Abstract
The transporter associated with antigen processing (TAP) is essential for the transport of antigenic peptides across the membrane of the endoplasmic reticulum. In addition, TAP interacts with major histocompatibility complex class I heavy chain (HC)/β2-microglobulin (β2-m) dimers. We have cloned a cDNA encoding a TAP1/2-associated protein (TAP-A) corresponding in size and biochemical properties to tapasin, which was recently suggested to be involved in class I–TAP interaction (Sadasivan, B., Lehner, P. J., Ortmann, B., Spies, T. & Cresswell, P. (1996) Immunity 5, 103–114). The cDNA encodes a 448-residue-long ORF, including a signal peptide. The protein is predicted to be a type I membrane glycoprotein with a cytoplasmic tail containing a double-lysine motif (-KKKAE-COOH) known to maintain membrane proteins in the endoplasmic reticulum. Immunoprecipitation with anti-TAP1 or anti-TAP-A antisera demonstrated a consistent and stoichiometric association of TAP-A with TAP1/2. Class I HC and β2-m also were coprecipitated with these antisera, indicating the presence of a pentameric complex. In pulse–chase experiments, class I HC/β2-m rapidly dissociated from TAP1/2-TAP-A. We propose that TAP is a trimeric complex consisting of TAP1, TAP2, and TAP-A that interacts transiently with class I HC/β2-m. In peptide-binding assays using cross-linkable peptides and intact microsomes, TAP-A bound peptides only in the presence of ATP whereas binding of peptides to TAP1/2 was ATP-independent. This suggests a direct role of TAP-A in peptide loading onto class I HC/β2-m dimer.
Major histocompatibility complex (MHC) class I molecules present antigenic peptides to CD8+ T cells (1–3). The majority of peptides found associated with class I molecules are derived from nuclear and cytosolic proteins, and they are generated largely by the proteasome complex (4, 5). Peptides are transported from the cytosol into the lumen of the endoplasmic reticulum (ER) by a peptide transporter, which is known as the transporter associated with antigen processing (TAP) (2, 3, 6). TAP consists of two subunits, TAP1 and TAP2, both members of the ATP-binding cassette transporters (3, 6). Transfection of TAP1 and TAP2 cDNAs into TAP-mutant cells restores class I assembly and surface expression, indicative of involvement of these molecules in antigen processing (3, 6). The critical role of TAP in class I antigen processing has been demonstrated by functional assays with isolated microsomes or with semipermeabilized cells (7–10). These studies have shown that TAP preferentially transports peptides of 8–15 residues in an ATP-dependent fashion (6). Peptide translocation occurs in two steps involving ATP-independent peptide-binding to TAP and peptide translocation across the ER membrane, which is ATP-dependent (11–13).
In addition to functioning as a peptide transporter, a physical association between TAP1 and class I heavy chain (HC)/β2-microglobulin (β2-m) dimer has been demonstrated (14, 15). The binding of class I HC/β2-m to TAP1 is not required for the peptide translocation, so the binding of peptides to class I molecules is thought to be facilitated by association of assembled HC/β2-m heterodimers with the TAP complex (14–16). Recently, it has been found that a point mutation of threonine 134 to lysine (T134K) in the HLA-A2.1 makes the HC incapable of interacting with the TAP complex (17, 18). This results in decreased cell surface expression of HLA-A2.1, as well as in the loss of capacity of newly synthesized class I HC/β2-m complex to load peptide in a TAP-dependent manner (17, 18). Moreover, direct delivery of peptide to the ER in a TAP-independent manner restores the ability of T134K, HLA-A2.1 to present antigenic peptide to peptide-specific cytotoxic T lymphocytes (17). These results suggested that TAP is not only required for peptide transport across the ER membrane but also for the assembly of peptide and class I HC/β2-m complex. A genetic defect has been demonstrated in a human mutant cell line, 721.220, causing the failure of class I to associate with TAP (19). This deficiency impairs the ability of the class I binding to peptides (19), suggesting the involvement of an additional molecule in the assembly of peptide and class I HC/β2-m. Furthermore, the association of a 48-kDa glycoprotein (tapasin) to TAP1/2 and calreticulin recently was demonstrated (1). Tapasin was not detected in 721.220 cells (1, 20), which suggests that tapasin is required for the physical association of MHC class I HC/β2-m dimers with either TAP1/2 or calreticulin.
Here, we have cloned the cDNA of a human TAP-associated protein (designated TAP-A), which has the same size as tapasin (1). The amino acid sequence indicates that it is a membrane glycoprotein with a double lysine motif known to serve as an ER retention signal. Pulse–chase experiments with antisera to TAP1 and TAP-A demonstrated the consistent presence of stoichiometric amounts of TAP-A and TAP1/2, indicating that TAP-A, TAP1, and TAP-2 form a trimeric complex. We also demonstrate that TAP-A can bind peptide in the presence of ATP.
