Abstract
The recurring translocation t(11;16)(q23;p13.3) has been documented only in cases of acute leukemia or myelodysplasia secondary to therapy with drugs targeting DNA topoisomerase II. We show that the MLL gene is fused to the gene that codes for CBP (CREB-binding protein), the protein that binds specifically to the DNA-binding protein CREB (cAMP response element-binding protein) in this translocation. MLL is fused in-frame to a different exon of CBP in two patients producing chimeric proteins containing the AT-hooks, methyltransferase homology domain, and transcriptional repression domain of MLL fused to the CREB binding domain or to the bromodomain of CBP. Both fusion products retain the histone acetyltransferase domain of CBP and may lead to leukemia by promoting histone acetylation of genomic regions targeted by the MLL AT-hooks, leading to transcriptional deregulation via aberrant chromatin organization. CBP is the first partner gene of MLL containing well defined structural and functional motifs that provide unique insights into the potential mechanisms by which these translocations contribute to leukemogenesis.
The t(11;16)(q23;p13.3) is a rare recurring translocation that has been described in 11 patients to date (1). All of these patients have therapy-related acute leukemia of myeloid or lymphoid phenotype, or myelodysplasia, after exposure to DNA topoisomerase II inhibitors (anthracyclines or epipodophyllotoxins) for treatment of a primary malignancy.
MLL (also called ALL1, Htrx, and HRX; refs. 2–6), which is located on chromosomal band 11q23, is involved in translocations with at least 40 different partner genes (7–9). These translocations result in acute leukemia, either lymphoblastic or myeloid/monocytic, with a close correlation between the specific translocation and a particular leukemia phenotype. MLL also is involved in translocations that occur secondary to therapy of a primary malignant disease with drugs that target DNA topoisomerase II and result in therapy-related acute myeloid leukemia or acute lymphoblastic leukemia (10–14). The t(11;16)(q23;p13.3) occurs only in therapy-related leukemia or myelodysplasia, in contrast to other MLL translocations such as the t(9;11), t(4;11), or t(11;19), which are seen primarily in de novo leukemia with no more than about 5–10% having leukemia occurring after treatment.
MLL codes for a very large protein, with a predicted molecular mass of 431 kDa (4–6). The protein contains several domains identified by homology to other proteins or by functional analysis. Three AT-hook DNA-binding domains near the amino terminus also are found in the high-mobility group proteins HMG-I(Y) (15). MLL contains a region of homology to mammalian DNA methyltransferases, transcriptional activation and repression domains, and a cysteine-rich region that forms three C4HC3 zinc fingers [plant homeodomain (PHD) or leukemia-associated-protein domains] (16–21). The PHD domain and the SET [Su(var)3-9 enhancer of zeste, and trithorax] domain at the carboxyl terminus are the regions most conserved with the Drosophila trithorax (trx) protein. Trx is required to maintain the proper expression of homeotic genes of the Bithorax and Antennapaedia complexes in Drosophila. Mice with a single disrupted Mll gene created by homologous recombination display bidirectional homeotic transformations and those with homozygous deletions die at embryonic day 10.5 (22). This is similar to changes observed in trx mutant Drosophila. It is thought that trx regulates homeotic expression at the level of chromatin organization by maintaining an “open” chromatin structure.
CBP previously has been mapped to chromosome band 16p13.3. Its genomic locus spans approximately 190 kb, is transcribed from centromere to telomere, and encodes a large protein of 2,442 amino acids (refs. 23–25, this manuscript). CBP is a global transcriptional coactivator involved in the regulation of various DNA binding transcription factors. CBP interacts with the cAMP response element-binding protein (CREB), which is activated as a result of phosphorylation by protein kinase A. CBP also binds to JUN and ATF1 in a phosphorylation-dependent manner, to YY1, FOS, MYB, and to the RelA (p65) subunit of NF-κB (26–30). CBP binds to P/CAF, a histone acetyltransferase that preferentially acetylates histone H3 in mononucleosomes (31). CBP also possesses intrinsic histone acetyltransferase activity and can acetylate all four core histones in nucleosomes (32, 33). Thus, CBP can serve as a multifunctional adaptor protein that coordinates signals from many sequence-specific activators to modulate transcription and/or cell cycle progression. It also may contribute directly to transcriptional regulation via targeted acetylation of chromatin.
