Abstract
High endothelial venules (HEV) are specialized postcapillary venules found in lymphoid organs and chronically inflamed tissues that support high levels of lymphocyte extravasation from the blood. One of the major characteristics of HEV endothelial cells (HEVEC) is their capacity to incorporate large amounts of sulfate into sialomucin-type counter-receptors for the lymphocyte homing receptor L-selectin. Here, we show that HEVEC express two functional classes of sulfate transporters defined by their differential sensitivity to the anion-exchanger inhibitor 4,4′-diisothiocyanostilbene-2,2′-disulfonic acid (DIDS), and we report the molecular characterization of a DIDS-resistant sulfate transporter from human HEVEC, designated SUT-1. SUT-1 belongs to the family of Na+-coupled anion transporters and exhibits 40–50% amino acid identity with the rat renal Na+/sulfate cotransporter, NaSi-1, as well as with the human and rat Na+/dicarboxylate cotransporters, NaDC-1/SDCT1 and NaDC-3/SDCT2. Functional expression studies in cRNA-injected Xenopus laevis oocytes showed that SUT-1 mediates high levels of Na+-dependent sulfate transport, which is resistant to DIDS inhibition. The SUT-1 gene mapped to human chromosome 7q33. Northern blotting analysis revealed that SUT-1 exhibits a highly restricted tissue distribution, with abundant expression in placenta. Reverse transcription–PCR analysis indicated that SUT-1 and the diastrophic dysplasia sulfate transporter (DTD), one of the two known human DIDS-sensitive sulfate transporters, are coexpressed in HEVEC. SUT-1 and DTD could correspond, respectively, to the DIDS-resistant and DIDS-sensitive components of sulfate uptake in HEVEC. Together, these results demonstrate that SUT-1 is a distinct human Na+-coupled sulfate transporter, likely to play a major role in sulfate incorporation in HEV.
Keywords: lymphocyte recirculation, L-selectin, sulfation, sulfate transporter
Patrolling the body in search for foreign antigens, lymphocytes continuously recirculate from blood, through lymphoid and other tissues, and back through the efferent lymphatics to the blood (1, 2). The first critical step in lymphocyte migration from circulation into lymphoid tissues is the adhesion of lymphocytes to specialized postcapillary vascular sites called high endothelial venules (HEV) (3). In contrast to the endothelial cells from other vessels, the endothelial cells of HEV have a plump, almost cuboidal, appearance, express specialized ligands for the lymphocyte homing receptor L-selectin, and are able to support high levels of lymphocyte extravasion from the blood (1, 3, 4). A unique characteristic of the HEV endothelium that has intrigued many investigators over the past 20 years is the capacity of HEV endothelial cells (HEVEC), both in humans and in rodents, to incorporate large amounts of inorganic sulfate (5). Interestingly, HEV-like vessels that develop in chronically inflamed nonlymphoid tissues exhibit a similar ability to incorporate high levels of radioactive sulfate, and this property of the HEV endothelium has been shown to be closely related to the concentration of lymphocytes in the perivascular infiltrates (6). More recently, the functional implication for this major biosynthetic activity of HEV and HEV-like vessels has been found when it was discovered that large amounts of sulfate are incorporated into sialomucin-type L-selectin counter-receptors (7–9). Sulfate residues were shown to be essential for recognition of HEV sialomucins GlyCAM1 and CD34 by L-selectin and HEV-specific monoclonal antibody MECA-79 (9–11), an antibody that blocks L-selectin-dependent lymphocyte adhesion to HEV in vitro and in vivo (12).
Genes important for sulfation of L-selectin ligands in HEV include the genes encoding PAPS synthetase, a bifunctional enzyme catalyzing synthesis of PAPS (3′-phosphoadenosine-5′-phosphosulfate), the activated sulfate donor used by all sulfotransferases (13), and sulfotransferases of two specificities, N-acetylglucosamine-6-O-sulfotransferase(s), and galactose-6-O-sulfotransferase(s) (14–16). Although the transfer of sulfate from PAPS to HEV sialomucins by sulfotransferases appears to be the most specific step in the pathway, sulfation of HEV sialomucins may also be controlled at earlier steps. For example, the observation that sulfate incorporation is the rate-limiting step for sulfation in cartilage (17, 18) suggests that sulfate transport could play a major role in the control of HEV sialomucins sulfation in vivo.
HEV sulfate transporter(s) have not yet been molecularly characterized. However, two families of vertebrate sulfate transporters, which play a role in sulfate incorporation in other tissues, have recently been defined. The prototype members of these families, SAT-1 and NaSi-1, were initially described in rats (19, 20). SAT-1 is a member of the superfamily of anion exchangers functioning as a Na+-independent sulfate transporter in rat hepatocytes and being sensitive to the anion-exchanger inhibitor 4,4′-diisothiocyanostilbene-2,2′-disulfonic acid (DIDS) (20). NaSi-1 is a member of the superfamily of Na+-coupled transporters functioning as a DIDS-resistant Na+-dependent sulfate transporter in rat kidney and small intestine (19). In humans, no Na+-dependent sulfate transporter has yet been found. In contrast, two members of the superfamily of anion exchangers, human DTD and DRA, have been described (17, 21), that contain 12 membrane-spanning domains and are sensitive to DIDS (21, 22). Both DTD and DRA have been implicated in the pathogenesis of human diseases. While DTD is mutated in three different chondrodysplasias (diastrophic dysplasia, atelosteogenesis type II, and achondrogenesis type IB) (17, 18), DRA is mutated in congenital chloride diarrhea (23). DTD mutations result in undersulfation of cartilage proteoglycans and thus lead to severe developmental defects of the skeleton (17, 18). It is unknown whether DTD and DRA are expressed in HEV.