MATERIALS AND METHODS
Cells.
T2, T1, CIR, Raji, and Daudi cells were obtained from the American Type Culture Collection. The B lymphoblastoid cell lines (LCL) were kindly provided by Maria Masucci (Karolinska Institute, Stockholm, Sweden). The cell lines were cultured in RPMI 1640 medium (GIBCO/BRL) supplemented with 10% heat-inactivated fetal bovine serum, 100 units/ml penicillin, 100 μg/ml streptomycin, and 2 mM glutamine at 37°C in a 5% CO2 atmosphere.
Antibodies.
A rabbit antiserum prepared against the C-terminal TAP1 peptide and rabbit antiserum R425, specific to both free and assembled HLA HCs, have been described (21). The anti-TAP-A antiserum was generated by immunization with a peptide (WASGLTPAQNCPR) derived from TAP-A. Anti-transferrin receptor (Tf-R) mAb was kindly provided by H. Garoff, Karolinska Institute. The antibodies were affinity-purified before use.
Metabolic Labeling, Immunoprecipitation, and Immunoblotting.
Cells were washed twice with PBS and incubated for 15 min at 37°C in methionine-free RPMI 1640 medium containing 3% dialyzed fetal bovine serum. Then, 0.2 mCi/ml [35S]-methionine (Amersham) was added, and the incubation was continued for 60 min. Where indicated, pulse-labeled cells were chased for different periods of time at 37°C in complete medium. At the end of the chase periods, cells were washed three times with ice-cold PBS and lysed in 1% digitonin (repurified from digitonin purchased from Sigma) lysis buffer containing 0.15 M NaCl, 25 mM Tris⋅HCl (pH 7.5), 1.5 μg/ml iodoacetamide, and a mixture of protease inhibitors (2 mM phenylmethylsulfonyl fluoride/10 μg/ml leupeptin/30 μg/ml aprotinin/10 μg/ml pepstatin). The cleared lysates were added to antibodies previously bound to protein A Sepharose beads (Pharmacia). After washing, the immunoprecipitates were analyzed by SDS/PAGE as described (22). For immunoblotting, aliquots of cell lysates were loaded onto a 10% SDS/PAGE. Proteins were transferred onto a nitrocellulose filter, which was probed with the anti-TAP-A antiserum at a dilution of 1:1000. Detection was performed according to Kaltoft et al. (23).
cDNA Cloning of TAP-A.
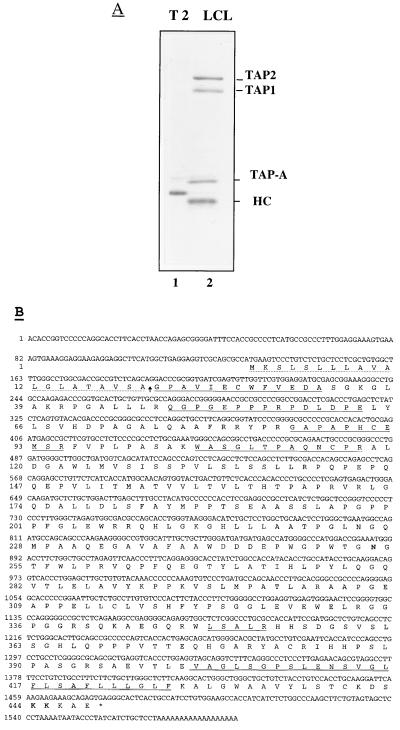
The TAP-A molecules were coprecipitated from LCL cells by anti-TAP1 antibody and resolved by SDS/PAGE. After visualization with Coomassie blue staining, the appropriate TAP-A band was excised and subjected to in-gel digestion with modified trypsin (Promega) (24). After isolation of generated fragments by narrow bore, reversed phase liquid chromatography, selected peptides were sequenced in an Applied Biosystems model 470A instrument. Mixtures of two degenerate DNA oligomers, 5′-TGGTT(CT)GTIGA(GA)GA(CT)GC-3′ and 5′-CAT(CT)TC(AG)CA(AG)TGIGGIGC-3′ corresponding to the amino acid sequences of the two TAP-A-derived peptides (NH2-WFVEDA-COOH and NH2-APHCEM-COOH, respectively), were synthesized with a DNA synthesizer (Applied Biosystems) and used as primers. Total RNA was extracted from LCL cells and amplified by reverse transcription-PCR. Amplified fragments were directly sequenced and subsequently used as probes to screen a cDNA library constructed into a λgt10 phage vector (CLONTECH) from poly(A)-selected RNA prepared from Daudi cells. Ten positive plaques contained overlapping inserts. These inserts were sequenced and reconstructed in a 1.6-kb fragment into a pGM3f(−) vector under T7 promoter. The predicted amino acid sequences underlined were identified by sequencing of peptide fragments derived from purified TAP-A (Fig. 1B).
