Abstract
The integrin family of cell surface receptors is strongly conserved in higher animals, but the evolutionary history of integrins is obscure. We have identified and sequenced cDNAs encoding integrin β subunits from a coral (phylum Cnidaria) and a sponge (Porifera), indicating that these proteins existed in the earliest stages of metazoan evolution. The coral βCn1 and, especially, the sponge βPo1 sequences are the most divergent of the “β1-class” integrins and share a number of features not found in any other vertebrate or invertebrate integrins. Perhaps the greatest difference from other β subunits is found in the third and fourth repeats of the cysteine-rich stalk, where the generally conserved spacings between cysteines are highly variable, but not similar, in βCn1 and βPo1. Alternatively spliced cDNAs, containing a stop codon about midway through the full-length translated sequence, were isolated from the sponge library. These cDNAs appear to define a boundary between functional domains, as they would encode a protein that includes the globular ligand-binding head but would be missing the stalk, transmembrane, and cytoplasmic domains. These and other sequence comparisons with vertebrate integrins are discussed with respect to models of integrin structure and function.
The integrins comprise a large family of cell surface receptors involved in diverse adhesion and signaling events in animals (1). In vertebrates, at least 20 different αβ heterodimeric integrins are important for processes such as embryonic morphogenesis, leukocyte migration, platelet aggregation, and regulation of cell proliferation and differentiation. Integrins in Drosophila melanogaster and Caenorhabditis elegans also are known to be required for a host of morphogenetic events (2, 3). While some integrins bind directly to other transmembrane proteins, most are receptors for extracellular matrix components.
Each integrin is composed of nonhomologous α and β subunits, both of which are transmembrane proteins of approximately 100 kDa (1). The large extracellular domains form a globular head, connected by stalks from each subunit to a pair of short cytoplasmic tails. Integrin polypeptides from nematodes, insects, and vertebrates display a remarkable degree of sequence conservation, which extends throughout the α and β subunits (2, 3). The worm βpat3, fly βPS, and most vertebrate β subunits contain 56 conserved cysteine residues, suggesting that the proteins all fold into a stereotypical structure. The high level of structural conservation may be a reflection of the fact that in response to extracellular ligand binding or intracellular signals integrins can undergo conformational changes that are propagated along the molecule (4). That is, extensive associations between the α and β subunits, as well as interactions with a host of other proteins inside and outside the cell, probably have placed significant selective constraints on integrin structure in higher animals.
Unlike the cadherins or immunoglobulin domain receptors, integrins are not part of a superfamily whose members contain repeating structural units arranged in various ways. Recently, however, an argument has been made that seven short repeats in the globular heads of integrin α subunits are folded into a β-propeller domain similar to that found in the β subunits of heterotrimeric G proteins (5). This β-propeller associates with a 180 amino acid structure known as the I domain in some α subunits, and the head of the integrin β subunits also appears to contain an I domain-like structure (6, 7), including a characteristic metal ion-dependent adhesion site (MIDAS). Natural mutations and experimental studies of β subunits have identified a number of specific I domain amino acid residues required for integrin ligand binding (7–16).
While progress is being made in defining integrin structure, and similarities to very distantly related proteins are becoming evident, very little information is available concerning the evolutionary origins of integrins as cell surface receptors. Previously, the most primitive organism from which an integrin-encoding gene had been isolated was the nematode C. elegans (3). There have been suggestions of integrin-like proteins in the cnidarian Hydra, based on immunological data and inhibition studies using Arg-Gly-Asp (RGD) peptides (17–19). In the most primitive metazoan phylum, the Porifera (sponges), there have been no good experimental suggestions of integrins, but some extracellular matrix proteins similar to those from vertebrates have been identified, including collagens and a tenascin-like molecule (20–24). With these facts in mind, we set out to identify genes encoding integrin-related proteins from primitive metazoans. Here, we report the isolation of cDNAs encoding integrin β subunits from two of the most primitive animal phyla, the Cnidaria (represented by the coral Acropora millepora) and the Porifera (represented by the marine sponge Ophlitaspongia tenuis). These sequences represent divergent members of what we term the “β1-class” of integrin subunits.
MATERIALS AND METHODS
cDNA Libraries.
The coral cDNA library was made from presettlement embryos of A. millepora. Fertilized eggs were collected from the surface of wading pools containing A. millepora colonies at Magnetic Island, Queensland, Australia (19° 08′S, 146° 50′E), and raised in bulk in mesh-covered containers suspended at sea until the embryos reached presettlement stage. Embryos were then placed in cryo tubes and immediately frozen in liquid nitrogen.
Specimens of the marine sponge O. tenuis were collected off Jervis Bay, New South Wales, Australia (35° 03′S, 150° 44′E). Each sponge was separated from the substratum, shaken off, and placed individually in a plastic bag. On returning to the surface the sponges were buried in dry ice until being stored in a −80°C freezer.
Total RNA was prepared from material ground in liquid nitrogen according to the method of Chomczynski and Sacchi (25). Poly(A)+ RNA was isolated using the poly(A) Tract mRNA isolation system (Promega). cDNA libraries were constructed using the Lambda ZAP system (Stratagene).