To further characterize the molecular mechanisms controlling lymphocyte extravasation through HEV, we have started to clone relevant genes involved in sulfation of L-selectin ligands (13). In this study, we report the molecular cloning and functional characterization of SUT-1, a sulfate transporter from human HEVEC. Sequence comparisons revealed that SUT-1 belongs to the family of Na+-coupled sulfate/dicarboxylate transporters. Functional analysis in Xenopus laevis oocytes showed that SUT-1 encodes a DIDS-resistant Na+-dependent sulfate transporter. In HEVEC, SUT-1 is coexpressed with the anion exchanger DTD. SUT-1 and DTD are likely to be two important mediators of the sulfate transport activities of human HEV in vivo.
Materials and Methods
Analysis of Sulfate Transport Activities in Cultured HEVEC.
HEVEC were isolated and cultured as described (24), and sulfate uptake assays were performed essentially as reported by Satoh et al. (22). The cells were washed three times in ion-free washing buffer (300 mM sucrose, 10 mM Hepes/Tris, pH 7.5) and then incubated in 100 μl of uptake buffer [149 mM salts, 1 mM [35S]sulfate (40 μCi/ml; 1 μCi = 37 kBq), 10 mM Hepes/Tris, pH 7.5] for 5 min at 37°C. Uptake was stopped by washing the cells three times with ice-cold washing buffer containing 5 mM K2SO4, and the cells were lysed in 100 μl of 1% Triton X-100. Cell-associated radioactivity was determined by scintillation counting.
cDNA Cloning and DNA Sequencing.
Screening of an HEV-derived cDNA library (25) was performed by moderate-stringency hybridization using as a probe a 32P-labeled human brain expressed sequence tag (EST55985, GenBank accession no. AA349378), homologous to rat Na+-dependent sulfate transporter NaSi-1 (19). Duplicate filter lifts (Amersham) from 20 plates, each containing 50,000 plaques, were hybridized at 42°C overnight in 50% formamide, 5× Denhardt’s solution, 5× standard saline/citrate (SSC), 0.5% SDS, 50 μg/ml tRNA, and 50 μg/ml herring sperm DNA. The membranes were then washed with 1× SSC/0.1% SDS at room temperature (twice for 15 min) and 42°C (twice for 15 min). Plaques from positive clones were picked, and the corresponding pBluescript II SK− phagemids were rescued by in vivo excision using Exassist helper phage (Stratagene). Five cDNA clones were thus obtained and sequenced by using a Sequenase DNA sequencing kit with Sequenase version 2.0 T7 DNA polymerase (Amersham). The 5′ part of the SUT-1 cDNA was obtained by rapid amplification of cDNA ends (RACE) using Marathon-ready placenta cDNAs (CLONTECH) and the gene-specific antisense primer SUT6 (5′-GGAAGCTGAAGAGGAACCAGGTGCC-3′). The two parts of the SUT-1 cDNA were then ligated at an internal PvuII site. The resulting full-length SUT-1 cDNA (2.9-kb cDNA) was further sequenced on both strands with custom oligonucleotide primers, synthesized on an Applied Biosystems synthesizer. The program blast (26) was used to compare SUT-1 nucleic and amino acid sequences with all sequences present in the National Center for Biotechnology Information (NCBI) nonredundant nucleic acid and protein databases. Protein sequences of Na+-coupled sulfate/dicarboxylate cotransporters were aligned with the program clustal w (27), and conserved regions were highlighted with boxshade. The hypothetical membrane topology model of human SUT-1 was predicted with the program TMPred from the ExPaSy World Wide Web molecular biology server (Geneva).
Northern Blot and Reverse Transcription (RT)-PCR Analysis.
For Northern blot analysis, blots of poly(A)+ RNA from multiple human tissues were hybridized according to the manufacturer’s instructions (CLONTECH). A 1.0-kb BamHI–PstI fragment encompassing the 3′ end of the human SUT-1 cDNA was 32P-labeled by random priming (GIBCO/BRL) and used as probe in the Northern blots. For RT-PCR analysis, the Smart PCR cDNA synthesis kit (CLONTECH) was first used to generate full-length cDNAs from 1 μg of total RNA of placenta or cultured human tonsil-derived HEVEC (24) that were 98% MECA-79-positive on day 2 (HEVEC-2D) and <1% MECA-79-positive by day 8 (HEVEC-8D). DTD (17), DRA (21), and SUT-1 sulfate transporters and PAPS synthetase 1 control (13) fragments were then selectively amplified with the following primers: DTD1, 5′-GTGTGGGCTCACTCATCACTACCTGG-3′; DTD2, 5′-TGGGATTGCACTGAGCCAGCAGAACC-3′; DRA1, 5′-GGTATCAGCACAGGGATTGTGGCCG-3′; DRA2, 5′-CCCAATGCTAAAGCTGCCAGGACGG-3′; SUT6, 5′-GGAAGCTGAAGAGGAACCAGGTGCC-3′; SUT14, 5′-CCGAGCAGCAGCAGCAAAGGTGTGG-3′; PAPS7, 5′-GGTGATGGAACAAGGAGATTGGCTG-3′; and PAPS8, 5′-GAGTGACTGGGTTAACAGCCTAAGC-3′. All PCRs were performed with 40 cycles (94°C, 30 s; 68°C, 2 min) in 25 μl of reaction buffer containing 200 μM dNTP, 10 pmol of each primer, 1 μl of diluted HEVEC and placenta cDNAs (1/5 dilution), and Advantage KlenTaq Polymerase Mix (CLONTECH).