CBP is the gene responsible for Rubinstein–Taybi syndrome (RTS), a developmental disorder characterized by facial anomalies, broad thumbs, broad big toes, mental retardation, and a propensity for development of malignancies (34, 35). Constitutional microdeletions, translocations, inversions, and point mutations in the CBP gene have been found in patients with RTS (24). Patients are heterozygous for these mutations leading to the proposition that haploinsufficiency may be the cause of the syndrome (24). The CBP gene also has recently been described as the translocation partner in the t(8;16) in which MOZ, a putative acetyltransferase located on 8p11, is fused to CBP (25). This translocation results in acute myeloid leukemia, both de novo and therapy related, classified mainly as FAB M4 or M5 (36, 37).
In this paper, we show that MLL is fused to CBP in therapy-related acute myeloid or lymphoid leukemia and myelodysplasia in the t(11;16), and propose mechanisms by which this fusion may contribute to leukemogenesis. A better understanding of the molecular processes affected by the fusion of these genes (MLL and CBP) may shed light on the role of MLL and its multiple other fusion partners in the development of leukemia.
MATERIALS AND METHODS
Southern Blot and Spot Blot Hybridization.
DNA was extracted from cryopreserved patient material, digested with restriction enzymes, electrophoresed on 0.8% agarose gels, and transferred onto nylon membranes. Hybridization was performed at 42°C with 32P-labeled cDNA probes. The probe used in this case was the 0.74-kb fragment that spans the MLL breakpoint cluster region (BCR) (exons 5–11) (8). The blots were washed at a final stringency of 1% saline sodium citrate buffer and 1% sodium dodecyl sulfate before autoradiography. p1 artificial chromosome (PAC) DNA and cosmid DNA also were spotted onto nylon membranes, and hybridization and radiolabeling with CBP cDNA probes were performed as described above. CBP cDNA probes used were a 1.1-kb fragment encoding amino acids 1–346, a 1.2-kb fragment encoding amino acids 722–1,120, and a 4.8-kb fragment encoding amino acid 1,120 through the 3′ untranslated region (UTR). Hybridization of PAC DNA with genomic probes spanning the 5′ end of the CBP gene (to map the extent of the PAC relative to the CBP genomic region) also was performed using similar conditions.
PAC/Yeast Artificial Chromosome (YAC) Screening.
PAC (Genome Systems–Down to the Well) and YAC libraries were screened using the PCR primers V16T1 (5′-AGC ACATCC CAG AAC AGA AAA TCAG-3′) and V16B2 (5′-CTGCACTGTTAAAGAAATCTCTTTGGGG-3′). Approximately 700 mega YACs, which comprise the chromosome 16 physical map, were grouped into 94 pools representing 94 overlapping bins from pter to qter. YACs, which make up positive pools, were screened individually to identify the positive YAC(s). PCR conditions for YAC screening were done as previously described (38).
Fluorescence in Situ Hybridization (FISH) Analysis.
FISH was performed as previously described (39). A mixture of cosmids c365F4, c444A4, c443G8, c388H4, c304A10, c312B2, c379G3, c330H2, c58E12, c307E6 (NAD) as well as cosmids TES2, 541E10-c23, TES 5, 541E10-C53, and 376E2-C1 (JB) were used for FISH on patient material.
MLL-CBP RT-PCR Assay.
Total RNA was extracted from cryopreserved patient material (patient 1) or from material previously processed for cytogenetic purposes (patient 2) using TriReagent (Molecular Research). First-strand cDNA was made from 5 μg of RNA, using the first-strand cDNA cycle kit (Invitrogen), according to manufacturer’s instructions. Two microliters of the reaction was used as template for nested RT-PCR using MLL exon 5 or 6 (forward) primers and CBP (reverse) primers for amplification of MLL-CBP fusion transcripts.