Figure 1.
Purification of TAP-A, its cDNA, and deduced amino acid sequences. (A) T2 (lane 1) or LCL (lane 2) cells were metabolically labeled with [35S]-methionine and lysed by 1% digitonin lysis buffer. The lysates were immunoprecipitated with anti-TAP1 antiserum and analyzed on a 10% SDS gel. The positions of TAP1, TAP2, TAP-A, and class I HC are indicated. (B) The predicted N-terminal signal sequence (discontinuous line), the five microsequenced peptides (continuous line), and the predicted C-terminal transmembrane domain (double line) are indicated. The anticipated signal peptidase cleavage site is marked with a arrow. The single N-glycosylation site at position Asn253 is in bold, as is the double lysine motif at positions 444 and 445.
Peptides and Peptide Modification.
All peptides were synthesized in a peptide synthesizer (Applied Biosystems model 431A) using conventional Fmoc chemistry. Peptides were subsequently purified by HPLC and dissolved in PBS. The HLA-B27 binding peptide of influenza A virus nucleoprotein NP383–391R389K (SRYWAIKTR) was covalently modified by coupling a phenyl azide with a nitro group (NP383–391-ANB-NOS) as described (12, 21). An aliquot (100 ng) of the peptide was labeled by chloramine T-catalyzed iodination (125I). The modification and labeling experiments were performed in the dark.
Preparation of Microsomes and Photo-Cross-Linking.
Microsomes from T1, T2, Raji, Daudi, CIR-B27, and LCL cells were prepared and purified according to Saraste et al. (25). For photo-cross-linking, 100 nM of 125I-NP383–391-ANB-NOS peptide was mixed with 20 μl of microsomes (concentration of 60 A280/ml) in RM buffer (250 mM sucrose/50 mM triethanolamine⋅HCl/50 mM KOAc/2 mM MgOAc2/1 mM DTT). This mixture was then kept for 10 min at 26°C. UV irradiation was subsequently carried out at 366 nm for 5 min on ice. Microsomal membranes were recovered by centrifugation through a 0.5-M sucrose cushion in RM buffer containing 1 mM cold peptide (unlabeled peptide without ANB-NOS modification). The microsomal membranes were washed once with cold RM buffer, lysed by 1% digitonin, and subjected to immunoprecipitation. Cross-linked microsomal proteins were immunoprecipitated with specific antiserum and analyzed by SDS/PAGE. Cross-linking reactions with 1 mM ATP were performed as described (21). For peptide competition, 100 nM of the 125I-NP383–391-ANB-NOS peptide was mixed with a 10-fold molar excess of unlabeled and unmodified NP383–391 peptide before the cross-linking reaction. The extractions of microsomes with high pH, high salt, and urea were performed as described (26).
Velocity Sedimentation Analysis.
The analysis was carried out essentially as described by Bonnerot et al. (27). In brief, lysates (0.8 ml) from digitonin-solubilized, metabolically labeled cells were loaded onto a 8–35% linear sucrose gradient (12 ml total volume) containing 0.3% (wt/vol) digitonin, 0.3 M NaCl, 1.5 μg/ml iodoacetamide, and 50 mM Tris⋅HCl (pH 7.5). Gradients were centrifuged for 16 h at 10°C in an SW41 rotor (Beckman) at 39,000 rpm. Gradients were collected from below into 15 fractions. Each fraction was subjected to immunoprecipitation by the broadly reactive anti-HLA antiserum R425. The human transferrin receptor, which served as an internal size marker (Mr 180 kDa), was detected by an anti-transferrin receptor mAb.
RESULTS AND DISCUSSION
cDNA Cloning of TAP-A.
