Abstract
The importance of CCAAT/enhancer binding proteins (C/EBPs) and binding sites for HIV-1 replication in primary macrophages, T cell lines and primary CD4+ T cells was examined. When lines overexpressing the C/EBP dominant-negative protein LIP were infected with HIV-1, replication occurred in Jurkat T cells but not in U937 promonocytes, demonstrating a requirement for C/EBP activators by HIV-1 only in promonocytes. Primary macrophages did not support the replication of HIV-1 harboring mutant C/EBP binding sites in the long terminal repeat but Jurkat, H9 and primary CD4+ T cells supported replication of wild-type and mutant HIV-1 equally well. Thus the requirement for C/EBP sites is also confined to monocyte/macrophages. The requirement for C/EBP proteins and sites identifies the first uniquely macrophage-specific regulatory mechanism for HIV-1 replication.
AIDS is characterized by the drastic depletion of CD4+ T cells due to cytopathic effects of HIV-1. However, T-cell-tropic syncytium-inducing viruses predominate late during the course of infection whereas nonsyncytium forming macrophage-tropic viruses (M-tropic) dominate early stages of infection (1, 2). The tropism of the virus is determined by the viral envelope glycoprotein gp120 that recognizes CD4 and a chemokine-like second receptor on the target cell (3–7). For M-tropic viruses CCR5 has been shown to be a second receptor for HIV-1 infection (4, 8–11) whereas T-cell-tropic syncytium-inducing viruses viruses utilize CXCR4 (fusin) as their second receptor (3). Recent studies demonstrated that individuals with mutations in their CCR5 genes are resistant to HIV-1 infection despite multiple sexual exposures (12–14), suggesting that M-tropic viruses are preferentially transmitted and that monocyte/macrophages play a central role in HIV-1 infection (15). Macrophages are also resistant to the cytopathic effects of HIV-1 in vitro and may serve as sites for virus replication late in AIDS when T cell numbers are low or following withdrawal of treatment with viral inhibitors (16–18). In addition, monocyte/macrophages are a source of inflammatory cytokines, such as interleukin 1 (IL-1), IL-6, and tumor necrosis factor α, which contribute to AIDS-related pathologies of the lungs, lymph nodes, skin, and central nervous system, and can activate HIV-1 in latently infected neighboring cells (19–21). Because of the key role played by monocyte/macrophages in AIDS, it is important to understand how HIV-1 replication is regulated in these cells.
After HIV-1 enters the host cell and proviral DNA becomes integrated into the host chromosome, transcription of viral RNA is dependent on cellular proteins (22, 23). Some previously identified cellular transcription factors that are required for HIV-1 transcription, such as Sp1 and NF-κB/rel, are ubiquitously expressed while others, such as GATA-3, ETS-1, and LEF-1, are lymphoid or T cell specific (24–30). However, cell type-specific transcription factors have not been identified that regulate HIV-1 in monocyte/macrophages. We have previously shown that CCAAT/enhancer binding proteins (C/EBP) are required for HIV-1 replication in monocytic cell lines (31), but their importance in regulating HIV-1 replication in primary macrophages or in CD4+ T cells is not known. In this study we present data showing that C/EBP proteins are specifically required for HIV-1 transcription in macrophages but not T cells.
MATERIALS AND METHODS
Cell Lines and Cells.
U937 promonocytes and H9 and Jurkat CD4+ T cell lines were cultured in RPMI medium 1640 supplemented with 10% fetal calf serum (FCS). U937 and Jurkat cell lines that overexpress liver-enriched inhibitory protein (LIP) were generated by cotransfecting 5 μg linearized cytomegalovirus-LIP construct and 1 μg pSV2-Neo plasmid (32, 33). Transfections were performed by pulsing cell suspension with 240 volts, 960 μF. Cells were resuspended in 5 ml of medium and allowed to recover for 24 hr before selection with 1 mg/ml G418. Clones were generated by limiting dilution and selection in RPMI 1640 medium supplemented with 10% FCS, 1 mg/ml G418. 293T cells were grown in Iscove’s modified Dulbecco’s medium supplemented with 10% FCS.
Primary macrophages were isolated by overlaying peripheral blood onto a Ficoll/Hypaque gradient followed by adherence to a plastic tissue culture plate. After 4 hr nonadherent cells were removed and adherent cells were cultured in RPMI 1640 medium with 10% FCS for an additional 5–7 days. This cell population was >96% macrophages as determined by specific-esterase staining and fluorescence staining for CD11b, CD54, and major histocompatibility complex class II. CD4+ cells were positively selected from the nonadherent mononuclear cells by staining the cells with biotin-conjugated-mouse anti-human CD4 antibody and streptavidin-conjugated magnetic beads. CD4+ T cells were isolated using a MACS VS+ Separation column and SuperMACS magnet (Miltenyi Biotec, Sunnyvale, CA). The positively selected cell population was >90% positive for CD3 and CD4 as determined by fluorescence staining.