For patient 1, primers used in the first round were 5′-GTCCAGAGCAGAGCAAACAGAAAAAAGTGGCTCCC-3′ (MLL exon 6) and 5′-ATAATATTCATCCCTGCTGTTGGC-3′ (CBP). In the second round, primers used were 5′-GCCCAAGTATCCCTGTAAAACAAAAACCAAAAG-3′ (MLL exon 6) and 5′-AGACTCGTACATGTCCCCTTCCAC-3′ (CBP). For patient 2, primers used in the first round were 5′-GGATCCTGCCCCAAAGAAAGCAGTAGTGAGCC-3′ (MLL exon 5) and 5′-AGGAATGGTACACAGCTGCTTCCC-3′ (CBP). In the second round, primers used were 5′-GCCAGCACTGGTCATCCCGCCT- CAG-3′ (MLL exon 5) and 5′-CAGCACAAAGTCTGTGGGGAAAA-3′ (CBP). PCR reactions were in a final volume of 50 μl with 0.4 μM each primer, 125 μM each dNTP, PCR buffer (10 mM Tris⋅HCl/1.5 mM MgCl2/0.1% Triton X), and 2 units of Taq polymerase. PCR amplification was performed for 35 cycles (94°C for 1 min, 60°C for 1 min, and 72°C for 2 min). PCR products were analyzed by electrophoresis on a 1% agarose gel. RT-PCR experiments to amplify the CBP-MLL fusion in patient 1 used identical conditions. Primers used in the first round were 5′-GTTCCAGATGCTGCTTCCAAAC-3′ (CBP) and 5′-GGGGGGTCTAGACCTGTGGACTCCATCTGCTGGAAT-3′ (MLL exon 7). In the second round primers used were 5′-AACAACTGTCGGAGCTTCTACGAG-3′ (CBP) and 5′-TTGGAGAGAGTGCTGAGGATGTTC-3′ (MLL exon7). The position of the CBP primers used in these experiments are indicated by arrows under the appropriate sequence in Fig. 4.
Figure 4.
Complete cDNA and amino acid sequence of human CBP. The arrowheads indicate the two different fusion junctions in the MLL-CBP chimeric cDNA. The three cysteine/histidine-rich regions are shown by circling the C and H residues. The second cysteine/histidine-rich region forms a C4HC3 finger, hence only the zinc coordinating residues are circled here. This C4HC3 finger is part of the minimal area, which contains the histone acetyltransferase (HAT) activity. This HAT-containing region is indicated by shading. The first boxed area (amino acids 1,106–1,170) corresponds to the bromodomain, and the second boxed area (amino acids 1,852–2,411) to the Q-rich carboxyl terminus. The PCR primers used for RT-PCR analysis are indicated by arrows under the appropriate sequence.
Subcloning and Sequencing of PCR Products.
PCR products were purified after electophoresis on agarose gels using the Qiaquick protocol (Qiagen) according to manufacturer’s instructions. Subcloning into the pcr 2.1 vector was performed using the TA cloning kit (Invitrogen). Dideoxy sequencing was performed using Sequenase version 2.0 (U.S. Biochemicals).
RESULTS
Genomic DNA Cloning of the t(11;16).
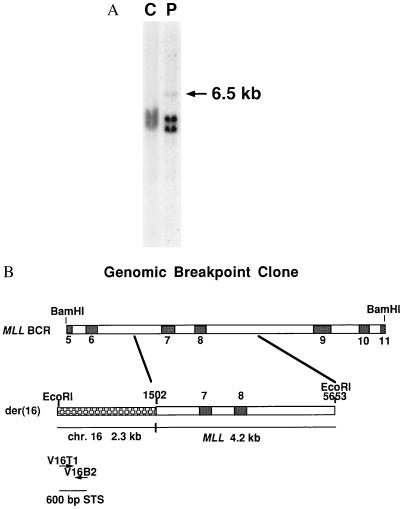
We previously had determined in two cases of t(11;16)(q23;p13) that MLL was the gene on chromosome 11 involved in the translocation (40, 41). A Southern blot analysis of DNA from one patient (40) digested with EcoRI demonstrated a 6.5-kb rearranged band using an MLL probe (Fig. 1A). To determine the partner gene on chromosome 16, a genomic breakpoint clone was obtained from a library of EcoRI-digested genomic DNA from this patient (patient 1). Partial sequencing of this clone and comparison to the MLL genomic breakpoint cluster region (BCR) sequence (42) revealed that it corresponded to the der(16) fusion product. It contained 4.2 kb of normal MLL sequence that ended within intron 6, fused to 2.3 kb of novel sequence (Fig. 1B). A nonrepetitive sequence within this region was hybridized to a somatic cell hybrid panel blot and was shown to be from chromosome 16 (data not shown). This sequence was used to design primers that amplify a 600-bp sequence tagged site (STS) (Fig. 1B) to use for PCR screening of genomic PAC and YAC libraries. A single PAC clone, S10622, isolated with these STS primers was used for FISH. The hybridization signal on normal chromosomes occurred in band 16p13.3, confirming the location of the translocation breakpoint to this band. The PAC was translocated to the derivative chromosome 11 [der(11)] in patient 1, but remained on the der(16) chromosome in patient 2 (1), implying that the genomic breakpoints in these two patients were likely to be at least 100 kb apart (the size of a PAC clone). The same 600-bp STS was used to screen Centre d’Etude Polymorphisme Humain (CEPH) mega YAC pools comprising a nearly complete physical map of human chromosome 16 (38). A single YAC clone (615F4) was identified from these pools, but was found to be chimeric and not useful as a probe for FISH. This YAC was located on the chromosome 16 physical map near the CBP gene based on previous STS content results (38), suggesting that CBP was a good candidate gene for the MLL fusion partner.