Using an anti-TAP1 antiserum, we consistently detected a coprecipitating complex containing TAP1, TAP2, MHC class I, and a 48-kDa protein, which we have designated TAP-A for TAP-associated protein (Fig. 1A, lane 2). The same results were obtained in four LCL cell lines with distinct HLA heterotypes (data not shown). To clone the cDNA of TAP-A, we purified the TAP-A protein from LCL cells by coprecipitation with anti-TAP1 antiserum. Several peptide fragments were obtained by trypsin digestion of the TAP-A protein, and degenerate oligonucleotide primers were synthesized based on the sequences of two fragments (see Materials and Methods). Using these primers, a 200-bp fragment was amplified from mRNA of LCL cells by reverse transcription-PCR. The amplified cDNA fragments were used as probes for screening a cDNA library prepared from Daudi cells. Among 10 positive clones, a full length clone with a 1.6-kb insert harbored an ORF encoding a polypeptide of 448 amino acids (Fig. 1B) with an estimated molecular mass of 47,563. The finding that five sequenced TAP-A peptides were contained in the deduced amino acid sequence confirmed that the cDNA encoded TAP-A (Fig. 1B). The predicted polypeptide sequence contains an N-terminal 20-amino acid signal peptide. This is based on predicted signal peptidase cleavage sites (28) and the fact that one sequenced peptide matches the predicted N terminus of the proposed mature protein. According to the hydrophobic profile (29) (data not shown), a 26-amino acid-long hydrophobic stretch (positions 402–427) is predicated to be a transmembrane domain. The predicted lumenal domain is 381 residues and contains an N-linked glycosylation consensus sequence (positions 253–255). The C-terminal end contains 21 amino acids, and its last five residues, KKKAE, contain a double-lysine motif, which mediates the retrieval of proteins back from the cis-Golgi and thus maintains membrane proteins in the ER (30, 31). TAP-A is complexed with TAP1/2 and MHC class I, so the strong ER retention signal of TAP-A may be important for maintaining TAP1/2 and immature class I molecules in the ER. Of interest, >10% of the deduced amino acids consist of prolines, indicative of interrupted helix structures in the protein. Based on the amino acid sequence, TAP-A has the property of a type I ER membrane glycoprotein and most likely corresponds to the recently described protein tapasin (1) (see below).
TAP-A Is an Integral Membrane Protein.
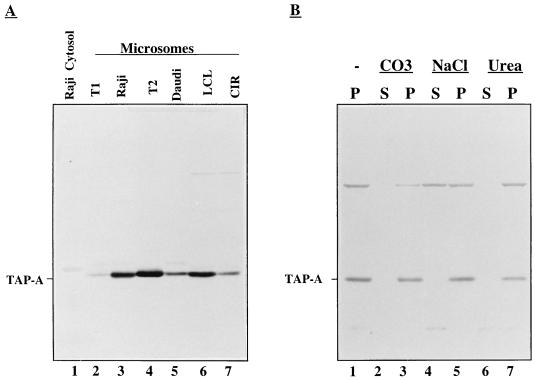
To further characterize the properties of TAP-A, we prepared an anti-TAP-A peptide antiserum (see Materials and Methods) and used it in immunoblotting of cytosolic and microsomal fractions from several cell lines (Raji, Daudi, T1, T2, LCL, and CIR). As shown in Fig. 2A, TAP-A was detected in the microsomal fraction from all cell lines but not in the cytosol from Raji cells. After extraction with sodium carbonate (pH 11.3), high salt, or urea, equal amounts were recovered from the membrane fractions whereas no protein was found in the soluble fractions (Fig. 2B). We thus conclude that TAP-A is an integral membrane protein in accordance with the conclusion deduced from the amino acid sequence.
Figure 2.
Analysis of membrane integration of TAP-A in purified microsomes. (A) Cytosol (Raji, lane 1) and microsomes (T1, Raji, T2, Daudi LCL, and CIR, lanes 2–7) were purified and solubilized in SDS-loading buffer. Proteins were separated on SDS-gel and transferred onto a nitrocellulose filter, which was probed with the anti-TAP-A antiserum. Position of TAP-A is indicated. (B) Microsomes purified from LCL cells were extracted by alkaline (CO3), high salt (NaCl), or urea as described in Materials and Methods. After extraction, supernatants (lanes 2, 4, and 6) and pellets (lanes 1, 3, 5, and 7) were analyzed by immunoblotting with anti-TAP-A antiserum.
TAP-A Is a Subunit of the TAP Complex.