In Vitro Transcription and Translation.
For in vitro transcription and translation experiments, the entire SUT-1 ORF was amplified by PCR and inserted into the pSP64 Poly(A) vector (Promega). For in vitro transcription, the pSP64-SUT-1 plasmid (5 μg) was linearized with EcoRI, and capped SUT-1 cRNA was synthesized by using SP6 RNA polymerase (Promega) in the presence of the capping analogue m7G(5′)ppp(5′)G (Roche Biochemicals) and RNasin ribonuclease inhibitor (Promega). Capped SAT-1 cRNA was similarly synthesized with T7 RNA polymerase and plasmid pSPORT1-SAT-1 (20) linearized with PvuII. In vitro translation experiments in rabbit reticulocyte lysate were performed according to the supplier’s instructions (Promega).
Injection into Xenopus Oocytes and Sulfate Transport Assay.
The handling of Xenopus oocytes and the sulfate transport assay were carried out as previously described (19–21). Oocytes were injected with 50 nl of water, with or without SUT-1 (or SAT-1) cRNA (200–500 ng/μl) and, after 2 days, uptake of [35S]sulfate (carrier free, Amersham) was measured in the presence or in the absence of Na+. Briefly, five to eight oocytes were washed twice in sulfate-free uptake solution (100 mM NaCl or 100 mM choline chloride, 2 mM KCl, 1 mM CaCl2, 1 mM MgCl2, and 10 mM Hepes/Tris, pH 7.5) and then incubated for 1 hr at 25°C in 500 μl of uptake solution containing [35S]sulfate (40 μCi/ml) and 1 mM K2SO4, in the presence or absence of 1 mM DIDS (Sigma). After the indicated time period, the uptake was stopped by washing the oocytes three times in ice-cold uptake solution supplemented with 5 mM K2SO4. Single oocytes were then lysed in 0.5 ml of 10% SDS, and the oocyte-associated radioactivity was determined in a Tri-Carb 2100 liquid scintillation counter (Packard).
Results
Human HEVEC Coexpress DIDS-Sensitive and DIDS-Resistant Sulfate Transport Activities.
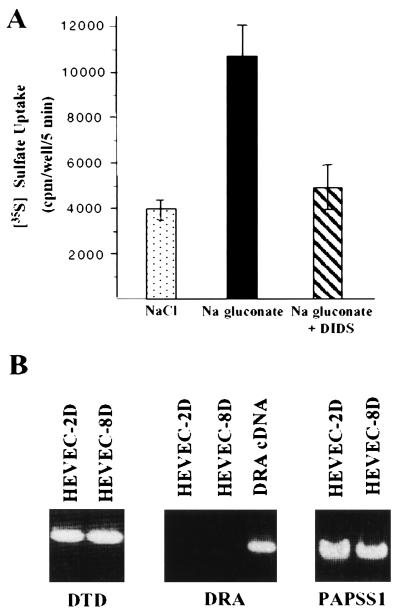
To explore the nature of sulfate transport in HEVEC, we performed sulfate incorporation assays in cultured cells. Confluent HEVEC cultures were incubated in [35S]sulfate uptake media containing NaCl or sodium gluconate, as described by Satoh et al. (22) for primary rat growth plate chondrocytes. In the presence of sodium gluconate, HEVEC exhibited significantly higher sulfate uptake activity than in NaCl buffer (Fig. 1A). To determine the relative contribution of the two functional classes of sulfate transporters (anion exchangers and Na+/sulfate cotransporters), we performed experiments with the anion-exchanger inhibitor DIDS. We found that DIDS (1 mM) inhibits 50% of the sulfate transport activity of cultured HEVEC (Fig. 1A), indicating that HEVEC express functional sulfate transporters of the anion-exchanger class. Since treatment with DIDS did not result in complete inhibition of sulfate transport, these data provided strong evidence that sulfate incorporation into HEVEC is mediated by both DIDS-sensitive anion exchangers and DIDS-resistant sulfate transporters. To identify the transporter(s) responsible for the DIDS-sensitive activity, we performed RT-PCR analysis of cultured HEVEC for the two known human DIDS-sensitive anion exchangers, DTD and DRA (Fig. 1B). We found that DTD, but not DRA, is expressed in HEVEC freshly purified from human tonsils (HEVEC-2D) and its expression is maintained after 8 days in culture (HEVEC-8D). These expression data suggested a major role for DTD in the DIDS-sensitive sulfate transport activity of HEVEC.
Figure 1.
HEVEC express both DIDS-resistant and DIDS-sensitive sulfate transport activities. (A) Analysis of sulfate incorporation into cultured HEVEC. Primary human tonsillar HEVEC were grown to confluency in 96-well plates. Uptake of 1 mM [35S]sulfate (40 μCi/ml) was measured for 5 min at 37°C in a Na+ chloride or Na+ gluconate buffer in the presence or absence of 1 mM DIDS. Values are the means ± SD for triplicate wells and are representative of at least two similar experiments. (B) HEVEC express the DIDS-sensitive sulfate transporter DTD. RT-PCR analysis of the expression of human sulfate transporters DTD and DRA in HEVEC cultured for 2 days (HEVEC-2D) or 8 days (HEVEC-8D) was performed as described in the text. Amplifications of DRA cDNA and PAPS synthetase 1 gene (PAPSS1) were used as controls in these experiments.