PCR Identification of Integrin cDNAs.
The general strategy for isolation of integrin genes was to amplify fragments from cDNAs or cDNA libraries by PCR using degenerate primers and a variety of thermal profiles. Candidate bands were gel-purified and sequenced with the primers used for amplification, to ascertain whether they represented bona fide integrin subunit genes.
Fragments of the coral gene initially were amplified with the forward primer 5′-GGITTC/TGGIIIITTC/TA/CTIGAC/TAA (corresponding to amino acid sequence GFGSFVDK) and the reverse primers 5′-CCICCC/TTCIGGIGCA/GTC (DAPEGG) and 5′-GCA/GTCA/GAAICCICCC/TTC (EGGFDA). The buffer contained 1.5 mM Mg2+, and Taq polymerase was used in all primary amplifications. Initially, we used a thermal profile with three cycles of annealing at 47°C and 30 cycles at 42°C.
The initial amplification of the sponge cDNA used a forward primer of 5′-GAC/TC/TTITAC/TTAC/TC/TTIATGGA (DLYYLMD) and a reverse primer of 5′-A/GTGA/GAAICCIGCA/GTCIGT (TDAGFH). These primers amplified a fragment in 2.5 mM Mg2+, but not in 1.25 mM Mg2+, perhaps because, as indicated by subsequent sequencing, there are two mismatches between the primer and cDNA sequences. For this gene, we used a thermal profile of five annealing cycles at 37°C (and a slow ramp to the elongation temperature) followed by 45 cycles at 42°C.
Cloning and Sequencing.
After sequencing indicated an integrin gene had been amplified, radiolabeled probes were made from amplified fragments and used to screen the phage libraries, using standard methods (26). Positive clones were recovered as inserts in Bluescript SK− plasmids and grown in DH5α cells. DNA was sequenced using both vector and internal primers with Applied Biosystems automatic sequencing machines. The coral sequences reported are from plasmid C11A5. Most of the sponge sequence is from SP10C, but because this plasmid is not full length, the 5′ end of the sequence is derived from a combination of plasmid SP10D (nucleotides encoding residues 3–34) and a PCR product (amplified with Pfu polymerase) derived from phage SP9B (encoding residues 1 and 2). Both strands were sequenced for each plasmid, but when scanning the SP10D plasmid for alternative splicing only one strand was sequenced.
An alternatively spliced exon found in plasmid SP10D inserts the sequence cagGTGAGTATACTGAACTATTATTATATCTCTCACAAGACTGTATGATTATTGTTGTGTAGtat (italics indicate flanking residues included in SP10C) beginning at nucleotide 1355 of the sequence in GenBank. We also found heterogeneity in the size of the 3′ untranslated region of the sponge clones. Nine nucleotide polymorphisms were found in SP10D relative to SP10C; these would be expected to change five amino acids, none in well conserved positions. Subsequent experiments showed that either splice variant could contain different polymorphisms, indicating that the inserted exon is not simply a cloning artifact that was amplified in the library.
The assignment of the N-terminal methionine for the coral protein is confirmed by the presence of stop codons in the 5′ untranslated region. For the sponge, there are no in-frame stops (or other AUG codons) in the 44 nucleotides 5′ to the presumed initiating AUG. However we are confident this is indeed the initiating codon, as we have sequenced 2,770 nucleotides exclusive of the 3′ poly(A), and the mRNA on Northern blots appears as a broad band of approximately 2.8 kb. Moreover, the N-terminal sequence satisfies the criteria for a signal sequence for a protein destined for the plasma membrane (as determined by prosort), and this is not true if potential residues encoded by the upstream nucleotides are included.
Sequence Analysis.
Sequences were aligned first with pileup in GCG9, followed by manual adjustments, ensuring that the 56 cysteines were aligned. Identity and similarity scores were determined using bestfit of optimal pairwise alignments, again manually adjusted to align the conserved cysteines.
Linkages for generating phylogenetic trees were constructed using paup 3.1.1. Different algorithms were run with different combinations of the data to assess the reliability of various groupings. The tree shown in Fig. 2 was generated with a heuristic algorithm, with randomly ordered sample addition. Data for other β subunits are from GenBank (3, 27–38).
Figure 2.
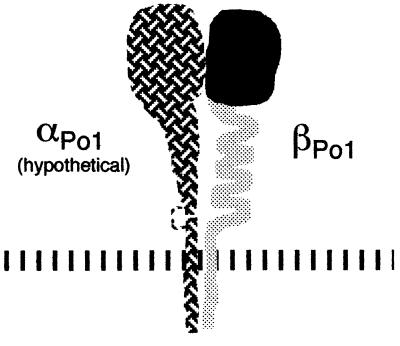
Expected βPo1 fragment generated by alternative RNA splicing. The protein would be expected to consist of the β ligand-binding domain (solid shading) without the stalk and cytoplasmic regions (light shading).