Immunoblots.
Whole cell extracts were prepared from cell lines by resuspending 1–2 × 106 cells in 200 μl of 2× SDS-running dye (0.2% bromophenol blue/4% SDS/100 mM Tris, pH 6.8/200 mM DTT/3 ng/ml aproptinin/2 ng/ml pepstatin A/1 ng/ml leupeptin/10 mM phenylmethylsulfonyl fluoride/30% glycerol). Nuclear and cytoplasmic extracts were prepared as described (34). Samples were loaded onto a SDS/10% polyacrylamide gel and the protein was transferred to 0.2 μ nitrocellulose filter by electroblotting. The filter was blocked with PBS, 5% nonfat milk, 0.2% Tween-20 prior to adding primary antibodies. Blots were probed with rabbit-anti-C/EBPβ, rabbit-anti-C/EBPα, rabbit-anti-C/EBPδ (Santa Cruz Biotechnology), or rabbit-anti-NF-IL-6 (35). Following incubation with primary antibody, the filters were washed with PBS and 0.2% Tween-20 three times before adding the second antibody, peroxidase-conjugated goat-anti-rabbit. The filter was washed several times with PBS and 0.2% Tween-20 and once with PBS. Proteins were visualized using the enhanced chemiluminescence detection system (Amersham) and quantitated using a densitometer.
Viruses and Infections.
The generation of pBS-mC2,C3 HXB2 virus clone has been described (31). mC2,C3 Ba-L was generated by replacing the BamHI–XbaI fragment [which contains the 3′ long-terminal repeat (LTR)] of pBS-HIV-1 Ba-L (36) with the BamHI–XbaI fragment from pBS-mC2,C3 HXB2. Infectious virus stocks were generated by CaPO4 transfection of virus constructs into 293T cells as described (37, 38). Virus packaging was standardized by reverse transcriptase (RT) activity that was performed 48 hr posttransfection. For infections, equal amounts of virus were added to 1 × 106 cells/ml cells for 5–8 hr. The medium was removed and replaced with fresh RPMI 1640 medium supplemented with 10% FCS. CD4+ T cells were stimulated for 24 hr with 10 ng/ml of phytohemagglutinin prior to infection and maintained in RPMI 1640 medium with 10% FCS and 50 units/ml IL-2 (Sigma). RT activity was monitored at various times postinfection and the cells were fed every 3–4 days.
To detect proviral DNA, genomic DNA was prepared from primary macrophages 4 weeks postinfection by lysing the cells with TES buffer (10 mM Tris, pH 8.0/400 mM NaCl/2 mM EDTA/1% SDS) and treating with 0.5 mg/ml proteinase K at 55°C. Genomic DNA (500 ng) was amplified by 30 cycles of PCR (94°C for 1 min, 53°C for 2 min, 72°C for 2 min) using the upstream primer 5′-GCCTGCATGGGATGGA-3′ and the downstream primer 5′-CCACTGCTAGAGATTTTCCAC-3′ (31). PCR products were detected by Southern blots using the mC3 oligonucleotide (5′-GCTGACATCGACAGCTGTACAAGGGAC-3′) as a probe. To detect all amplified LTR sequences, the mC3 probe was allowed to hybridize for 4 hr at 25°C in 1.5× standard saline phosphate/EDTA (0.23 M NaCl/1.5 × 10−2 M NaH2PO4/1.5 × 10−3 M EDTA), 10% polyethelene glycol, 7% SDS, and 0.1 mg/ml salmon sperm DNA. Initially filters were washed three times at 25°C in 0.2× standard saline citrate (SSC; 0.3 M sodium chloride/0.03 M sodium citrate, pH 7), 0.1% SDS, whereas PCR products with C/EBP mutations were specifically detected by more stringent washes at 55°C (31). A PCR product containing the mutated sites could not be generated by contaminating DNA from the transfections since the mC2,C3 construct lacks sequences that bind the downstream primer (31).
Reverse Transcriptase Assays.
Supernatant from infected cells (10 μl) was added to 60 mM Tris, 24 mM DTT, 7 mM MgCl2, 75 mM NaCl, 6 μg/ml oligodG, 12 μg/ml polyrC, 0.06% Nonidet P-40, and 10 μCi (1 Ci = 37 GBq) 32P-αdGTP in a final volume of 50 μl and incubated at 37°C for 1 hr. Five microliters of this reaction mixture was then transferred to DEAE paper and washed twice in 2× SSC for 15 min at 22°C (39). RT activity was quantified using a PhosphorImager.