Molecular Cloning of SUT-1, a Member of the Na+-Coupled Transporter Family.
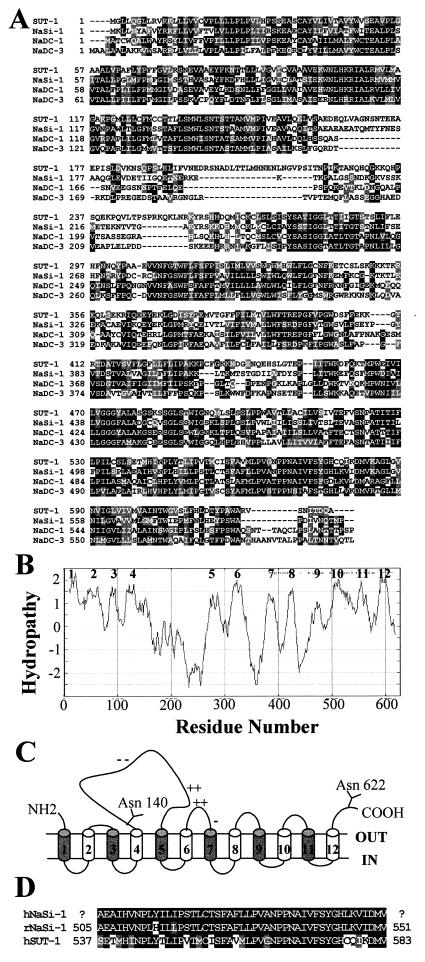
In an attempt to identify the transporter responsible for the DIDS-resistant sulfate transport activity, possible expression in HEVEC of novel human sulfate transporters of the Na+-coupled transporter family was examined. Screening of duplicate filter lifts from an HEVEC cDNA library (25), by moderate stringency hybridization with a human infant brain EST probe homologous to the rat Na+-dependent sulfate transporter NaSi-1, resulted in the identification of five positive cDNA clones corresponding to a unique mRNA species. Since the longest cDNA did not appear to contain the 5′ end, we performed 5′ RACE-PCR to obtain a full-length 2.9-kb cDNA. This 2.9-kb cDNA contains a single ORF of 1881 nt that translates into a putative 627-amino acid protein, tentatively designated SUT-1 for sulfate transporter 1 (Fig. 2A). The ORF is flanked by a long 5′-untranslated sequence (678 nt) and a 3′-untranslated sequence containing a consensus polyadenylation signal (AATAAA). The SUT-1 protein sequence contains three potential N-glycosylation sites (Asn-84, Asn-140, and Asn-622) and a leucine zipper motif (residues 8–29) that could participate in homodimerization of the transporter. Hydropathy analysis of the primary amino acid sequence shows that the protein is highly hydrophobic, with 12 putative transmembrane domains (Fig. 2B). When human SUT-1 is modeled to accommodate two of the three putative N-glycosylation sites toward the extracytoplasmic side (the third site is in a transmembrane region), both the amino terminus and the carboxyl terminus of the protein are directed toward the extracellular side of the cell membrane (Fig. 2C). This is, however, only a hypothetical model, and further experimentation is needed to confirm the membrane topology of human SUT-1.
Figure 2.
Predicted amino acid sequence of human SUT-1. (A) Amino acid sequence alignment of human SUT-1 with other members of the family of Na+-coupled anion transporters, rat Na+/sulfate cotransporter NaSi-1 (GenBank no. U08031), human Na+/dicarboxylate cotransporter NaDC-1 (GenBank no. U26209), and rat Na+/dicarboxylate cotransporter NaDC-3/SDCT2 (GenBank no. AF081825). Regions of identity (dark shading) and similarity (light shading) are indicated. Dashed lines represent gaps introduced to align sequences. (B) Hydropathy analysis of the deduced SUT-1 amino acid sequence by the algorithm of Kyte and Doolittle with a window of 19. The 12 potential membrane-spanning domains are indicated by numbers. (C) Hypothetical membrane topology model of human SUT-1 based on hydropathy analysis. The numbering of the 12 putative transmembrane domains corresponds to B. Clusters of charged amino acids (+ or −) and potential N-glycosylation sites (Y) are indicated. (D) Comparison of amino acids 537–583 of human SUT-1 with homologous sequences from rat NaSi-1 (amino acids 505–551) and a putative human NaSi-1 ortholog coded by EST GenBank no. AI491922.