RESULTS AND DISCUSSION
As detailed in Materials and Methods, we used degenerate PCR primers to amplify fragments of integrin β subunit cDNAs from a coral and a marine sponge. Adult corals are known to harbor numerous symbionts and other visitors, and to avoid these contaminants the cDNA library was constructed from staged embryos. A probe from the isolated plasmid identifies a single band of approximately 3.8 kb on A. millepora Northerns. This band is present at all embryonic stages examined and in unfertilized eggs (D. Miller, personal communication).
Sponges also can harbor commensal organisms, and large quantities of pure embryonic tissue were not available to us. However, the species investigated is fan-shaped and 2–3 mm thick and lacks the pores and cavities frequented by commensals in more massive sponges. Each individual was shaken to dislodge external commensals during collection. Other observations also indicate that the gene isolated does not derive from a rare contaminating organism. For example, Northern blots of O. tenuis mRNA confirm that the 2.8-kb transcript identified is abundant (not shown), and during the cDNA library screening positive plaques were present at a high frequency of roughly 1 in 2,000.
Sequence Analysis.
Upon conceptual translation, both cDNAs encode proteins that are obviously integrin β subunits (Fig. 1). The coral protein (βCn1, for the first β from the phylum Cnidaria) comprises 792 amino acids, whereas there are 838 in the sponge subunit (βPo1, signifying the phylum Porifera). All of the major structural features of integrin β subunits (signal sequence, putative I domain, cysteine-rich stalk, transmembrane, and cytoplasmic domains) are conserved in both sequences, including the 56 cysteines characteristic of most β subunits. While the coral and sponge proteins are different from other β subunits, there is no indication that they are specialized proteins such as vertebrate β8 and Drosophila βν. That is, the sequences are consistent with the idea that these proteins represent major tissue integrins of these organisms, in the same way that vertebrate β1 and Drosophila βPS are common in their respective organisms (see below).
Figure 1.
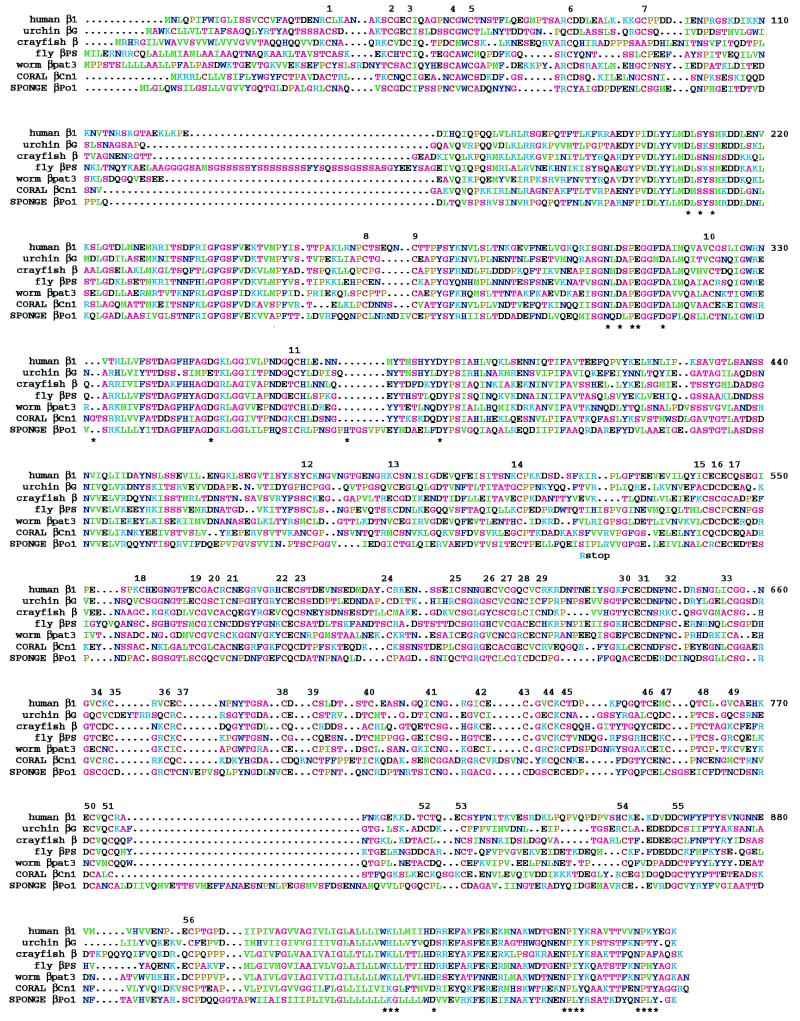
Alignment of amino acid sequences of coral βCn1 and sponge βPo1 with β1-like integrin subunits. Numbers above the sequences indicate the 56 conserved cysteines, and numbers to the right of each block denote the position in the matrix. Residues mentioned in the text are indicated by ∗ below the block. The amino acid change and stop codon in the alternatively spliced sponge cDNAs are indicated below the block at position 521. The transmembrane domain is the largely green region around position 920. No attempt was made to align the signal sequences before the first conserved cysteine. The color scheme used to indicate amino acid similarity is: yellow, proline; red, cysteine; magenta, alanine, glycine, serine, and threonine; blue, asparagine, histidine, phenylalanine, tryptophan, and tyrosine; cyan, arginine and lysine; green, isoleucine, leucine, methionine, and valine, and black, aspartic acid, glutamic acid, and glutamine.
