more advanced lesions demonstrates the utility of MSP-ISH, but is preliminary and must be confirmed in a larger number of samples.
The MSP-ISH results for p16 inactivation in cervical carcinoma are particularly intriguing, demonstrating that hypermethylation of the CpG island in the p16 promoter occurs in subpopulations of the dysplastic cells of low-grade SIL, the earliest recognizable form of squamous cell dysplasia. Moreover, p16 hypermethylation was invariably present in the severely dysplastic cells of a high-grade SIL and in the invasive cells of squamous cell cancers. This suggests that there is a natural selection process for mildly dysplastic cells with reduced expression of p16 and promoter hypermethylation in cases that evolve to high-grade lesions and invasive tumors. This statement must be tempered by the realization that p16 inactivation was also detected in low-grade SILs that contained nononcogenic HPV types. This suggests that p16 inactivation is involved, in general, with hyperproliferative processes that do not always progress to cancer, but appears to provide a permissive event that may predispose cells to respond to further oncogenic molecular events.
Most importantly, inactivation of p16 occurred in dysplastic cells of low-grade SILs that were distinct from those that contained either viral E6 or E7 transcripts and, presumably, had markedly reduced activities of p53 and Rb proteins, respectively. It has been observed that the E6 and E7 proteins allow cells to escape separate mortality checkpoints mediated by each of these proteins (19, 20). In fact, mammary cells transfected with E6, but not those transfected with E7, develop hypermethylation of p16 as they bypass the mortality checkpoint mediated by Rb (22). It has been debated whether, during the natural infection associated with the initiation and evolution of cervical carcinomas, blockage of Rb and p53 activities is partial or complete as the tumors evolve. Thus, it is not clear whether HPV infection alone can account for complete progression of these cancers. Our results with MSP-ISH now help resolve this question. It would appear that the viral inhibition alone may not be severe enough to ensure progression and that a more complete inactivation event, transcriptional inactivation of p16 in association with hypermethylation, arises and gets selected for with time. This pressure to select for inactivated p16 may be integrally related to the finding that cells with hypermethylated copies of the gene appear to have lost expression of E7 over time. The origin of the hypermethylated p16 could even relate to the original presence of HPV, because viruses have been associated with gene hypermethylation (23).
In summary, we describe an assay to examine the methylation of individual gene sequences in specific cells and to directly assess the consequences for gene expression. This approach has been used to dissect the complex evolution during tumor progression of the abnormal methylation and concomitantly altered gene expression for the important tumor suppressor gene p16. MSP-ISH should allow a similar dissection for the methylation dynamics now associated with abnormal inactivation of other key genes in cancer. Moreover, other important questions concerning methylation of CpG islands can now be addressed with the MSP-ISH approach. For example, the technique could help in defining when the normal protection from methylation, characteristic of most of these sequences in adult cells, arises during embryogenesis, and the developmental timing in specific cell types for involvement of CpG island methylation in gene imprinting and mammalian X chromosome inactivation. Finally, MSP-ISH now offers possibilities for diagnostic approaches at the cellular level. Sensitive detection of genetic disorders involving aberrant patterns of CpG island methylation, such as the fragile X syndrome, and diseases involving abnormalities of gene imprinting can be considered, as well as sensitive detection of cancer cells in clinical specimens.
Acknowledgments
This work was supported by a grant from the Lewis Foundation (to G.J.N.), and by National Institutes of Health Grants CA43318 and SPORE P50CA58184 (to S.B.B. and J.G.H.) and Grant ES08801 (to S.A.B.). J.G.H. is a V Foundation Scholar. S.B.B. and J.G.H. receive research funding and are entitled to sales royalties from Oncor, which is developing products related to research described in this paper. The terms of this arrangement have been reviewed and approved by The Johns Hopkins University in accordance with its conflict of interest policies.