RESULTS
C/EBP Activators Are Required for HIV-1 Infection in Monocytic Cell Lines but Not T Cell Lines.
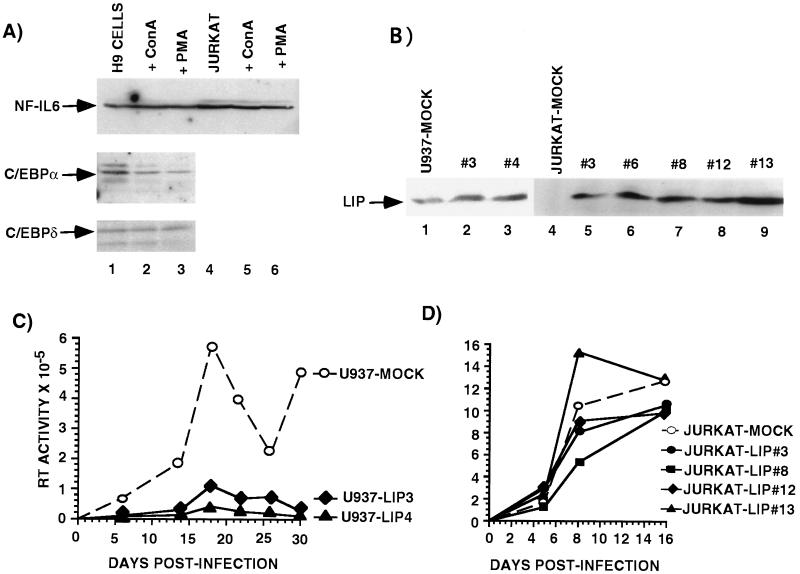
Activation of monocytes has previously been shown to cause ≈15-fold induction of one member of the C/EBP family, NF-IL-6 (40, 41). To determine the expression pattern of NF-IL-6 and other C/EBP proteins in T cells we used immunoblots to assay whole cell, cytoplasmic, and nuclear extracts from Jurkat and H9 cells before and after activation (Fig. 1A). Jurkat and H9 T cell lines express several C/EBP family transcriptional activators including C/EBPα, NF-IL-6, and C/EBPδ and these factors are present primarily in the nuclear fraction (Fig. 1A and data not shown). However, none of these C/EBP proteins are induced upon activation with phorbol myristate acetate or concanavalin A (Fig. 1A; ref. 42).
Figure 1.
C/EBP activators are required to establish HIV-1 infection in monocytic cells but not in T cell lines. (A) Western blot analysis of whole cell extracts prepared from H9 T cells (lanes 1–3) and Jurkat T cells (lanes 4–6) cultured in the absence or presence of 10 ng/ml of concanavalin A (lanes 2 and 5) or phorbol myristate (lanes 3 and 6). (B) Immunoblots of U937 and Jurkat mock-transfected cells (lanes 1 and 4) and U937-LIP overexpressing cell lines (lanes 2 and 3), and Jurkat-LIP overexpressing cell lines (lanes 5–9). (C) U937-mock-transfected cells (○) or U937-LIP overexpressing cells (solid symbols) were infected with HIV-1 HXB2 and at various times supernatants from cell cultures were assayed for RT activity. These data are from a single experiment that is representative of three independent experiments. Each data point represents the mean of three independent infections. (D) Jurkat-mock-transfected cells (○) or Jurkat-LIP-overexpressing cells (solid symbols) were infected with HIV-1 HXB2, and at various times supernatants were assayed for RT activity. This is a single experiment that is representative of three independent experiments. Each data point represents the mean of three independent infections.