Comparison of the SUT-1 predicted protein sequence with sequence databases revealed significant homologies with the rat renal Na+/sulfate cotransporter NaSi-1 (49% identity), the human Na+/dicarboxylate cotransporter NaDC-1 (41% identity), and the rat Na+/dicarboxylate cotransporter NaDC-3/SDCT2 (38% identity) (Fig. 2A). This analysis also identified four putative proteins of Caenorhabditis elegans (SP:Q93655, SP:P46556; SP:Q21339; and SP:P32739) that exhibit 25–30% identity with human SUT-1. In contrast, no homologous sequence was found in plants or yeasts. In addition, database comparisons revealed that SUT-1 and the Na+-coupled sulfate/dicarboxylate transporter family are not related to any of the other transporter superfamilies, especially not to other cloned Na+/solute cotransport systems (Na+/Pi, Na+/HCO3−, Na+/Cl−, and Na+/glucose cotransporters). These sequence analyses strongly suggested that the isolated SUT-1 HEV cDNA encodes a distinct human member of the family of Na+-coupled sulfate/dicarboxylate transporters. However, these sequence comparisons did not allow us to conclude that this transporter was a human equivalent of the rat Na+/sulfate cotransporter NaSi-1 because we found evidence in the dbEST database for the existence of a putative human NaSi-1 ortholog, coded by EST no. AI491922. This putative human NaSi-1 exhibits 95% identity with rat NaSi over 47 residues, whereas human SUT-1 has only 63% identity with rat NaSi-1 in the same region (Fig. 2D). Although a slightly higher level of homology with NaSi-1 (49%) than NaDC-1 (41%) suggested that SUT-1 was a sulfate transporter, it remained equally possible that this human protein was involved in transport of dicarboxylates or other anions, rather than sulfate.
SUT-1 Is a Na+-Dependent Sulfate Transporter.
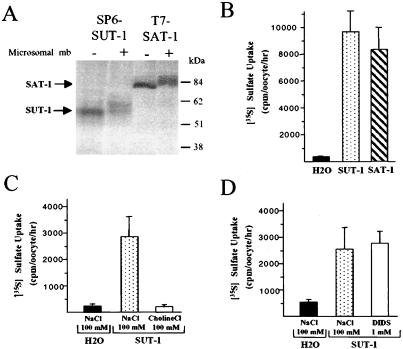
To determine whether the isolated SUT-1 cDNA encoded a bona-fide sulfate transporter, we performed functional expression studies in Xenopus laevis oocytes. First, expression constructs for SUT-1 and the rat sulfate transporter SAT-1 (used as a control) were analyzed by in vitro translation in rabbit reticulocyte lysate. The primary translation products of human SUT-1 and rat SAT-1 had apparent molecular masses of 55 kDa and 80 kDa, respectively, which were shifted to 60 kDa and 85 kDa in the presence of pancreatic microsomes, suggesting some core glycosylation (Fig. 3A). We then analyzed sulfate uptake into Xenopus oocytes after injection of SUT-1 (or control SAT-1) cRNA. We found that injection of SUT-1 (or SAT-1) cRNA into oocytes led to >30-fold stimulation of sulfate uptake, compared with water-injected oocytes, which displayed only low [35S]sulfate uptake (Fig. 3B). Replacement of Na+ by choline in the uptake buffer abolished sulfate transport, demonstrating that SUT-1 is a Na+-dependent sulfate transporter (Fig. 3C). To further characterize SUT-1 cRNA-induced sulfate uptake, we studied its sensitivity to DIDS. We observed no effect of DIDS on the SUT-1-mediated Na+/sulfate cotransport activity (Fig. 3D), confirming that SUT-1 encodes a Na+-coupled transport system and not an anion exchanger. Collectively, these data indicate that SUT-1 is a human Na+/sulfate cotransporter.
Figure 3.
Expression and functional characterization of human SUT-1 in Xenopus laevis oocytes. (A) In vitro translation of human SUT-1 and rat SAT-1 cDNAs. pSP64-SUT-1 (SP6-SUT-1) and pSPORT-SAT-1 (T7-SAT-1) expression vectors were subjected to in vitro translation in the presence (+) or absence (−) of pancreatic microsomal membranes by using a rabbit reticulocyte lysate translation system. The translation products were analyzed directly by SDS/10% polyacrylamide gel electrophoresis and autoradiography. (B) SUT-1-mediated sulfate uptake in cRNA-injected oocytes. Oocytes were injected with 50 nl of water, with or without SUT-1 (or SAT-1) cRNA (25 ng per oocyte). Two days after injection, 1-hr uptake of 1 mM [35S]sulfate was measured at 25°C in the presence of Na+ (100 mM NaCl). (C) Na+ dependency of SUT-1-mediated sulfate uptake. Sulfate uptake in SUT-1 cRNA-injected oocytes (10 ng per oocyte) was measured in the presence (100 mM NaCl) or absence (100 mM choline chloride) of Na+. (D) DIDS resistance of SUT-1-mediated sulfate uptake. Sulfate uptake in SUT-1 cRNA-injected oocytes (10 ng per oocyte) was measured in the presence (1 mM DIDS) or absence (100 mM NaCl) of DIDS. In B–D, data are shown as means ± SD for five oocytes per condition and are representative of at least two similar experiments.
Chromosomal Localization and Tissue Distribution of Human SUT-1.
The chromosomal location of the SUT-1 gene was determined in two different homology searches. Comparison of the SUT-1 sequence with the National Center for Biotechnology Information (NCBI) database of Sequence Tagged Sites resulted in the identification of a human sequence tag SHGC-11392 (GenBank accession no. G14541) corresponding to nucleotides 2579–2833 of the SUT-1 cDNA. SHGC-11392 is located on human chromosome 7 at Genomic Database locus D7S509, which maps to 7q33 close to 7q32. We confirmed these mapping data by searching the NCBI UniGene collection for ESTs identical to the SUT-1 cDNA sequence. We identified the UniGene cluster Hs. 9098 that contains two different ESTs identical to SUT-1 (A005R34, Cda1bf12) mapping between markers D7S500 and D7S509 on chromosome 7 region q33. Together, these data indicate that human SUT-1 is located at 7q33 close to 7q32.