Figure 1.
Genomic cloning of the t(11;16). (A) Southern blot analysis of DNA to confirm MLL rearrangement. DNA from patient 1 with a t(11;16) (P) or from placental control (C) was digested with EcoRI, electrophoresed, and then hybridized with an MLL 0.74-kb BamHI cDNA probe. The 6.5-kb rearranged band is indicated by the arrow. The germ-line bands are 4.6 kb and 4.3 kb. The rearranged fragment is faint because only a small portion of the probe hybridizes to this fragment. (B) Schematic diagram of the 6.5-kb genomic breakpoint clone. The 2.3-kb novel sequence on chromosome 16 (chr 16) is fused to a 4.2-kb MLL sequence and corresponds to the der(16) chromosome. MLL exons are indicated by numbered boxes. The indicated nucleotide positions in MLL (nt) correspond to the genomic MLL BCR with nucleotide 1 as the BamHI site at the centromeric end of the BCR (40). V16T1 and V16B2 are primers derived from sequence on chromosome 16, and they amplify a 600-bp STS.
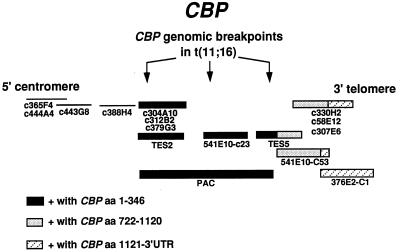
CBP cDNA clones were used as probes to determine whether the PAC contained any CBP coding sequence. A probe that codes for the amino end of the protein (a 1.1-kb PstI fragment containing amino acids 1–346) hybridized to the PAC, whereas probes that span the more carboxyl regions (a 1.2-kb EcoRI fragment that spans amino acids 722–1,120 and a 4.8-kb fragment that codes for amino acid 1,121 through the 3′ UTR) did not hybridize to the PAC. This indicated that the PAC isolated using the genomic breakpoint STS contained CBP coding sequences corresponding to the amino terminus of CBP (Fig. 2).
Figure 2.
Schematic diagram of cosmids and PAC that span CBP. A PAC and cosmids that span CBP were used for FISH analysis of patients with a t(11;16). A probe that codes for the amino end of CBP (a 1.1-kb PstI fragment encoding amino acids 1–346) hybridized to the PAC, whereas probes that span the more carboxyl regions (a 1.2-kb EcoRI fragment that spans amino acids 722–1,120 and a 4.8-kb fragment that codes for amino acid 1,121 through the 3′ UTR) did not hybridize to the PAC. The cosmids that hybridized to the CBP probes are indicated. The variability in CBP genomic breakpoints based on results obtained by FISH experiments with PAC DNA (described in ref. 1), is depicted by the arrows. The breakpoints in patients 1 and 2, from whom the translocation junctions were cloned, are identified by the most centromeric and the most telomeric arrows, respectively. Other patients described in reference 1 had a breakpoint in the middle of the PAC.
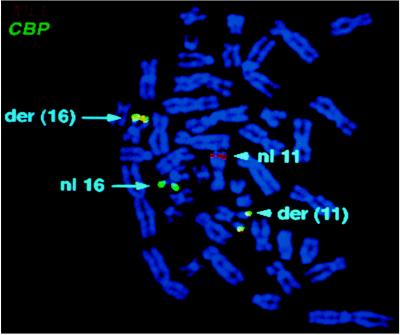
Because CBP exons were contained in the genomic PAC clone, we next obtained cosmids that spanned the CBP gene region and used them for FISH analysis of patient material to determine whether CBP was involved in the t(11;16). Using a mixture of CBP cosmids, a signal was obtained on both the der(11) and der(16) chromosomes, confirming that CBP is split in the t(11;16) (Fig. 3).