To test if C/EBP proteins are required to establish HIV-1 infection and replication in monocytic or T cell lines, stably transfected lines that expressed ectopic LIP, a C/EBP dominant-negative protein, were established. U937-LIP promonocytic lines expressed 2- to 4-fold more LIP than mock-transfected controls and Jurkat-LIP T cell lines expressed high levels of LIP whereas controls expressed no detectable LIP (Fig. 1B). LIP overexpression did not significantly alter the growth rate of either U937 or Jurkat cells (data not shown). In addition, transient transfection experiments with a C/EBP-dependent reporter confirmed that LIP had dominant-negative activity in the stable cell lines (data not shown). These cell lines were infected with HIV-1 HXB2. The ability of virus to replicate was monitored at various times postinfection by measuring RT activity in the culture medium. The ability of the LIP-overexpressing U937 cell lines to support virus replication was significantly reduced (Fig. 1C). Five days postinfection these cells produced 5-fold less virus, and by 2 weeks the amount of virus produced by the LIP lines was reduced by more than 17-fold compared with mock-transfected U937 controls (Fig. 1C). These data are consistent with previous experiments that showed that LIP can inhibit HIV-1 LTR activity and provirus transcription in monocytic cell lines (31, 40). In contrast to the U937 cell lines, the mock-transfected and LIP-overexpressing Jurkat cell lines were capable of supporting efficient HIV-1 replication. After 16 days postinfection there was no significant difference in the amount of virus produced by any of the Jurkat cell lines (Fig. 1D). These data suggest that functional C/EBP activators are required for establishment of HIV-1 infection in monocytic cell lines but not in T cell lines.
C/EBP Sites Are Not Required for HIV-1 Replication in T Cell Lines.
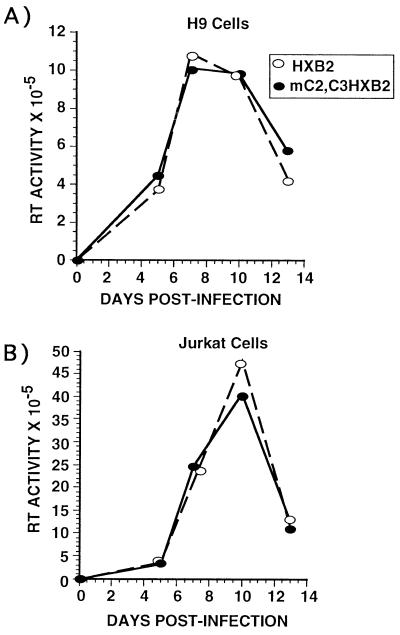
Previous experiments (31) showed that HIV-1 provirus harboring mutations in the two high affinity C/EBP sites in the LTR was unable to replicate in monocytic cell lines. We tested whether C/EBP sites were required for HIV-1 to replicate in Jurkat and H9 T cell lines. Jurkat or H9 cells were infected with either wild-type HXB2 or HXB2 containing site-directed mutations in the two high affinity C/EBP sites in the 3′ LTR (31). The C/EBP mutations will be copied into the 5′ LTR following infection and reverse transcription, generating a HIV-1 provirus with mutant C/EBP sites in both 5′ and 3′ LTRs (31). There was no difference in the ability of the wild-type and mutant viruses to replicated in Jurkat or H9 cell lines (Fig. 2) demonstrating that C/EBP sites are not required for efficient virus replication in T cell lines, further suggesting a minimal role for C/EBP proteins in regulating the expression of HIV-1 in T cells.
Figure 2.
C/EBP sites are not required for HIV-1 replication in T cells. (A) H9 cells or (B) Jurkat cells were infected with wild-type HIV-1 HXB2 (○) or mC2,C3 HIV-1 HXB2 (•). At various times postinfection supernatants were collected and RT activity was measured. This is a single experiment that is representative of three individual experiments. Each data point represents the mean of four independent infections.
C/EBP Sites Are Required for HIV-1 Replication in Primary Macrophages but Not in CD4+ T Cells.
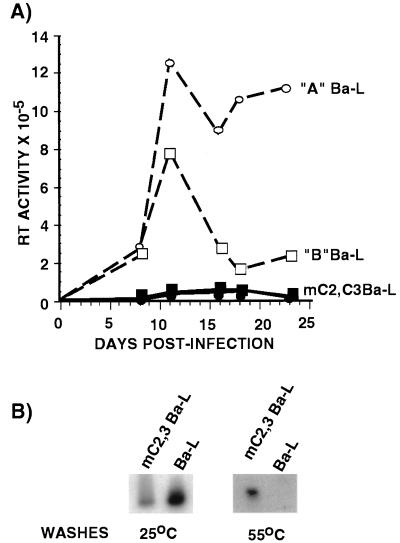
Since transformed cell lines may not be representative of normal primary cells, we tested whether primary macrophages and lymphocytes were capable of supporting the replication of HIV-1 harboring mutant C/EBP sites. Peripheral blood macrophages were enriched from a mononuclear cell population by adherence to plastic tissue culture dishes and cultured for 5–7 days prior to infection. The purified cell population was >96% macrophages as determined by staining for specific-esterase activity and fluorescence staining for CD11b, CD54, and major histocompatibility complex class II whereas <2% of the cells were positive for the T cell marker CD3 (data not shown). These cells were infected with a chimeric HIV-1 molecular clone encoding the M-tropic envelope protein from Ba-L (36) and a wild-type 3′ LTR or a 3′ LTR containing mutations in the high affinity C/EBP sites (31). As shown in Fig. 3A, the wild-type virus was able to replicate in primary macrophages over an extended period time. However, the ability of HIV containing the C/EBP mutations to replicate was severely reduced. Twenty-four days postinfection the primary macrophages infected with the mC2,C3 Ba-L produced 90% less virus compared with the cells infected with wild-type virus (Fig. 3A). The lack of replication of the mutant virus was not due to its inability to infect primary macrophages because mutant proviral sequences were detected in macrophages 4 weeks postinfection by a previously described PCR assay (Fig. 3B; ref. 31). These data are consistent with above results showing a requirement for C/EBP proteins for HIV-1 infection in monocytic cell lines and also support previous studies demonstrating that C/EBP proteins and binding sites are required for HIV-1 replication and are necessary for activity of the HIV-1 LTR in monocytic cell lines (31, 40).