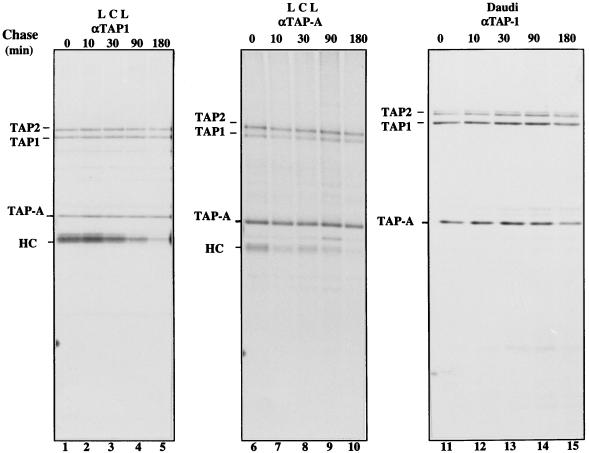
From recent reports, tapasin is detected in complexes with TAP1/2 in β2-m-deficient Daudi cells, in complexes with calreticulin and the MHC class I HC/β2-m dimer in TAP mutant .174 cells, and in complexes with calreticulin, TAP1/2, and MHC class I HC/β2-m in LCL cell lines (1). Like calnexin, calreticulin is a lumenal ER protein and a member of the lectin-like proteins that recognize partially trimmed, monoglucosylated glycan chains on newly synthesized glycoproteins (32, 33). Recently, it was reported that calnexin and calreticulin are associated with distinct forms of MHC class I molecules (1, 34). A key question is whether TAP-A (or tapasin) functions as a chaperone, like calreticulin, or is selectively required for the TAP-dependent antigen processing. We determined the difference in stability between TAP-A and TAP1/2 complexes on one hand and MHC class I and TAP complexes on the other by measuring the off-rates of TAP-A and MHC class I from TAP1/2. After different chase periods, digitonin lysates of LCL or the β2-m-deficient cell line Daudi were immunoprecipitated with anti-TAP1 antiserum. MHC class I molecules dissociated rapidly from the TAP complex in LCL cells (Fig. 3, lanes 1–5). In contrast, TAP-A was present in immunoprecipitates in roughly stoichiometric amounts with TAP1/2 throughout the chase and did not dissociate from TAP1/2 even after a 3-h chase (Fig. 3, lanes 1–5). TAP-A also was stably associated with TAP1/2 in Daudi cells, in which TAP does not interact with MHC class I (Fig. 3, lanes 11–15). With anti-TAP1 antiserum, two closely migrating species of TAP2 were detected in Daudi cells (Fig. 3, lanes 11–15) for unknown reasons. To exclude the possibility that anti-TAP-1 antiserum cross-reacted with TAP-A, we prepared an anti-TAP-A antiserum. This antiserum specifically detected TAP-A in both immunoprecipitation and Western blot assays (Fig. 3, lanes 6–10 and Fig. 2, A and B). The same pulse–chase experiments with LCL cells were repeated, and the digitonin lysates of each chase point were immunoprecipitated with anti-TAP-A antiserum. The reciprocal immunoprecipitation of both anti-TAP1 and anti-TAP-A antisera gave the same results (Fig. 3, lanes 1–5 and 6–10). Thus, we conclude that, in normal cells, TAP1, TAP2, and TAP-A form a trimeric complex. It has been reported that tapasin is not required for peptide translocation across the ER membrane (19), so the function of TAP-A may be related to the assembly of peptide and the MHC class I HC/β2-m dimer.
Figure 3.
Pulse–chase analysis of the association of class I or TAP-A with TAP1/2 in LCL and Daudi cells. LCL (lanes 1–10) and Daudi cells (lanes 11–15) were labeled for 1 h with [35S]-methionine and chased in an excess of unlabeled methionine for the indicated periods of time before lysis with 1% digitonin. Aliquots of LCL lysates were immunoprecipitated with anti-TAP-1 (lanes 1–5) or anti-TAP-A (lanes 6–10) antiserum. The lysates extracted from Daudi cells were precipitated with anti-TAP1 antiserum (lanes 11–15). The positions of TAP1, TAP2, TAP-A, and MHC class I HC are indicated.
MHC Class I Molecules Associate with Monomeric Forms of the TAP1/2-TAP-A Trimers.
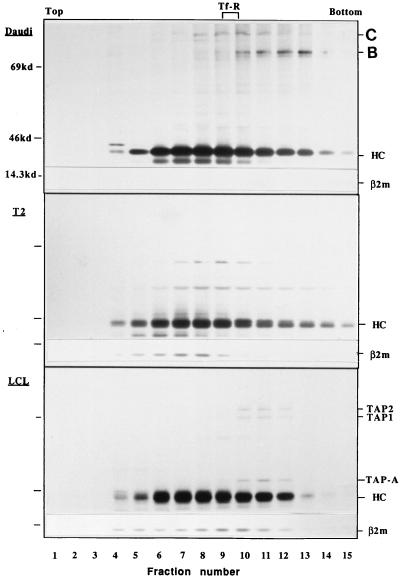
To determine the size of class I-TAP1/2-TAP-A complexes, we subjected digitonin-lysed Daudi, T2, and LCL cell extracts to velocity sedimentation analysis on 8–35% sucrose gradients in the presence of iodoacetamide and digitonin (Fig. 4). The class I HCs were recovered by precipitation with the R425 antibody, which binds to all forms of MHC class I molecules. A crude estimation of the size of the class I species was obtained by comparison with the sedimentation rate of the transferrin receptor (Mr = 180 kDa for the native dimeric species; ref. 27), which was recovered in fractions 9–10. The sedimentation patterns of class I HC from Daudi and T2 cells were very similar. However, there was a clear difference in that HC from Daudi cells sedimenting between fractions 7 and 14 (corresponding roughly to Mr of 100–500 kDa) were complexed with two proteins 80 and 90 kDa in size (Fig. 4, Top, lanes 7–14). These species were identified as Bip and calnexin, respectively (unpublished data). These chaperones were not clearly detected in association with HC from T2 cells in the corresponding fractions (Fig. 4). Association of MHC class I with calreticulin has been reported in T2 cells (1). In this report, calreticulin itself was not observed by autoradiography because of the long half-life of this protein (1). This may explain why we could not identify calreticulin in MHC class I immunoprecipitates in T2 cells. The HC bands recovered from LCL cells in fractions 10–12 had a rather sharp appearance on the SDS gel whereas those in fractions 5–9 were fuzzy. The latter probably represent terminally glycosylated glycans whereas the former are incompletely trimmed, nonsialylated ones. Only these HC species were complexed with TAP trimeric complexes (Fig. 4, lanes 10–12). Again, similar amounts of TAP-A, TAP1, and TAP2 molecules were recovered in the precipitated complex (Fig. 4, lanes 10–12), which sedimented at a position corresponding to ≈200–250 kDa as compared with the transferrin receptor marker. We thus conclude that the class I HC/β2-m dimer associates with the TAP1/2-TAP-A trimer to form a pentameric complex.