Figure 3.
FISH analysis of a t(11;16) patient with MLL and CBP probes. Metaphase cell from a t(11;16) patient labeled with MLL (chromosome 11 in red) and CBP (chromosome 16 in green). The two derivative chromosomes show a fusion (yellow) signal.
Complete Human CBP cDNA Sequence.
Although the sequence of murine CBP is available (excluding the UTRs), only partial sequences corresponding to its human counterpart have been reported. We previously have deposited a partial human CBP cDNA sequence extending from within the 5′ UTR to amino acid 405 (U47741). Additionally, a preliminary human protein sequence of the first 1,267 amino acids has been published (43). The GenBank entry (S39162) for a human CBP protein is actually a chimera of human and murine sequences (human from 1–1,267 and murine from 1,268–2,440). Because three different human disorders are now known to be associated with mutations within CBP, we decided to sequence the entire human cDNA to obtain an unambiguous sequence (Fig. 4). This sequence was redeposited as an update of U47741. A series of overlapping CBP cDNAs from a U937 cDNA library were sequenced using an Applied Biosystems prism DNA sequencer. Human CBP encodes a protein of 2,442 amino acids, which is highly conserved with its mouse counterpart (95% identity, 96.5% similarity). Comparison of our human CBP sequence with mouse CBP (S66385) and p300 (a functional homologue of CBP) suggests that amino acids 1,418–1,429 of murine CBP are frameshifted. The human cDNA sequence was used to design multiple sets of nested primers to identify the RT-PCR junction fragments between MLL and CBP.
RT-PCR Analysis of t(11;16) Patient Material.
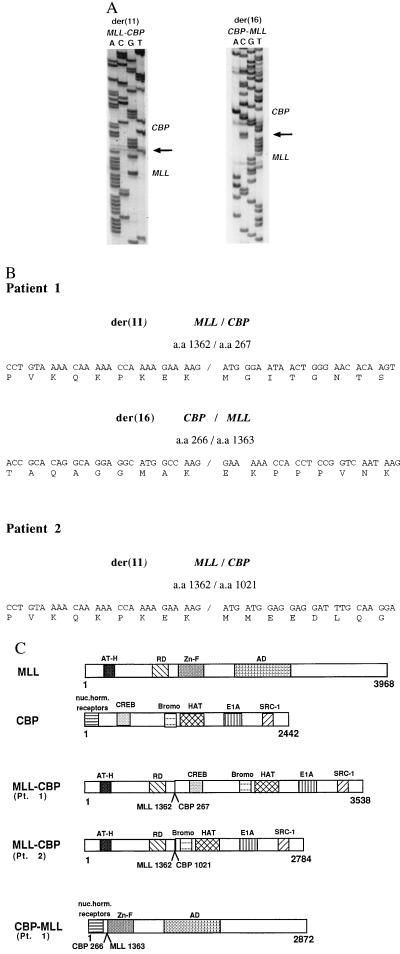
To determine where within MLL and CBP the break occurs, and whether the translocation results in an in-frame fusion of MLL to CBP, we used RT-PCR to amplify the cDNA junction from two patients. Oligonucleotide primers were designed from exons 5 and 6 of MLL and from the CBP gene (Fig. 4). Patient 1 had a more centromeric break in CBP at the genomic level, while patient 2 had a more telomeric break as determined by FISH experiments with PAC DNA (Fig. 2) (1). Amplification of cDNA from patient 1 resulted in a 1,169-bp fragment after nested RT-PCR with MLL exon 6 (forward) primers and CBP (reverse) primers (data not shown). A 768-bp fragment was obtained on amplification of cDNA from patient 2 with MLL exon 5 (forward) primers and more telomeric CBP (reverse) primers. The PCR fragments were subcloned and sequenced. Analysis of the sequence of these chimeric transcripts revealed an in-frame fusion between MLL codon 1,362 and CBP codon 267 in patient 1 (Fig. 5). In patient 2, MLL codon 1,362 was fused to CBP codon 1,021. Use of CBP (forward primers) and MLL exon 7 (reverse primers) to amplify the CBP-MLL fusion transcript produced from the der(16) chromosome in patient 1 resulted in a 665-bp fragment. Sequencing of this fragment showed an in-frame fusion of CBP codon 266 to MLL codon 1,363 (Fig. 5).