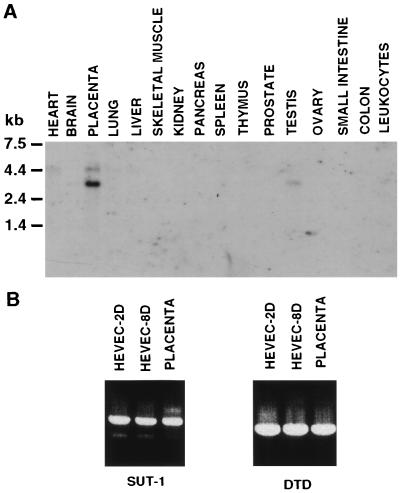
To determine the tissue distribution of SUT-1 mRNA, we performed Northern blot analysis of 16 different adult human tissues (Fig. 4A). A strong 2.9-kb mRNA band, corresponding to the size of the SUT-1 cDNA (2900 bp), was detected predominantly in placenta, in addition to a longer transcript of 4.5 kb. Although expressed at lower levels, the 2.9-kb and 4.5-kb mRNAs were also detected in testis and heart, respectively. The restricted tissue distribution of SUT-1 mRNA could also be deduced from the presence in dbEST and TIGR EST databases of very few SUT-1 ESTs coming from only two different tissue sources, infant brain (6 ESTs) and placenta (3 ESTs). The presence of SUT-1 mRNAs or ESTs in very few tissues suggests that SUT-1 has a highly restricted tissue distribution in the human body.
Figure 4.
Analysis of SUT-1 mRNA expression in human tissues and cultured HEVEC. (A) Northern blot. Each lane contains 2 μg of poly(A)+ RNA isolated from the indicated human tissues. The blot was hybridized, under high-stringency conditions, with a 32P-labeled SUT-1 cDNA probe, and exposed at −70°C for 72 hr. (B) RT-PCR analysis of the expression of SUT-1 in HEVEC cultured for 2 days (HEVEC-2D) or 8 days (HEVEC-8D) was performed as described in the text. Placenta tissue sample and amplification of DTD were used as controls.
To analyze SUT-1 mRNA expression in human tonsillar HEVEC, we performed RT-PCR analysis using oligonucleotides specific for DTD as control (Fig. 4B). We found that SUT-1 is expressed in tonsillar HEVEC cultured for 2 days (HEVEC-2D) or 8 days (HEVEC-8D), at levels comparable with those found in placenta. These results indicate that HEVEC coexpress transcripts for the two functional classes of human sulfate transporters, the Na+-dependent sulfate transporter SUT-1 and the Na+-independent anion exchanger DTD.
Discussion
Incorporation of large amounts of sulfate into functional L-selectin ligands is a major biosynthetic activity of HEV, which contributes significantly to the uniqueness of the HEV ligands. Therefore, characterization of the genes and molecular mechanisms involved in sulfation of L-selectin counter-receptors will provide a better understanding of the mechanisms controlling the recruitment and migration of lymphocytes through HEV. In this study, we report the molecular cloning and functional characterization of a sulfate transporter, designated SUT-1, that is expressed in human HEV. Analysis of the sequence and predicted secondary structure revealed that SUT-1 is a member of the Na+-coupled sulfate/dicarboxylate transporter family. To our knowledge, SUT-1 is the first Na+/sulfate cotransporter to be described in humans. Using a Xenopus laevis oocyte expression system, we demonstrated that SUT-1-mediated sulfate uptake is absolutely dependent on Na+ but resistant to inhibition by the potent anion-exchanger inhibitor DIDS. In addition, we showed that SUT-1 and the Na+-independent DIDS-sensitive anion exchanger DTD are coexpressed in HEVEC, supporting the possibility that these two sulfate transporters are important mediators of the DIDS-resistant and DIDS-sensitive components of sulfate uptake in HEVEC.
SUT-1 Is a Distinct Human Na+-Dependent Sulfate Transporter.
SUT-1 exhibits significant similarities (40–50% identity) with rat NaSi-1 and two other members of the Na+-coupled sulfate/dicarboxylate transporter family, human and rat NaDC-1/SDCT1 (28) and rat NaDC-3/SDCT2 (29, 30). Despite its homology with NaSi-1, human SUT-1 appears to be a distinct member of this family rather than the ortholog of rat NaSi-1. We found evidence in the dbEST database for the existence of a putative human NaSi-1 ortholog that exhibits significantly better homology with rat NaSi-1 (95% identity over 47 residues) than SUT-1 (63% identity with rat NaSi-1 in the same region). In addition, SUT-1 and rat NaSi-1 have very different expression patterns. NaSi-1 is specifically expressed in kidney and small intestine (19), whereas SUT-1 mRNA is found predominantly in placenta (in addition to HEVEC) but is not expressed in kidney and small intestine. Therefore, similarly to the Na+-coupled dicarboxylate transporters, for which at least two different members have been defined in rat (NaDC-1 and NaDC-3), multiple Na+-coupled sulfate transporters may exist in a single species. Confirmation of this prediction will require molecular characterization of the SUT-1 ortholog in rat or cloning the full-length cDNA corresponding to the human NaSi-1 ortholog, partly defined by the EST identified in this study (Fig. 2D).