Figure 3.
C/EBP sites are required for HIV-1 replication in human primary macrophages. (A) Mononuclear cells isolated from peripheral blood obtained from two healthy donors (donor A, circles; donor B, squares) were infected with either wild-type HIV-1 HXB2 containing a Ba-L envelope (open symbols) or mC2,C3 HIV-1 HXB2 with Ba-L envelope (solid symbols). Supernatants from infected cells were collected at various times and RT activity was measured. These data are a single experiment that is representative of three individual experiments. Each data point represents the mean of three independent infections. (B) Detection of mC2,C3 Ba-L provirus in primary macrophages by PCR. Genomic DNA was prepared from primary macrophages 4 weeks after infection with wild-type Ba-L or mutant Ba-L. Provirus sequences were amplified by PCR as described (31). Specific HIV-1 LTR sequences were detected by Southern blots using the mC3 oligonucleotide. All amplified LTR sequences were detected following washes at 25°C whereas PCR products with C/EBP mutations were specifically detected by more stringent washes at 55°C (31).
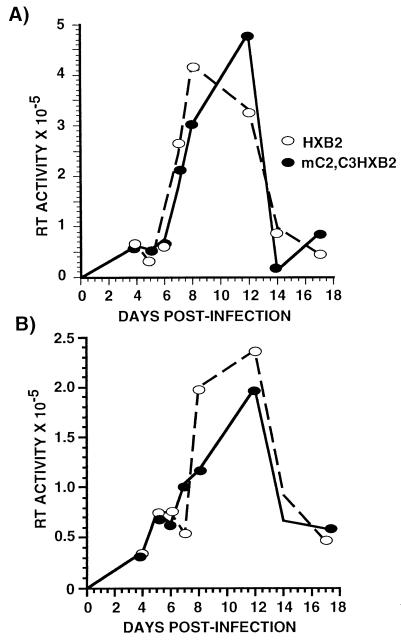
We also tested whether primary CD4+ T cells from peripheral blood were capable of supporting the replication of HIV-1 containing C/EBP mutations. CD4+ T cells were positively selected using magnetic beads from a population of mononuclear cells depleted of adherent cells. Following selection the cell population was >90% positive for CD3+ CD4+ cells as determined by fluorescence-activated cell sorter staining (data not shown). Prior to infection the cells were stimulated for 24 hr with phytohemagglutinin and then cultured in the presence of IL-2 for the duration of the experiment. As shown in Fig. 4 there was no significant difference in the ability of the wild-type or mutant HIV to replicate in T cells. Both viruses replicated with similar kinetics and there was <2-fold differences in the levels of wild-type or mutant viruses at all time points (Fig. 4). Therefore, primary T cells, unlike primary macrophages, do not require C/EBP sites for HIV-1 replication, supporting the conclusion that C/EBP proteins are not necessary for virus transcription in T cells.
Figure 4.
C/EBP sites are not required for HIV-1 replication in primary CD4+ T cells. CD4+ T cells were positively selected and stimulated for 24 hr with 10 ng/ml of phytohemagglutinin prior to infection and maintained in medium with 50 units/ml IL-2. Cells were infected with equal amounts of wild-type HXB2 (open symbols) or HXB2 harboring C/EBP mutations (solid symbols). These data are representative of four individual experiments. Each data point represents the mean of four independent infections.