Figure 5.
MLL is fused in-frame to CBP in the t(11;16). (A) Sequence of the der(11) MLL-CBP fusion junction and the der(16) CBP-MLL fusion junction amplified from patient 1 with the t(11;16) by RT-PCR. The arrows indicate the breakpoint junctions. (B) Sequence of the der(11) MLL-CBP breakpoint junction and the der(16) CBP-MLL breakpoint junction from patient 1 and the der(11) MLL-CBP breakpoint junction from patient 2. All fusions are in-frame with no gain or loss of nucleotides at the cDNA level. (C) Schematic diagram of the consequences of the t(11;16) including the MLL-CBP and the CBP-MLL fusion proteins and the normal MLL and CBP proteins. AT-H, AT-hook DNA binding domain; RD, transcriptional repression domain; Zn-F, the C4HC3 zinc cluster (PHD domain); AD, transcriptional activation domain; Bromo, bromodomain; nuc. horm. receptors, nuclear hormone receptor binding domain; SRC-1, steroid receptor coactivator-1 binding domain; HAT, histone acetyltransferase activity; E1A, E1A binding activity.
DISCUSSION
Cloning of the t(11;16) breakpoint of therapy-related acute leukemia has demonstrated that MLL is fused to the CBP gene (Fig. 5C). The breaks in MLL fall within the previously described 8.3-kb genomic BCR (8, 44). The location of the breaks within CBP span a very large genomic region that consists mainly of one intron. It is intriguing that, to date, all cases of t(11;16) occur secondary to treatment for a primary malignancy with drugs that target DNA topoisomerase II (1). We previously have noted the association of prior treatment with topoisomerase II-targeting drugs, particularly the epipodophyllotoxins, and therapy-related acute myeloid leukemias with MLL translocations (11). We and others also have identified a unique strong in vivo topoisomerase II cleavage site within the MLL BCR that is induced by treatment of cells with either VP16, VM26, or doxorubicin (45, 46). It may be that the genomic region of CBP that is involved in the therapy-related acute myeloid leukemias with a t(11;16) is especially accessible to rearrangements with MLL after exposure to drugs that target DNA topoisomerase II. It will be informative to map the location of drug-induced cleavage sites within CBP, and to determine whether the DNA only becomes accessible to fusion with MLL after drug treatment.
Translocations involving in-frame fusion of CBP to another gene result in acute leukemia [both the t(11;16) described in this report and the t(8;16)]. In the first patient studied here, the break in CBP occurs after amino acid 266. Interestingly, this is the same region of CBP that is fused to MOZ in the t(8;16) that also results in acute myeloid leukemia (25). Patients with t(8;16) are predominantly classified as FAB M4 (myelomonocytic) or M5 (monocytic), similar to the majority of patients with acute myeloid leukemia involving MLL. In the second patient studied here, however, the break in CBP occurs after amino acid 1,021, just upstream of the bromodomain and histone acetyltransferase domain. In contrast, deletions or point mutations of this gene result in RTS (24). Translocations also have been observed in RTS, but it is not known whether they result in novel fusion messages (24). The mutations that occur in RTS are constitutional, therefore are present in all tissues, whereas the CBP translocations that result in leukemia occur somatically in the hematopoietic precursor cells. In both cases only one allele of CBP is affected. This strongly suggests that a novel function, due to activity contributed by the fusion of CBP to MLL or to MOZ, can result in leukemia. Alternatively there may be three events involved, including haploinsufficiency caused by loss of one functional allele of both MLL and CBP, and gain of function with the fusion product. General genomic instability of this region is revealed by the inversions, deletions, and translocations found in patients with RTS and also by the t(8;16) and t(11;16).