Comparisons of specific sequence motifs with other β subunits reveals varying degrees of conservation. A few specific sequence comparisons are worthy of mention. A number of residues of the I domain region of β1, β2, β3, or β6 have been shown by experimental or natural mutations to be important for ligand binding (7–16). In addition to the DXSXS motif in the MIDAS domain (positions 208–212; for easy reference, all amino acid residue numbers are the positions in the alignment in Fig. 1), residues shown to be critical include Asn-305, Asp-307, Pro-309, Glu-310, Asp-314, Asp-351, and Asp-388, and at least some of these are thought also to be involved in coordination of a divalent cation in a ligand-binding pocket (6, 7, 39). All of these residues are conserved in βCn1 and βPo1.
The membrane distal region of the cytoplasmic domains of most β subunits contains one or two NPXY motifs (beginning at positions 958 and 970). This region generally has been shown to be important in various assays of integrin function or protein association (reviewed in ref. 40), and studies of missense mutations have directly implicated the NPXY motifs in integrin function (16, 41–44). Both of the NPXYs are found in both βPo1 and βCn1. However, the basic residue that invariably follows the first NPXY in other β subunits is a Gln in βCn1.
The membrane-proximal cytoplasmic sequence WKLLXXXHD (positions 929–937) is conserved in most β subunits and has been shown to contain residues important for associations with the conserved GFFKR and nearby sequences found in α subunits (16, 45). In particular, the terminal Asp-937 residue of the WKLLXXXHD motif of β3 has been shown to interact with the Arg of the αIIb GFFKR; this Asp is conserved in both βCn1 and βPo1, lending support to the supposition that these proteins function in association with α subunits. Interestingly, however, the upstream WK/RL sequence, at the cytoplasmic side of the transmembrane domain, is replaced by I/LKG in βCn1 and βPo1. The similarity of the two new sequences suggests that this region remains functionally important, and it will be interesting to compare the sequences of the corresponding α subunits in this region.
Such sequence comparisons of divergent, but functionally similar, integrins will take on added interest as more high-resolution structural information becomes available for β subunits. Recently, a model has been proposed for the β subunit I domain-like structure (7). Although the resolution of this model does not allow many specific predictions to be made, it is noteworthy that the insertion of eight residues at position 373 in βPo1, and the shorter insertion of two amino acids at position 332 in βCn1, are consistent with the model, as both inserts fall in predicted loops between a β strand and an α helix.
Cysteine Spacings Are Not Conserved.
The most noteworthy difference between the βCn1 and βPo1 sequences compared with those from higher animals is in the cysteine-rich stalk that connects the I domain-like ligand binding region to the plasma membrane. In most β subunits, this domain contains four repeats in which the spacings between cysteines are generally well conserved. In both βCn1 and βPo1, all of the cysteines are present, but the intercysteine spacings are much more variable in repeats 3 and 4. For example, between the 34th and 48th cysteines, 10 of the 14 Cys-Cys spacings are absolutely conserved in all previously reported β subunits (or at least in those that contain the relevant cysteines, as some of these are missing in β4, β8 or βν). In six of these 10 sites either βCn1 or βPo1 contains extra residues relative to the others, and the stereotyped CXCXXCXC motifs of repeats 3 and 4 are disrupted. In addition, βPo1 contains a unique insert of approximately 37 residues between cysteines 51 and 52.
If one assumes that the repeats of the stalk region arose from duplications of an original unique structural domain, then the β subunits of higher animals, where the repeat structure is more strongly conserved, probably reflect the ancestral state more closely than do βCn1 or βPo1. Independent divergence from the original repeat structures in the latter proteins also is suggested by the fact that the coral and sponge intercysteine spacings are not similar to one another. Of course, this raises the question of why the integrins of higher animals have been constrained to maintain the spacings, while the others have been free to vary. Conformation-sensitive antibodies that recognize membrane proximal regions of the extracellular domain have been isolated for other β subunits (46–50), and it is likely that α-β contacts will be important throughout the stalk. Because it is generally more difficult for selection to change many components simultaneously, one hypothesis is that the structure of the stalk became more constrained when β subunits began to associate with multiple α subunits.
A β Subunit Fragment From the Sponge.
In the course of analyzing sponge cDNAs, we found that some contained an alternatively spliced exon of 59 base pairs. When translated in frame, one amino acid is changed relative to the intact βPo1 (Fig. 1, position 521), and the second and fifth new codons are stops, generating a protein missing the C-terminal 400 amino acids of βPo1. This protein would contain the globular ligand-binding domain, but would be missing the stalk, transmembrane, and cytoplasmic domains of a typical integrin β subunit (Fig. 2). While we have no direct evidence that such a fragment is actually present in sponges, the abundance of the cDNA (3 of 10 isolates from the library) indicates that this protein is very likely to be made.