Figure 4.
Velocity sedimentation analysis on sucrose gradients of MHC class I and its associated proteins expressed in LCL, Daudi, and T2 cells. T2, Daudi and LCL cells were labeled for 1 h with [35S]-methionine and solubilized in 1% digitonin lysis buffer in the presence of 1.5 μg/ml iodoacetamide. Lysates were fractionated by sedimentation on 8–35% linear sucrose gradients, as described in Materials and Methods. Each gradient was subdivided into 15 fractions from which class I molecules were immunoprecipitated with the anti-class I antibody, R425, reactive with both free and assembled class I HC. The position of the transferrin receptor used as a sedimentation standard is indicated. The positions of the class I HC, TAP1, TAP2, TAP-A, and β2-m, as well as molecular markers, are indicated. The protein species (80 and 90 kDa) in fractions 7–14 of Daudi cells (lanes 7–14) are Bip (B) and calnexin (C), respectively.
ATP-Dependent Interaction of an Antigenic Peptide with TAP-A in Intact Microsomes.
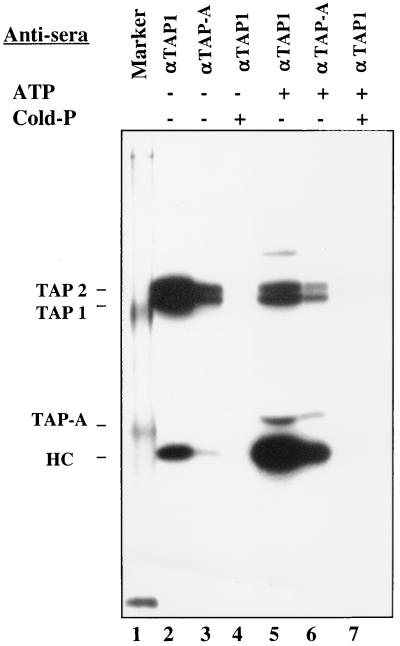
To elucidate the involvement of TAP-A in the processing of antigenic peptides, a photo-cross-linker modified and 125I-labeled HLA-B27 binding peptide (NP383–391, SRYWAIKTR) was used to examine the possible interaction of peptide with TAP1/2, TAP-A, and class I molecules in intact microsomes purified from HLA-B27-transfected CIR cells. After UV cross-linking, microsomes were collected by centrifugation and lysed in 1% digitonin. The cleared lysates were divided into aliquots and precipitated with anti-TAP1 or anti-TAP-A antiserum. Peptide-cross-linked TAP1 and TAP2 were readily detected in the absence of ATP by both anti-TAP1 and anti-TAP-A antisera whereas only a relatively small amount of peptide was cross-linked to class I under these conditions (Fig. 5, lanes 2 and 3). This may be due to the leakage of a small fraction of microsomes. Addition of ATP to the cross-linking reaction resulted in decreased binding of peptide to TAP1/2 (Fig. 5, lanes 5 and 6) due to an increased off-rate (21). In contrast, a large amount of class I molecules were cross-linked (Fig. 5, lanes 5 and 6), a finding in agreement with published results showing ATP-dependent peptide translocation (11–13). In addition, peptide-cross-linked TAP-A was only detected in the presence of ATP (Fig. 5, lanes 5 and 6). These interactions were competed by cold and unmodified peptide (Fig. 5, lanes 4 and 7). It has been reported that tapasin is not required for peptide translocation (19), so the ATP-dependent peptide-binding may be anticipated to occur after peptides have been translocated into the ER lumen and may be related to the peptide loading onto MHC class I molecules.
Figure 5.