The function of MLL is unknown; however, because of its homology to Drosophila trx, it is thought to be critical for maintaining the proper expression of homeotic genes. Evidence to support this hypothesis is found in recent experiments that demonstrate altered Hox gene expression and bidirectional homeotic transformations in Mll-mutant mice (22). Some information is known about several functional domains of MLL that suggest potential mechanisms of action. In the t(11;16), the resultant fusion product expressed from the der(11) chromosome contains the AT-hooks, the methyltransferase homology domain, and the repression domain of MLL and the carboxyl portion of CBP, whereas there is loss of the cysteine-rich C4HC3 PHD domains (20) and the activation domain of MLL in the der(11) product (18). What is also likely to be critical in generating the leukemia phenotype are the novel domains of CBP that are juxtaposed to MLL sequences. The der(11) fusion product in patients with the more centromeric DNA break, would lose only the domain of CBP that binds nuclear hormone receptors, whereas in patients with the more telomeric break, the CREB binding domain is also lost. The shorter MLL-CBP fusion presumably would not retain the ability to act as a transcriptional coadaptor in the cAMP- and mitogen-responsive signaling pathways, or in the nuclear hormone pathways. However, it should maintain its function as an adaptor protein that coordinates cell cycle progression with NF-κB transcriptional regulation. Moreover, the fusion proteins presumably would retain CBP’s ability to acetylate all four core histones, whether free or in nucleosomes. Targeted histone acetylation could contribute to promoter activation by changing or disrupting the repressive chromatin structure (31, 47). The bromodomain also would be retained, which is thought to be important in protein-protein interactions as well as modulation of chromatin because of its conservation in the SWI/SNF2 complex, that appears to modulate the chromatin of the genes it regulates (48, 49). All of these functions would be brought to the amino terminal portion of MLL, that presumably would retain its ability to bind AT-rich DNA, perhaps redirecting the chromatin modification and coadaptor functions of CBP to inappropriate genomic regions.
None of the other partner genes of MLL cloned to date has been characterized in as much detail structurally or functionally as CBP. There is strong evidence, though, that these partner genes are critical in leukemogenesis. Almost all the breaks occur in the same region in MLL yet the leukemia phenotype varies depending on the partner gene involved in a particular translocation. Moreover, Mll-AF9 chimeras, created by homologous recombination develop acute myeloid leukemia, similar to patients with the t(9;11), whereas Mll-myc chimeras do not (50). A recent report has shown that ELL, the partner gene of MLL in the t(11;19)(q23;p13.3) encodes an RNA polymerase II elongation factor (51). This provides another example (besides CBP) of a partner gene of MLL that is involved in gene regulation, although its mechanism of action has not yet been elucidated. It will be important to study other partner genes of MLL to determine whether they possess any domains functionally analogous to some or all of the multiple domains contributed by CBP to the MLL/CBP fusion. It will be particularly important to determine whether MLL or any of its partner genes possess histone acetyltransferase activity. A similar type of functional activity could be present in the various genes involved even though they do not contain a conserved motif that is apparent in the amino acid or DNA sequence (32). This may generate a more global or unifying hypothesis with regard to the role of the partner gene(s) in the many translocations in which MLL is involved.
Acknowledgments
We thank Drs. Michelle Le Beau, Diane Roulston, and Richard Larson for cytogenetic analysis and clinical information on patient 1. We thank Stephen Hunger and Christopher Denny for helpful advice regarding the paper. This work was supported by grants from the National Institutes of Health (CA42557) (J.D.R.), NCHGR HG00299 and NIH-NCI CA17575 (D.H.); a Special Fellowship from the Leukemia Society of America (J.B.); the U.S. Department of Energy under contract W7405-ENG-35 (N.A.D.); and the Wilhelm Sander Stiftung (B.S.).
ABBREVIATIONS
- CREB
cAMP-response element binding protein
- CBP
CREB binding protein
- RTS
Rubinstein–Taybi syndrome
- UTR
untranslated region
- PHD
plant homeodomain
- PAC
p1 artificial chromosome
- YAC
yeast artificial chromosome
- FISH
fluorescence in situ hybridization
- STS
sequence-tagged site
- BCR
breakpoint cluster region
Note Added in Proof
Since submitting this paper, we have learned that two other groups also have recently demonstrated that MLL and CBP are the genes involved in the 11;16 translocation (52, 53). We also have learned that another group recently sequenced the human CBP cDNA (54).
Footnotes
Data deposition: The human CBP sequence reported on in this paper has been deposited in the GenBank database (accession no. U47741).