There is one previous report of a similarly truncated integrin subunit, a 60-kDa protein that arises from an alternatively spliced β3 mRNA (51). Translation of the β3C protein stops within four amino acids of the position of the C-terminus of the truncated βPo1 protein, although the sequences indicate that these alternative splicing events are not homologous. Both truncations lie just upstream of the cysteine-rich stalk, and the coincidence in the positions of the stops in β3C and βPo1 suggests that this short stretch of residues following the 14th cysteine defines the limit of the ligand-binding head of the β subunit.
Phylogenetic Analyses.
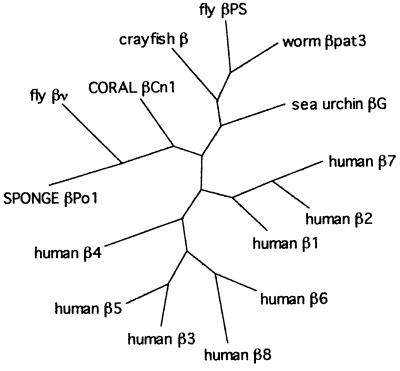
Fig. 3 shows a phylogenetic tree of the sequenced integrin β subunits, constructed using the paup heuristic parsimony algorithm. The tree is unrooted, because there are no β sequences known that are expected to have diverged before the sponge gene. Except for the unexpected position of the Drosophila βν protein, discussed below, the most parsimonious β subunit tree is consistent with the accepted divergence of sponges and cnidarians early in metazoan evolution.
Figure 3.
Tree showing relationships of β subunit sequences, generated using paup heuristic parsimony algorithm. The tree is unrooted; see text for details.
The integrin sequences do not permit the reconstruction of a rigorous phylogeny; for example, while a bootstrapping algorithm yields a tree similar to that shown, many of the bootstrap values are less than 50. However, we have analyzed the sequences using a variety of tree-constructing algorithms and various pair-wise comparisons, and we find no good indication for an early, conserved divergence in β protein sequences. The data available suggest that the radiation of β subunits seen in vertebrates is a relatively late evolutionary event, probably occurring in the deuterostome lineage and possibly within the chordates. To make this conclusion firmly more invertebrate β subunits need to be examined. Partial sequences of two additional sea urchin β subunits, βL and βC, have been reported (28), and their complete sequences might be especially informative in this regard, considering the close proximity of echinoderms and vertebrates in the phylogenetic tree.
Functional studies suggest that β1 is the best candidate for the “primal” vertebrate β subunit; it associates with an unusually large array of α subunits and is the subunit most commonly involved in fundamental morphogenetic events (1, 52). The Drosophila βPS and C. elegans βpat3 proteins show a high degree of sequence similarity to β1, associate with multiple α subunits, and also are characterized by widespread involvement in developmental processes (2, 3). For these reasons, we propose to consider these proteins to be representatives of a “β1-class” subunit. This is not meant to imply that these proteins are strict orthologues; rather, we use the term to distinguish these proteins from those, such as the Drosophila βν, that appear to have diverged as they have acquired specialized functions. While we know little about the expression patterns of βCn1 and βPo1, their sequences are consistent with their inclusion in the β1-class. Perusal of the similarity scores in Table 1, and comparisons of specific sequence motifs, indicate that the sponge and coral proteins are at least as much like β1 as they are similar to other invertebrate proteins. Indeed, the remarkable similarity of β1 to all of the invertebrate β subunits suggests that β1 may have changed relatively little over the past 500 million years. Because β1 associates with a large number of α subunits (1), its structure may have been constrained to a greater degree than that of most integrin subunits.
Table 1.
Pairwise comparisons of β subunit amino acid sequences
| Coral βCn1 | Worm βpat3 | Crayfish β | Fly βPS | Urchin βG | Human β1 | |
|---|---|---|---|---|---|---|
| Sponge | 48 | 45 | 45 | 43 | 44 | 49 |
| βPol | 38 | 35 | 37 | 35 | 35 | 38 |
| Coral | 51 | 48 | 48 | 51 | 54 | |
| βCn1 | 41 | 42 | 41 | 43 | 45 | |
| Worm | 50 | 55 | 51 | 53 | ||
| βpat3 | 41 | 47 | 42 | 43 | ||
| Crayfish | 54 | 50 | 52 | |||
| β | 44 | 41 | 43 | |||
| Fly | 54 | 54 | ||||
| βPS | 46 | 47 | ||||
| Urchin | 54 | |||||
| βG | 45 |
Scores for % similarity (top number) and % identity (bottom number) for human β1 and invertebrate integrin β subunit sequences. For each comparison, alignments from pileup (GCG9) were manually adjusted to ensure that the 56 conserved cysteines were aligned, and scores then were calculated by bestfit. Signal sequences upstream of the initial conserved cysteine were not included in the analysis.