Relatively little information is available regarding the structure of Na+-coupled sulfate/dicarboxylate transporters, but members of this family are likely to exhibit strong structural similarities. On the basis of hydropathy analysis, which predicts that SUT-1 contains 12 hydrophobic regions long enough to span the membrane as α-helices, we propose a membrane topology model of human SUT-1 with 12 putative transmembrane domains. This model predicts an extracellular location of both amino and carboxyl termini. A similar model has recently been proposed for rat NaDC-3 (30). In contrast, a different model with only 11 transmembrane domains has been suggested for NaDC-1 and SDCT2/NaDC-3 (28, 29). The two models agree on the extracellular location of the carboxyl terminus, which contains one or two consensus sites for N-glycosylation. One of these sites is known to be glycosylated in NaDC-1 (28), suggesting that the corresponding site in SUT-1 (Asn-622) might also be utilized. The size increase observed after in vitro translation of the SUT-1 cDNA in the presence of microsomes (Fig. 3A) is consistent with this possibility. However, elucidation of the membrane topology and secondary structure of SUT-1 as well as other members of the Na+-coupled sulfate/dicarboxylate transporter family will require further studies.
Northern blot analysis, EST database searches, and RT-PCR analysis revealed expression of SUT-1 mRNA in placenta, in addition to HEVEC. The preferential expression of SUT-1 in placenta suggests that SUT-1 might be involved in Na+-coupled sulfate transport into the placental trophoblast, and thus participate in the active transport of sulfate from mother to fetus across the placenta, to serve as “metabolic fuel” for fetal utilization. A similar placental function has recently been proposed for the high-affinity dicarboxylate transporter NaDC-3, another member of the Na+-coupled sulfate/dicarboxylate transporter family (30).
SUT-1-Mediated Sulfate Uptake and Sulfation of L-Selectin Ligands in HEV.
The capacity of the HEV endothelium to incorporate large amounts of sulfate probably contributes significantly to the extensive sulfation of O-linked carbohydrates from HEV sialomucins. Sulfate incorporation into HEVEC is the first essential step in the HEV sulfation pathway, providing high levels of inorganic sulfate to PAPS synthetase (13) for PAPS production. PAPS is then transferred to the Golgi apparatus by PAPS translocase for use by the recently characterized HEV N-acetylglucosamine-6-O- and galactose-6-O-sulfotransferases (14–16). By modulating sulfate incorporation into HEV, SUT-1 and DTD could play a major role in sulfation of L-selectin counter-receptors and regulation of lymphocyte migration through HEV in organized lymphoid tissues and through HEV-like vessels at sites of chronic inflammation. The coexpression in HEVEC of the two functional classes of sulfate transporters—i.e., Na+-coupled transporters (SUT-1) and anion exchangers (DTD)—may allow efficient sulfate incorporation under various physiological conditions because these two classes of transporters have very different modes of regulation (19, 20). No immunological phenotype has yet been described in diastrophic dysplasia patients, suggesting that another sulfate transporter such as SUT-1 might compensate for the absence of DTD expression in HEVEC. In agreement with this possibility, we found that cultured HEVEC exhibit considerable sulfate incorporation activity in the presence of the anion-exchanger inhibitor DIDS, which is known to block DTD function completely (22). Further studies will be required to determine the relative contribution of SUT-1 and DTD to sulfate incorporation into HEV in vivo. Also, the possibility that HEVEC may express other “SUT-1-like” or “DTD-like” sulfate transporters should also be explored. Nevertheless, our results demonstrate that SUT-1 is a distinct Na+-coupled sulfate transporter that is likely to play a key role in sulfation of L-selectin counter-receptors in HEV.
Acknowledgments
We are grateful to Benoit Roger (Institut de Pharmacologie et de Biologie Structurale, Toulouse) for kindly providing Xenopus oocytes. We thank Drs. Bruno Hagenbuch and Peter J. Meier (University Hospital, Zurich, Switzerland) for their gift of rat SAT-1 cDNA. Special thanks go to members of the vascular biology laboratory (Institut de Pharmacologie et de Biologie Structurale, Toulouse) for helpful discussions. This work was supported by grants from Fondation de France, Centre National de la Recherche Scientifique, Région Midi-Pyrénées, Association pour la Recherche sur le Cancer, the Research Council of Norway, and the Norwegian Cancer Society.
Abbreviations
- DIDS
4,4′-diisothiocyanostilbene-2,2′-disulfonic acid
- EST
expressed sequence tag
- HEV
high endothelial venules
- HEVEC
HEV endothelial cells
- PAPS
3′-phosphoadenosine-5′-phosphosulfate
- RT-PCR
reverse transcription–PCR
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF169301).