Binding of peptide to TAP1, TAP2, TAP-A, and MHC class I in purified microsomes. The 125I-NP383–391-ANB-NOS peptide was mixed with microsomes purified from CIR-B27 cells in the absence (lanes 2–4) or presence of ATP (lanes 5–7). In lanes 4 and 7, the 125I-NP383–391-ANB-NOS peptide was mixed with a 10-fold molar excess of unlabeled and unmodified NP-383–391. The mixtures were incubated for 10 min at 26°C and then transferred to ice and exposed to UV light for 5 min to allow for cross-linking. After cross-linking, microsomes were collected by centrifugation and lysed in 1% digitonin buffer. The cleared lysates were precipitated with anti-TAP1 (lanes 2, 4, 5, and 7) or anti-TAP-A (lanes 3 and 6) antiserum. The immunoprecipitates were analyzed on an SDS gel. Position of TAP1, TAP2, TAP-A, and class I HC are indicated.
The requirement for molecules other than TAP1/2 in TAP-dependent antigen processing became apparent from studies of a mutant cell line, 721.220, in which MHC class I surface expression is deficient, seemingly because of lack of formation of TAP-class I complexes (19). Indeed, a 48-kDa (termed “tapasin”) glycoprotein was found in complexes with TAP1/2, MHC class I HC/β2-m dimer, and calreticulin (1). Tapasin, which seems to be absent from 721.220 cells (1, 20), was suggested to form a bridge between the MHC class I HC and TAP or between HC and calreticulin (1). Recently, it was reported that free β2-m binds to TAP molecules (20). Based on the present and published results, we suggest the possibility that, after dimerization of HC/β2-m, TAP-A binds to the peptide binding site and thereby may prevent binding of irrelevant peptide. When translocated peptides have high affinity for binding to MHC class I, they could displace TAP-A, allowing for the formation of mature peptide-loaded class I. This hypothesis could also explain the functional role of the association of TAP-A (tapasin) with calreticulin and MHC class I HC/β2-m dimers. It could prevent the binding of irrelevant, low affinity peptides until brought into contact with high affinity peptides after MHC class I molecules have bound to TAP. The role of a dilysine motif at the C terminus of TAP-A may be to retain TAP-associated immature MHC class I in the ER or to recycle it back from the intermediate compartment/cis-Golgi until the class I HC/β2-m dimer and peptide have been assembled.
Acknowledgments
We thank Dr. B. Widegren for discussion throughout the cloning of TAP-A. We also thank C. Raynoschek, K. Svensson, and C. Wernstedt for excellent technical support. This study was partially supported by grants from the Swedish Medical Research Council (B96-16X-06498) and the Swedish Cancer Society (0609-B96).
ABBREVIATIONS
- MHC
major histocompatibility complex
- TAP
transporter associated with antigen processing
- HC
heavy chain
- LCL
lymphoblastoid cell lines
- ER
endoplasmic reticulum
- β2-m
β2-microglobulin
Footnotes
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession no. Y13582).
References
- 1.Sadasivan B, Lehner P J, Ortmann B, Spies T, Cresswell P. Immunity. 1996;5:103–114. doi: 10.1016/s1074-7613(00)80487-2. [DOI] [PubMed] [Google Scholar]
- 2.Yewdell J W, Bennink J R. Adv Immunol. 1992;52:1–123. doi: 10.1016/s0065-2776(08)60875-5. [DOI] [PubMed] [Google Scholar]
- 3.Heemels M-T, Ploegh H L. Annu Rev Biochem. 1995;64:463–491. doi: 10.1146/annurev.bi.64.070195.002335. [DOI] [PubMed] [Google Scholar]
- 4.Rammensee H-G. Curr Opin Immunol. 1995;7:85–96. doi: 10.1016/0952-7915(95)80033-6. [DOI] [PubMed] [Google Scholar]
- 5.Goldberg A L, Rock K L. Nature (London) 1992;357:375–379. doi: 10.1038/357375a0. [DOI] [PubMed] [Google Scholar]
- 6.Koopmann J-O, Hämmerling G J, Momburg F. Curr Opin Immunol. 1997;9:80–88. doi: 10.1016/s0952-7915(97)80163-x. [DOI] [PubMed] [Google Scholar]
- 7.Heemels M-T, Schumacher T N M, Wonigeit K, Ploegh H L. Science. 1993;262:2059–2063. doi: 10.1126/science.8266106. [DOI] [PubMed] [Google Scholar]