The two sequences presented here represent the most divergent β1-class proteins; this is especially true for the sponge. Based on 18S rRNA data, there has been disagreement as to whether cnidarians such as coral are monophyletic with higher metazoans (53, 54). However, as more sequence data have become available, cnidarians appear to fall very much in the metazoan mainstream (e.g., ref. 55), and the high degree of sequence similarity between βCn1 and human β1 (Table 1) is in agreement with this assignment. The relationship of the Porifera to other metazoans is more uncertain, but while the sponge sequence is significantly more divergent, even here there is no evidence for a polyphyletic origin.
While it must be remembered that integrin sequences are not optimal for phylogenetic reconstructions, there are two surprises in the tree. The first is the anomalous position of Drosophila βν between the sponge and coral sequences. This most likely results from the fact that parsimony analyses can break down when attempting to place a highly divergent sequence. Most probably βν is not an orthologue of the other invertebrate sequences, but a functionally specialized and highly derived protein (27). In fact, βν is not especially like any of the other β subunits, and in pairwise comparisons βν is more similar to the crayfish β (46%) and human β1 (45%), than it is to the sponge and coral proteins (both 43%).
The other unexpected finding is the placement of the C. elegans βpat3 between two arthropod sequences. There appears to be an unusually high degree of similarity between the C. elegans and Drosophila sequences, as opposed to a particularly high degree of divergence in the crayfish. For example, in pairwise comparisons the crayfish β is most similar to fly βPS, as expected. In explanation, we can only note that both the derivation of arthropod lineages, and nematodes generally, have proven to be enigmatic in other molecular phylogenetic studies (53, 56).
Integrin-like proteins now have been shown to occur in the most primitive metazoan phyla. How deep in the evolutionary tree did integrins arise? The yeast genome is now fully sequenced, and no protein with obvious homology to an integrin β subunit has been uncovered. On the other hand, there have been numerous functional and immunological suggestions that integrin-like proteins are present in higher plants and fungi (57–64).
Acknowledgments
We thank David Miller for much advice, and members of his lab and Peter Harrison for assistance in obtaining the coral embryos. Jeremy Weinman and Chris Parish provided help in obtaining and advice concerning the sponge material. Roy Parker, Richard Maleszka, Scott Selleck, and Adrian Gibbs kindly allowed us the use of their PCR machines, and we are especially indebted to Richard Maleszka for invaluable help in our library fishing. Cameron McCrae and Skip Vaught’s gang made sequencing virtually painless. Pat O’Grady, Lisa Nagy, and Sam Ward educated D.B. on phylogenies, and David Miller, Robert Burke, and Torbjorn Holmblad kindly provided information before publication. Mike Graner, Tom Bunch, and Mark Ginsberg criticized the manuscript, and Scott Selleck helped with figures. This work was supported by National Institutes of Health Grant GM42474.
Note Added in Proof
Recently, the sequence of an integrin α subunit also has been reported from a sponge (65).
Footnotes
References
- 1.Hynes R O. Cell. 1992;69:11–25. doi: 10.1016/0092-8674(92)90115-s. [DOI] [PubMed] [Google Scholar]
- 2.Brown N H. BioEssays. 1994;15:383–390. doi: 10.1002/bies.950150604. [DOI] [PubMed] [Google Scholar]
- 3.Gettner S N, Kenyon C, Reichardt L F. J Cell Biol. 1995;129:1127–1141. doi: 10.1083/jcb.129.4.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Humphries M J. Cur Opin Cell Biol. 1996;8:632–640. doi: 10.1016/s0955-0674(96)80104-9. [DOI] [PubMed] [Google Scholar]
- 5.Springer T A. Proc Natl Acad Sci USA. 1997;94:65–72. doi: 10.1073/pnas.94.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee J, Rieu P, Arnaout M, Liddington R. Cell. 1995;80:631–638. doi: 10.1016/0092-8674(95)90517-0. [DOI] [PubMed] [Google Scholar]