Abbreviations
- MSP
methylation specific PCR
- MSP-ISH
MSP in situ hybridization
- HPV
human papillomavirus
- Rb
retinoblastoma
- CIS
carcinoma in situ
- SIL
squamous intraepithelial lesion
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Lock L F, Melton D W, Caskey C T, Martin G R. Mol Cell Biol. 1986;6:914–924. doi: 10.1128/mcb.6.3.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Razin A, Cedar H. Cell. 1994;77:473–476. doi: 10.1016/0092-8674(94)90208-9. [DOI] [PubMed] [Google Scholar]
- 3.Baylin S B, Herman J G, Graff J R, Vertino P M, Issa J P. Adv Cancer Res. 1998;72:141–196. [PubMed] [Google Scholar]
- 4.Herman J G, Latif F, Weng Y, Lerman M I, Zbar B, Liu S, Samid D, Duan D S, Gnarra J R, Linehan W M, Baylin S B. Proc Natl Acad Sci USA. 1994;91:9700–9704. doi: 10.1073/pnas.91.21.9700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Herman J G, Merlo A, Mao L, Lapidus R G, Issa J P J, Davidson N E, Sidransky D, Baylin S B. Cancer Res. 1995;55:4525–4530. [PubMed] [Google Scholar]
- 6.Merlo A, Herman J G, Mao L, Lee D J, Gabrielson E, Burger P C, Baylin S B, Sidransky D. Nat Med. 1995;1:686–692. doi: 10.1038/nm0795-686. [DOI] [PubMed] [Google Scholar]
- 7.Kane M F, Loda M, Gaida G M, Lipman J, Mishra R, Goldman H, Jessup J M, Kolodner R. Cancer Res. 1997;57:808–811. [PubMed] [Google Scholar]
- 8.Herman J G, Umar A, Polyak K, Graff J R, Ahuja N, Issa J P J, Markowitz S, Willson J K V, Hamilton S R, Kinzler K W, et al. Proc Natl Acad Sci USA. 1998;95:6870–6875. doi: 10.1073/pnas.95.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cunningham J M, Christensen E R, Tester D J, Kim C Y, Roche P C, Burgart L J, Thibodeau S N. Cancer Res. 1998;58:3455–3460. [PubMed] [Google Scholar]
- 10.Herman J G, Graff J R, Myohanen S, Nelkin B D, Baylin S B. Proc Natl Acad Sci USA. 1996;93:9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frommer M, McDonald L E, Millar D S, Collis C M, Watt F, Grigg G W, Molloy P L, Paul C L. Proc Natl Acad Sci USA. 1992;89:1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gonzalgo M L, Jones P A. Nucleic Acids Res. 1997;25:2529–2531. doi: 10.1093/nar/25.12.2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clark S J, Harrison J, Frommer M. Nat Genet. 1995;10:20–27. doi: 10.1038/ng0595-20. [DOI] [PubMed] [Google Scholar]
- 14.Nuovo G J. PCR in Situ Hybridization: Protocols and Applications. Philadelphia: Lippincott; 1997. [Google Scholar]
- 15.Nuovo G J, Gallery F, MacConnell P, Becker J, Bloch W. Am J Pathol. 1991;139:1239–1244. [PMC free article] [PubMed] [Google Scholar]
- 16.Nuovo G J, MacConnell P B, Simsir A, Valea F, French D L. Cancer Res. 1995;55:267–275. [PubMed] [Google Scholar]
- 17.Nuovo G J. Int J Cancer. 1997;71:1056–1060. doi: 10.1002/(sici)1097-0215(19970611)71:6<1056::aid-ijc23>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 18.Belinsky S A, Nikula K J, Palmisano W A, Michels R, Saccomanno G, Gabrielson E, Baylin S B, Herman J G. Proc Natl Acad Sci USA. 1998;95:11891–11896. doi: 10.1073/pnas.95.20.11891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dyson N, Howley P M, Munger K, Harlow E. Science. 1989;243:934–937. doi: 10.1126/science.2537532. [DOI] [PubMed] [Google Scholar]
- 20.Scheffner M, Werness B A, Huibregtse J M, Levine A J, Howley P M. Cell. 1990;63:1129–1136. doi: 10.1016/0092-8674(90)90409-8. [DOI] [PubMed] [Google Scholar]
- 21.Wong D J, Barrett M T, Stoger R, Emond M J, Reid B J. Cancer Res. 1997;57:2619–2622. [PubMed] [Google Scholar]
- 22.Foster S A, Wong D J, Barrett M T, Galloway D A. Mol Cell Biol. 1998;18:1793–1801. doi: 10.1128/mcb.18.4.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mikovits J A, Young H A, Vertino P, Issa J P, Pitha P M, Turcoski-Corrales S, Taub D D, Petrow C L, Baylin S B, Ruscetti F W. Mol Cell Biol. 1998;18:5166–5177. doi: 10.1128/mcb.18.9.5166. [DOI] [PMC free article] [PubMed] [Google Scholar]