- 8.Neefjes J J, Momburg F, Hämmerling G J. Science. 1993;261:769–771. doi: 10.1126/science.8342042. [DOI] [PubMed] [Google Scholar]
- 9.Androlewicz M J, Anderson K S, Cresswell P. Proc Natl Acad Sci USA. 1993;90:9130–9134. doi: 10.1073/pnas.90.19.9130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shepherd J C, Schumacher T N M, Ashton-Rickardt P G, Imaeda S, Ploegh H L, Janeway C A, Tonegawa S. Cell. 1993;74:577–584. doi: 10.1016/0092-8674(93)80058-m. [DOI] [PubMed] [Google Scholar]
- 11.Androlewicz M J, Cresswell P. Immunity. 1994;1:7–14. doi: 10.1016/1074-7613(94)90004-3. [DOI] [PubMed] [Google Scholar]
- 12.Wang P, Raynoschek C, Svensson K, Ljunggren H-G. J Biol Chem. 1996;271:24830–24835. doi: 10.1074/jbc.271.40.24830. [DOI] [PubMed] [Google Scholar]
- 13.van Endert P M, Tampe R, Meyer T H, Tisch R, Bach J-F, McDevitt H O. Immunity. 1994;1:491–500. doi: 10.1016/1074-7613(94)90091-4. [DOI] [PubMed] [Google Scholar]
- 14.Such W-K, Cohen-Doyle M F, Fruh K, Wang K, Peterson P A, Williams D B. Science. 1994;264:1322–1326. doi: 10.1126/science.8191286. [DOI] [PubMed] [Google Scholar]
- 15.Ortmann B, Androlewicz M J, Cresswell P. Nature (London) 1994;368:864–867. doi: 10.1038/368864a0. [DOI] [PubMed] [Google Scholar]
- 16.Androlewicz M J, Ortmann B, van Endert P M, Spies T, Cresswell P. Proc Natl Acad Sci USA. 1994;91:12716–12720. doi: 10.1073/pnas.91.26.12716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peace-Brewer A L, Tussey L G, Matsui M, Li G, Quinn D G, Frelinger J A. Immunity. 1996;4:505–514. doi: 10.1016/s1074-7613(00)80416-1. [DOI] [PubMed] [Google Scholar]
- 18.Lewis J W, Neisig A, Neefjes J, Elliott T. Curr Biol. 1996;6:873–883. doi: 10.1016/s0960-9822(02)00611-5. [DOI] [PubMed] [Google Scholar]
- 19.Grandea A G, Androlewicz M J, Athwal R S, Geraghty D E, Spies T. Science. 1995;270:105–108. doi: 10.1126/science.270.5233.105. [DOI] [PubMed] [Google Scholar]
- 20.Solheim J C, Harris M R, Kindle C S, Hansen T H. J Immunol. 1997;158:2236–2241. [PubMed] [Google Scholar]
- 21.Wang P, Gyllner G, Kvist S. J Immunol. 1996;157:213–220. [PubMed] [Google Scholar]
- 22.Kvist S, Hamann U. Nature (London) 1990;348:446–448. doi: 10.1038/348446a0. [DOI] [PubMed] [Google Scholar]
- 23.Kaltoft M-B, Koch C, Uerkvitz W, Hendil K B. Hybridoma. 1992;11:507–517. doi: 10.1089/hyb.1992.11.507. [DOI] [PubMed] [Google Scholar]
- 24.Hellman U, Wernstedt C, Gonez J, Heldin C-H. Anal Biochem. 1995;224:451–455. doi: 10.1006/abio.1995.1070. [DOI] [PubMed] [Google Scholar]
- 25.Saraste J, Palade G E, Farquhar M G. Proc Natl Acad Sci USA. 1986;83:6425–6429. doi: 10.1073/pnas.83.17.6425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Howell K E, Palade G E. J Cell Biol. 1982;92:822–832. doi: 10.1083/jcb.92.3.822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bonnerot C, Marks M S, Cosson P, Robertson E J, Bikoff E K, Germain R N, Bonifacino J S. EMBO J. 1994;13:934–936. doi: 10.1002/j.1460-2075.1994.tb06338.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.von Heijne G. J Mol Biol. 1985;184:99–105. doi: 10.1016/0022-2836(85)90046-4. [DOI] [PubMed] [Google Scholar]
- 29.Kyte J, Doolittle R F. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 30.Nilsson T, Jackson M, Peterson P A. Cell. 1989;58:707–718. doi: 10.1016/0092-8674(89)90105-0. [DOI] [PubMed] [Google Scholar]
- 31.Jackson M R, Nilsson T, Peterson P A. EMBO J. 1990;9:3153–3162. doi: 10.1002/j.1460-2075.1990.tb07513.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Helenius A. Mol Biol Cell. 1994;5:253–255. doi: 10.1091/mbc.5.3.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hammond C, Helenius A. Curr Opin Cell Biol. 1995;7:523–525. doi: 10.1016/0955-0674(95)80009-3. [DOI] [PubMed] [Google Scholar]
- 34.van leeuwen J E, Kearse K P. Proc Natl Acad Sci USA. 1996;24:13997–4001. doi: 10.1073/pnas.93.24.13997. [DOI] [PMC free article] [PubMed] [Google Scholar]