- 7.Tozer E C, Liddington R C, Sutcliffe M J, Smeeton A H, Loftus J L. J Biol Chem. 1996;271:21978–21984. doi: 10.1074/jbc.271.36.21978. [DOI] [PubMed] [Google Scholar]
- 8.Loftus J C, O’Toole T E, Plow E F, Glass A, Frelinger A L F, Ginsberg M H. Science. 1990;249:915–918. doi: 10.1126/science.2392682. [DOI] [PubMed] [Google Scholar]
- 9.Takada Y, Ylanne J, Mandelman D, Puzon W, Ginsberg M H. J Cell Biol. 1992;119:913–921. doi: 10.1083/jcb.119.4.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bajt M L, Ginsberg M H, Frelinger A L, III, Berndt M C, Loftus J C. J Biol Chem. 1992;267:3789–3794. [PubMed] [Google Scholar]
- 11.Bajt M L, Loftus J C. J Biol Chem. 1994;269:20913–20919. [PubMed] [Google Scholar]
- 12.Bajt M, Goodman T, McGuire S. J Biol Chem. 1995;270:94–98. doi: 10.1074/jbc.270.1.94. [DOI] [PubMed] [Google Scholar]
- 13.Huang X, Chen A, Agrez M, Sheppard D. Am J Respir Cell Mol Biol. 1995;13:245–251. doi: 10.1165/ajrcmb.13.2.7626292. [DOI] [PubMed] [Google Scholar]
- 14.Kamata T, Puzon W, Takada Y. Biochem J. 1995;305:945–951. doi: 10.1042/bj3050945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Puzon-McLaughlin W, Takada Y. J Biol Chem. 1996;271:20438–20443. doi: 10.1074/jbc.271.34.20438. [DOI] [PubMed] [Google Scholar]
- 16.Baker E K, Tozer E C, Pfaff M, Shattil S J, Loftus J C, Ginsberg M H. Proc Natl Acad Sci USA. 1997;94:1973–1978. doi: 10.1073/pnas.94.5.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ziegler U, Stidwell R P. Exp Cell Res. 1992;202:281–286. doi: 10.1016/0014-4827(92)90076-k. [DOI] [PubMed] [Google Scholar]
- 18.Zhang X, Sarras M P., Jr Development (Cambridge, UK) 1994;120:425–432. doi: 10.1242/dev.120.2.425. [DOI] [PubMed] [Google Scholar]
- 19.Agbas A, Sarras M P., Jr Cell Adhes Commun. 1994;2:59–73. doi: 10.3109/15419069409014202. [DOI] [PubMed] [Google Scholar]
- 20.Labat-Robert J, Robert L, Auger C, Lethias C, Garrone R. Proc Natl Acad Sci USA. 1981;78:6261–6265. doi: 10.1073/pnas.78.10.6261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Diehl-Seifert B, Kurelec B, Zahn R K, Dorn A, Jericevic B, Uhlenbruck G, Muller W E. J Cell Sci. 1985;79:271–285. doi: 10.1242/jcs.79.1.271. [DOI] [PubMed] [Google Scholar]
- 22.Exposito J Y, Ouazana R, Garrone R. Eur J Biochem. 1990;190:401–406. doi: 10.1111/j.1432-1033.1990.tb15589.x. [DOI] [PubMed] [Google Scholar]
- 23.Exposito J Y, Le Guellec D, Lu Q, Garrone R. J Biol Chem. 1991;266:21923–21928. [PubMed] [Google Scholar]
- 24.Humbert-David N, Garrone R. Eur J Biochem. 1993;216:255–260. doi: 10.1111/j.1432-1033.1993.tb18140.x. [DOI] [PubMed] [Google Scholar]
- 25.Chomczynski P, Sacchi N. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 26.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1992. [Google Scholar]
- 27.Yee G H, Hynes R O. Development (Cambridge, UK) 1993;118:845–858. doi: 10.1242/dev.118.3.845. [DOI] [PubMed] [Google Scholar]
- 28.Marsden M, Burke R D. Dev Biol. 1997;181:234–245. doi: 10.1006/dbio.1996.8451. [DOI] [PubMed] [Google Scholar]
- 29.Argraves W S, Suzuki S, Arai H, Thompson K, Pierschbacher M D, Ruoslahti E. J Cell Biol. 1987;105:1183–1190. doi: 10.1083/jcb.105.3.1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kishimoto T K, O’Connor K, Lee A, Roberts T M, Springer T A. Cell. 1987;48:681–690. doi: 10.1016/0092-8674(87)90246-7. [DOI] [PubMed] [Google Scholar]
- 31.Fitzgerald L A, Steiner B, Rall S C, Lo S S, Phillips D R. J Biol Chem. 1987;262:3936–3939. [PubMed] [Google Scholar]
- 32.Suzuki S, Naitoh Y. EMBO J. 1990;9:757–763. doi: 10.1002/j.1460-2075.1990.tb08170.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ramaswamy H, Hemler M E. EMBO J. 1990;9:1561–1568. doi: 10.1002/j.1460-2075.1990.tb08275.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sheppard D, Rozzo C, Starr L, Quaranta V, Erle D J, Pytela R. J Biol Chem. 1990;265:11502–11507. [PubMed] [Google Scholar]
- 35.Erle D J, Ruegg C, Sheppard D, Pytela R. J Biol Chem. 1991;266:11009–11016. [PubMed] [Google Scholar]
- 36.Moyle M, Napier M A, McLean J W. J Biol Chem. 1991;266:19650–19658. [PubMed] [Google Scholar]