DISCUSSION
These studies demonstrate that the C/EBP proteins and their binding sites are necessary for HIV-1 replication in macrophages but not in T cells thus identifying a macrophage-specific requirement for HIV-1 transcription. NF-IL-6 is the only C/EBP family protein that is induced upon activation of macrophages (40, 41) and it activates transcription of many endogenous genes whose products are important for monocyte/macrophage function including cytokines IL-1β, IL-6, IL-8, tumor necrosis factor α, granulocyte-colony stimulating factor, and the chemokine MIP1α (43). Thus HIV-1 takes advantage of the mechanism used by monocyte/macrophages to coordinate the transcription of a number of genes that are highly expressed in macrophages. Furthermore, many of the cytokines produced by macrophages can further up-regulate virus replication by inducing NF-κB and NF-IL-6 that have been demonstrated to interact synergistically (25, 44–46). Therefore, NF-IL-6 appears to play a central role in an autostimulatory pathway involving monocyte/macrophages, cytokines, and HIV-1 infection.
We have also shown that C/EBP proteins are not induced upon activation of T cells nor are C/EBP proteins required for replication of HIV-1 in T cells. We assume that other transcription factors that are present in T cells but absent from monocyte/macrophages, such as GATA-3, ETS-1 or LEF-1, functionally replace NF-IL-6 in T cells (26, 28, 47). The complexity and redundancy of activator binding sites in the HIV-1 LTR apparently ensures that the virus can take advantage of the transcriptional machinery present in different host cells.
Our data show that C/EBP sites are functionally important only in monocyte/macrophages and are not required in T cells. Analysis of the 5′ LTR sequences of M-tropic and T cell-tropic syncytium-inducing viruses isolates of HIV-1 shows that the two high affinity C/EBP sites at −170 bp and −110 bp are present in both types of viruses. Although DNA binding sites in the LTR do not affect the tropism of the virus, which is determined by the V3 loop of envelope protein gp120 (4), or the ability of the virus to infect monocytic cells (Fig. 3B) they could be important for disease progression by influencing replication in a particular target cell or tissue. Examples in which cis elements can affect virus replication and tissue expression are provided by HIV, simian immunodeficiency virus, and Moloney murine leukemia viruses (47–52). The work presented here is the first to identify transcriptional activators that are uniquely required for HIV-1 replication in monocyte/macrophages and not in T cells. Agents that target C/EBP activators might, therefore, be important for controlling HIV-1 replication in monocyte/macrophages following virus transmission and during late stages of AIDS following T cell depletion or drug therapy.
Acknowledgments
We thank Drs. R. Connor, J. Luban, G. Siu, and S. Lederman for critical reading of the manuscript and for useful discussions. A.J.H. is a Special Fellow of the Leukemia Society of America. This work was also supported by National Institutes of Health Grant GM29361 to K.L.C.
ABBREVIATIONS
- FCS
fetal calf serum
- LTR
long terminal repeat
- RT
reverse transcriptase
- M-tropic
macrophage-tropic viruses
- LIP
liver-enriched inhibitory protein
- C/EBP
CCAAT/enhancer binding protein
- IL-1
interleukin 1
References
- 1.Schuitemaker H, Koot M, Kootstra N A, Dercksen M W, de Goede R E, van Steenwijk R P, Lange J M, Schattenkerk J K, Miedema F. J Virol. 1992;66:1354–1360. doi: 10.1128/jvi.66.3.1354-1360.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Connor R I, Ho D D. J Virol. 1994;68:4400–4408. doi: 10.1128/jvi.68.7.4400-4408.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Feng Y, Broder C C, Kennedy P E, Berger E A. Science. 1996;272:872–877. [Google Scholar]