References
- 1.Marchesi V T, Gowans J L. Proc R Soc London B. 1964;159:283–290. doi: 10.1098/rspb.1964.0002. [DOI] [PubMed] [Google Scholar]
- 2.Butcher E C, Picker L J. Science. 1996;272:60–66. doi: 10.1126/science.272.5258.60. [DOI] [PubMed] [Google Scholar]
- 3.Girard J P, Springer T A. Immunol Today. 1995;16:449–457. doi: 10.1016/0167-5699(95)80023-9. [DOI] [PubMed] [Google Scholar]
- 4.Anderson A O, Anderson N D. Immunology. 1976;31:731–748. [PMC free article] [PubMed] [Google Scholar]
- 5.Andrews P, Milsom D W, Ford W L. J Cell Sci. 1982;57:277–292. doi: 10.1242/jcs.57.1.277. [DOI] [PubMed] [Google Scholar]
- 6.Freemont A J. J Pathol. 1988;155:225–230. doi: 10.1002/path.1711550308. [DOI] [PubMed] [Google Scholar]
- 7.Lasky L A, Singer M S, Dowbenko D, Imai Y, Henzel W J, Grimley C, Fennie C, Gillett N, Watson S R, Rosen S D. Cell. 1992;69:927–938. doi: 10.1016/0092-8674(92)90612-g. [DOI] [PubMed] [Google Scholar]
- 8.Baumhueter S, Singer M S, Henzel W, Hemmerich S, Renz M, Rosen S D, Lasky L A. Science. 1993;262:436–438. doi: 10.1126/science.7692600. [DOI] [PubMed] [Google Scholar]
- 9.Imai Y, Lasky L A, Rosen S D. Nature (London) 1993;361:555–557. doi: 10.1038/361555a0. [DOI] [PubMed] [Google Scholar]
- 10.Hemmerich S, Butcher E C, Rosen S D. J Exp Med. 1994;180:2219–2226. doi: 10.1084/jem.180.6.2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shailubhai K, Streeter P R, Smith C E, Jacob G S. Glycobiology. 1997;7:305–314. doi: 10.1093/glycob/7.2.305. [DOI] [PubMed] [Google Scholar]
- 12.Streeter P R, Rouse B T, Butcher E C. J Cell Biol. 1988;107:1853–1862. doi: 10.1083/jcb.107.5.1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Girard J P, Baekkevold E S, Amalric F. FASEB J. 1998;12:603–612. doi: 10.1096/fasebj.12.7.603. [DOI] [PubMed] [Google Scholar]
- 14.Kimura N, Mitsuoka C, Kanamori A, Hiraiwa N, Uchimura K, Muramatsu T, Tamatani T, Kansas G S, Kannagi R. Proc Natl Acad Sci USA. 1999;96:4530–4535. doi: 10.1073/pnas.96.8.4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bistrup A, Bhakta S, Lee J K, Belov Y Y, Gunn M D, Zuo F R, Huang C C, Kannagi R, Rosen S D, Hemmerich S. J Cell Biol. 1999;145:899–910. doi: 10.1083/jcb.145.4.899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hiraoka N, Petryniak B, Nakayama J, Tsuboi S, Suzuki M, Yeh J C, Izawa D, Tanaka T, Miyasaka M, Lowe J B, Fukuda M. Immunity. 1999;11:79–89. doi: 10.1016/s1074-7613(00)80083-7. [DOI] [PubMed] [Google Scholar]
- 17.Hastbacka J, de la Chapelle A, Mahtani M M, Clines G, Reeve-Daly M P, Daly M, Hamilton B A, Kusumi K, Trivedi B, Weaver A, et al. Cell. 1994;78:1073–1087. doi: 10.1016/0092-8674(94)90281-x. [DOI] [PubMed] [Google Scholar]
- 18.Superti-Furga A, Hastbacka J, Wilcox W R, Cohn D H, van der Harten H J, Rossi A, Blau N, Rimoin D L, Steinmann B, Lander E S, Gitzelmann R. Nat Genet. 1996;12:100–102. doi: 10.1038/ng0196-100. [DOI] [PubMed] [Google Scholar]
- 19.Markovich D, Forgo J, Stange G, Biber J, Murer H. Proc Natl Acad Sci USA. 1993;90:8073–8077. doi: 10.1073/pnas.90.17.8073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bissig M, Hagenbuch B, Stieger B, Koller T, Meier P J. J Biol Chem. 1994;269:3017–3021. [PubMed] [Google Scholar]
- 21.Silberg D G, Wang W, Moseley R H, Traber P G. J Biol Chem. 1995;270:11897–11902. doi: 10.1074/jbc.270.20.11897. [DOI] [PubMed] [Google Scholar]
- 22.Satoh H, Susaki M, Shukunami C, Iyama K, Negoro T, Hiraki Y. J Biol Chem. 1998;273:12307–12315. doi: 10.1074/jbc.273.20.12307. [DOI] [PubMed] [Google Scholar]
- 23.Hoglund P, Haila S, Socha J, Tomaszewski L, Saarialho-Kere U, Karjalainen-Lindsberg M L, Airola K, Holmberg C, de la Chapelle A, Kere J. Nat Genet. 1996;14:316–319. doi: 10.1038/ng1196-316. [DOI] [PubMed] [Google Scholar]
- 24.Baekkevold E S, Jahnsen F L, Johansen F E, Bakke O, Gaudernack G, Brandtzaeg P, Haraldsen G. Lab Invest. 1999;79:327–336. [PubMed] [Google Scholar]
- 25.Girard J P, Springer T A. Immunity. 1995;2:113–123. doi: 10.1016/1074-7613(95)90083-7. [DOI] [PubMed] [Google Scholar]
- 26.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 27.Thompson J D, Higgins D G, Gibson T J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pajor A M. Annu Rev Physiol. 1999;61:663–682. doi: 10.1146/annurev.physiol.61.1.663. [DOI] [PubMed] [Google Scholar]
- 29.Chen X, Tsukaguchi H, Chen X Z, Berger U V, Hediger M A. J Clin Invest. 1999;103:1159–1168. doi: 10.1172/JCI5392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kekuda R, Wang H, Huang W, Pajor A M, Leibach F H, Devoe L D, Prasad P D, Ganapathy V. J Biol Chem. 1999;274:3422–3429. doi: 10.1074/jbc.274.6.3422. [DOI] [PubMed] [Google Scholar]