- 37.MacKrell A, Blumberg B, Haynes S R, Fessler J H. Proc Natl Acad Sci USA. 1988;85:2633–2637. doi: 10.1073/pnas.85.8.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Holmblad T, Thornqvist P, Soderhall K, Johansson M W. J Exp Zool. 1997;277:255–261. [PubMed] [Google Scholar]
- 39.Qu A, Leahy D J. Proc Natl Acad Sci USA. 1995;92:10277–10281. doi: 10.1073/pnas.92.22.10277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dedhar S, Hannigan G. Cur Opin Cell Biol. 1996;8:657–669. doi: 10.1016/s0955-0674(96)80107-4. [DOI] [PubMed] [Google Scholar]
- 41.Reszka A A, Hayashi Y, Horwitz A F. J Cell Biol. 1992;117:1321–1330. doi: 10.1083/jcb.117.6.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.O’Toole T E, Ylanne J, Culley B M. J Biol Chem. 1995;270:8553–8558. doi: 10.1074/jbc.270.15.8553. [DOI] [PubMed] [Google Scholar]
- 43.Ylanne J, Huuskonen J, O’Toole T E, Ginsberg M H, Virtanen I, Gahmberg C G. J Biol Chem. 1995;270:9550–9557. doi: 10.1074/jbc.270.16.9550. [DOI] [PubMed] [Google Scholar]
- 44.Tahiliani P D, Singh L, Auer K L, LaFlamme S E. J Biol Chem. 1997;272:7892–7898. doi: 10.1074/jbc.272.12.7892. [DOI] [PubMed] [Google Scholar]
- 45.Hughes P E, Diaz-Gonzales F, Leong L, Wu C, McDonald J A, Shattil S J, Ginsberg M H. J Biol Chem. 1996;271:6571–6574. doi: 10.1074/jbc.271.12.6571. [DOI] [PubMed] [Google Scholar]
- 46.Neugebauer K M, Reichardt L F. Nature (London) 1991;350:68–71. doi: 10.1038/350068a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shih D-T, Edelman J M, Horwitz A F, Grunwald G B, Buck C A. J Cell Biol. 1993;122:1361–1371. doi: 10.1083/jcb.122.6.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bazzoni G, Shih D T, Buck C A, Hemler M E. J Biol Chem. 1995;270:25570–25577. doi: 10.1074/jbc.270.43.25570. [DOI] [PubMed] [Google Scholar]
- 49.Wilkins J A, Li A, Ni H, Stupack D G, Shen C. J Biol Chem. 1996;271:3046–3051. [PubMed] [Google Scholar]
- 50.Faull R J, Wang J, Leavesley D I, Puzon W, Russ G R, Vestweber D, Takada Y. J Biol Chem. 1996;271:25099–25106. doi: 10.1074/jbc.271.41.25099. [DOI] [PubMed] [Google Scholar]
- 51.Djaffar I, Chen Y-P, Creminon C, Maclouf J, Cieutat A-M, Gayet O, Rosa J-P. Biochem J. 1994;300:69–74. doi: 10.1042/bj3000069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hynes, R. O. (1996) Dev. Biol. 402–412. [DOI] [PubMed]
- 53.Lake J A. Proc Natl Acad Sci USA. 1990;87:763–766. doi: 10.1073/pnas.87.2.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Field K G, Olsen G J, Lane D J, Giovannoni S J, Ghiselin M T, Raff E C, Pace N R, Raff R A. Science. 1988;239:748–753. doi: 10.1126/science.3277277. [DOI] [PubMed] [Google Scholar]
- 55.Berghammer H, Hayward D, Harrison P, Miller D J. Gene. 1996;178:195–197. doi: 10.1016/0378-1119(96)00353-8. [DOI] [PubMed] [Google Scholar]
- 56.Sidow A, Thomas W K. Curr Biol. 1994;4:596–603. doi: 10.1016/s0960-9822(00)00131-7. [DOI] [PubMed] [Google Scholar]
- 57.Marcantonio E E, Hynes R O. J Cell Biol. 1988;106:1765–1772. doi: 10.1083/jcb.106.5.1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schindler M, Meiners S, Cheresh D A. J Cell Biol. 1989;108:1955–1965. doi: 10.1083/jcb.108.5.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Quatrano R S, Brian L, Aldridge J, Schultz T. Development Suppl (Cambridge, UK) 1991;1:11–16. [PubMed] [Google Scholar]
- 60.Wayne R, Staves M P, Leopold A C. J Cell Sci. 1992;101:611–623. doi: 10.1242/jcs.101.3.611. [DOI] [PubMed] [Google Scholar]
- 61.Zhu J-K, Shi J, Singh U, Wyatt S E, Bressan R A, Hasegawa P M, Carpita N C. Plant J. 1993;3:637–646. [PubMed] [Google Scholar]
- 62.Kaminskyj S G, Heath I B. J Cell Sci. 1995;108:849–856. doi: 10.1242/jcs.108.2.849. [DOI] [PubMed] [Google Scholar]
- 63.Correa A, Jr, Staples R C, Hoch H C. Protoplasma. 1997;194:91–102. [Google Scholar]
- 64.Gens J S, Reuzeau C, Doolittle K W, McNally J G, Pickard B G. Protoplasma. 1997;194:215–230. doi: 10.1007/BF01882029. [DOI] [PubMed] [Google Scholar]
- 65.Pancer Z, Kruse M, Müller I, Müller W E G. Mol Biol Evol. 1997;14:391–398. doi: 10.1093/oxfordjournals.molbev.a025775. [DOI] [PubMed] [Google Scholar]