- 4.Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath P D, Wu L, Mackay C R, LaRosa G, Newman W, Gerard N, Gerard C, Sodroski J. Cell. 1996;85:1135–1148. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 5.Fouchier R, Groenink M, Kootstra N, Tersmette M, Huisman H, Miedema F, Schuitemaker H. J Virol. 1992;66:3183–3187. doi: 10.1128/jvi.66.5.3183-3187.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Trkola A, Dragic T, Arthos J, Binley J M, Olson W C, Allaway G P, Cheng-Mayer C, Robinson J, Maddon P J, Moore J P. Nature (London) 1996;384:184–187. doi: 10.1038/384184a0. [DOI] [PubMed] [Google Scholar]
- 7.Wu L, Gerard N P, Wyatt R, Choe H, Parolin C, Ruffing N, Borsetti A, Cardoso A A, Desjardin E, Newman W, Gerard C, Sodroski J. Nature (London) 1996;384:179–183. doi: 10.1038/384179a0. [DOI] [PubMed] [Google Scholar]
- 8.Dragic T, Litwin V, Allaway G P, Martin S R, Huang Y, Nagashima K A, Cayanan C, Maddon P J, Koup R A, Moore J P, Paxton W A. Nature (London) 1996;381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 9.Doranz B J, Rucker J, Yi Y, Smyth R J, Samson M, Peiper S C, Parmentier M, Collman R G, Doms R W. Cell. 1996;85:1149–1158. doi: 10.1016/s0092-8674(00)81314-8. [DOI] [PubMed] [Google Scholar]
- 10.Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, DiMarzio P, Marmon S, Sutton R, Hill C, Davis C, Peiper S, Schall T, Littman D, Landau N. Nature (London) 1996;381:661–667. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 11.Alkhatib G, Combadiere C, Broder C C, Feng Y, Kennedy P E, Murphy P M, Berger E A. Science. 1996;272:1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- 12.Liu R, Paxton W A, Choe S, Ceradini D, Martin S R, Horuk R, MacDonald M E, Stuhlmann H, Koup R A, Landau N R. Cell. 1996;86:367–377. doi: 10.1016/s0092-8674(00)80110-5. [DOI] [PubMed] [Google Scholar]
- 13.Samson M, Libert F, Doranz B J, Rucker J, Liesnard C, Farber C M, Saragosti S, Lapoumeroulie C, Cognaux J, Forceille C, Muyldermans G, Verhofstede C, Burtonboy G, Georges M, Imai T, Rana S, Yi Y, Smyth R J, Collman R G, Doms R W, Vassart G, Parmentier M. Nature (London) 1996;382:722–725. doi: 10.1038/382722a0. [DOI] [PubMed] [Google Scholar]
- 14.Dean M, Carrington M, Winkler C, Huttley G A, Smith M W, Allikmets R, Goedert J J, Buchbinder S P, Vittinghoff E, Gomperts E, Donfield S, Vlahov D, Kaslow R, Saah A, Rinaldo C, Detels R, O’Brien S J. Science. 1996;273:1856–1862. doi: 10.1126/science.273.5283.1856. [DOI] [PubMed] [Google Scholar]
- 15.Zhu T, Mo H, Wang N, Nam D S, Cao Y, Koup R A, Ho D D. Science. 1993;261:1179–1181. doi: 10.1126/science.8356453. [DOI] [PubMed] [Google Scholar]
- 16.Fauci A S. Science. 1988;239:480–484. doi: 10.1126/science.3277274. [DOI] [PubMed] [Google Scholar]
- 17.Ho D D, Neumann A U, Perelson A, Chen W, Leonard J, Markowitz M. Nature (London) 1995;373:123–126. doi: 10.1038/373123a0. [DOI] [PubMed] [Google Scholar]
- 18.Perelson A S, Neumann A U, Markowitz M, Leonard J M, Ho D D. Science. 1996;271:1582–1586. doi: 10.1126/science.271.5255.1582. [DOI] [PubMed] [Google Scholar]
- 19.Fauci A. Science. 1993;262:1011–1018. doi: 10.1126/science.8235617. [DOI] [PubMed] [Google Scholar]
- 20.Gartner S, Markovits P, Markovitz B M, Kaplan M H, Gallo R C, Popovic M. Science. 1986;233:215–219. doi: 10.1126/science.3014648. [DOI] [PubMed] [Google Scholar]
- 21.Koyanagi Y, O’Brien W, Zhao J Q, Golde D W, Gasson J, Chen I S Y. Science. 1988;241:1673–1675. doi: 10.1126/science.241.4873.1673. [DOI] [PubMed] [Google Scholar]
- 22.Gaynor R. AIDS. 1992;6:347–363. doi: 10.1097/00002030-199204000-00001. [DOI] [PubMed] [Google Scholar]
- 23.Vaishnav Y, Wong-Staal F. Annu Rev Biochem. 1991;60:577–630. doi: 10.1146/annurev.bi.60.070191.003045. [DOI] [PubMed] [Google Scholar]
- 24.Nabel G, Baltimore D. Nature (London) 1987;326:711–713. doi: 10.1038/326711a0. [DOI] [PubMed] [Google Scholar]
- 25.Griffin G E, Leung K, Folks T M, Kunkel S, Nabel G J. Nature (London) 1989;339:70–73. doi: 10.1038/339070a0. [DOI] [PubMed] [Google Scholar]
- 26.Yang Z, Engel J D. Nucleic Acids Res. 1993;21:2831–2836. doi: 10.1093/nar/21.12.2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Waterman M, Fischer W, Jones K. Genes Dev. 1991;5:656–669. doi: 10.1101/gad.5.4.656. [DOI] [PubMed] [Google Scholar]
- 28.Sheridan P, Sheline C, Cannon K, Voz M, Pazin M, Kadonaga J, Jones K. Genes Dev. 1995;9:2090–2104. doi: 10.1101/gad.9.17.2090. [DOI] [PubMed] [Google Scholar]
- 29.Moses A, Ibanez C, Gaynor R, Ghazal P, Nelson J. J Virol. 1994;68:298–307. doi: 10.1128/jvi.68.1.298-307.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jones K A, Kadonaga J T, Luciw P A, Tjian R. Science. 1986;232:755–759. doi: 10.1126/science.3008338. [DOI] [PubMed] [Google Scholar]
- 31.Henderson A, Connor R, Calame K. Immunity. 1996;5:91–101. doi: 10.1016/s1074-7613(00)80313-1. [DOI] [PubMed] [Google Scholar]
- 32.Cooper C, Johnson D, Roman C, Avitahl N, Tucker P, Calame K. J Immunol. 1992;149:3225–3231. [PubMed] [Google Scholar]
- 33.Morgenstern J P, Laud H. Nucleic Acids Res. 1990;18:3587–3596. doi: 10.1093/nar/18.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Screiber E, Matthias P, Muller M M, Schaffner W. Nucleic Acids Res. 1989;17:6419. doi: 10.1093/nar/17.15.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cooper C, Berrier A, Roman C, Calame K. J Immunol. 1994;153:5049–5058. [PubMed] [Google Scholar]
- 36.Connor R, Chen B, Choe S, Landau N. Virology. 1995;206:936–944. doi: 10.1006/viro.1995.1016. [DOI] [PubMed] [Google Scholar]
- 37.Heinzel S S, Krysan P J, Carlos M P, DuBridge R B. J Virol. 1988;62:3738–3746. doi: 10.1128/jvi.62.10.3738-3746.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pear W S, Nolan G P, Scott M L, Baltimore D. Proc Natl Acad Sci USA. 1993;90:8392–8396. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goff S, Traktman P, Baltimore D. J Virol. 1981;38:239–248. doi: 10.1128/jvi.38.1.239-248.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Henderson A, Zou X, Calame K. J Virol. 1995;69:5337–5344. doi: 10.1128/jvi.69.9.5337-5344.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Natsuka S, Akira S, Nishio Y, Hashimoto S, Sugita T, Ishiki H, Kishimoto T. Blood. 1992;79:460–466. [PubMed] [Google Scholar]
- 42.Davydov I, Krammer P, Li-Weber M. J Immunol. 1995;155:5273–5279. [PubMed] [Google Scholar]
- 43.Akira S, Kishimoto T. Immunol Rev. 1992;127:25–50. doi: 10.1111/j.1600-065x.1992.tb01407.x. [DOI] [PubMed] [Google Scholar]
- 44.LeClair K P, Blanar M A, Sharp P A. Proc Natl Acad Sci USA. 1992;89:8145–8149. doi: 10.1073/pnas.89.17.8145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stein B, Cogswell P C, Baldwin A S. Mol Cell Biol. 1993;13:3964–3974. doi: 10.1128/mcb.13.7.3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ruocco M, Chen X, Ambrosino C, Dragonetti E, Liu W, Mallardo M, De Falco G, Palmieri C, Franzoso G, Quinto I, Venuta S, Scala G. J Biol Chem. 1996;271:22479–22486. doi: 10.1074/jbc.271.37.22479. [DOI] [PubMed] [Google Scholar]
- 47.Zhang L, Huang Y, Yuan H, Chen B K, Ip J, Ho D D. J Virol. 1997;71:1651–1656. doi: 10.1128/jvi.71.2.1651-1656.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dollard S C, Gummuluru S, Tsang S, Fultz S, Dewhurst S. J Virol. 1994;68:7800–7809. doi: 10.1128/jvi.68.12.7800-7809.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Speck N, Renjifo B, Golemis E, Fredrickson T, Hartley J, Hopkins N. Genes Dev. 1990;4:233–242. doi: 10.1101/gad.4.2.233. [DOI] [PubMed] [Google Scholar]
- 50.Ilyinskii P O, Desrosiers R C. J Virol. 1996;70:3118–3126. doi: 10.1128/jvi.70.5.3118-3126.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ross E, Buckler-White A, Rabson A, Englund G, Martin M. J Virol. 1991;65:4350–4358. doi: 10.1128/jvi.65.8.4350-4358.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Leonard J, Parrott C, Buckler-White A J, Turner W, Ross E K, Martin M A, Rabson A B. J Virol. 1989;63:4919–4924. doi: 10.1128/jvi.63.11.4919-4924.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]