References
- 1.Rowley J D, Reshmi S, Sobulo O, Musvee T, Anastasi J, Raimondi S, Schneider N R, Barredo J C, Schlegelberger B, Behm F, Doggett N A, Borrow J, Zeleznik-Le N. Blood. 1997;90:535–541. [PubMed] [Google Scholar]
- 2.Ziemin-van der Poel S, McCabe N R, Gill H J, Espinosa R, III, Patel Y, Harden A, Rubinelli P, Smith S D, Le Beau M M, Rowley J D, Diaz M O. Proc Natl Acad Sci USA. 1991;88:10735–10739. doi: 10.1073/pnas.88.23.10735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cimino G, Moir D T, Canaani O, Williams K, Crist W M, Katzav S, Cannizzaro L, Lange B, Nowell P C, Croce C M, Canaani E. Cancer Res. 1991;51:6712–6714. [PubMed] [Google Scholar]
- 4.Djabali M, Selleri L, Parry P, Bower M, Young B D, Evans G A. Nat Genet. 1992;2:113–118. doi: 10.1038/ng1092-113. [DOI] [PubMed] [Google Scholar]
- 5.Tkachuk D C, Kohler S, Cleary M L. Cell. 1992;71:691–700. doi: 10.1016/0092-8674(92)90602-9. [DOI] [PubMed] [Google Scholar]
- 6.Gu Y, Nakamura T, Alder H, Prasad R, Canaani O, Cimino G, Croce C M, Canaani E. Cell. 1992;71:701–708. doi: 10.1016/0092-8674(92)90603-a. [DOI] [PubMed] [Google Scholar]
- 7.Rowley J D. In: Seminars in Cancer Biology. Rabbitts T H, editor. London: Academic; 1993. pp. 377–385. [Google Scholar]
- 8.Thirman M J, Gill H J, Burnett R C, Mbangkollo D, McCabe N R, Kobayashi H, Ziemin-van der Poel S, Kaneko Y, Morgan R, Sandberg A A, Chaganti R S K, Larson R A, Le Beau M M, Diaz M O, Rowley J D. N Engl J Med. 1993;329:909–914. doi: 10.1056/NEJM199309233291302. [DOI] [PubMed] [Google Scholar]
- 9.Bernard O A, Berger R. Genes Chromosomes Cancer. 1995;13:75–85. doi: 10.1002/gcc.2870130202. [DOI] [PubMed] [Google Scholar]
- 10.Hunger S P, Tkachuk D C, Amylon M D, Link M P, Carroll A J, Welborn J L, Willman C L, Cleary M L. Blood. 1993;81:3197–3203. [PubMed] [Google Scholar]
- 11.Gill-Super H J, McCabe N R, Thirman M J, Larson R A, Le Beau M M, Pedersen-Bjergaard J, Preben P, Diaz M O, Rowley J D. Blood. 1993;82:3705–3711. [PubMed] [Google Scholar]
- 12.Felix C A, Lange B J, Hosler M R, Fertala J, Bjornsti M A. Cancer Res. 1995;55:4287–4292. [PubMed] [Google Scholar]
- 13.Domer P H, Head D R, Renganathan N, Raimondi S C, Yang E, Atlas M. Leukemia. 1995;9:1305–1312. [PubMed] [Google Scholar]
- 14.Pedersen-Bjergaard J, Rowley J D. Blood. 1994;83:2780–2786. [PubMed] [Google Scholar]
- 15.Reeves R, Nissen M S. J Biol Chem. 1990;265:8573–8582. [PubMed] [Google Scholar]
- 16.Domer P H, Fakharzadeh S S, Chen C-S, Jockel J, Johansen L, Silverman G A, Kersey J H, Korsmeyer S J. Proc Natl Acad Sci USA. 1993;90:7884–7888. doi: 10.1073/pnas.90.16.7884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ma Q, Alder H, Nelson K K, Chatterjee D, Gu Y, Nakamura T, Canaani E, Croce C M, Siracusa L D, Buchberg A M. Proc Natl Acad Sci USA. 1993;90:6350–6354. doi: 10.1073/pnas.90.13.6350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zeleznik-Le N J, Harden A M, Rowley J D. Proc Natl Acad Sci USA. 1994;91:10610–10614. doi: 10.1073/pnas.91.22.10610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Prasad R, Yano T, Sorio C, Nakamura T, Rallapalli R, Gu Y, Leshkowitz D, Croce C M, Canaani E. Proc Natl Acad Sci USA. 1995;92:12160–12164. doi: 10.1073/pnas.92.26.12160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schindler U, Beckmann H, Cashmore A R. Plant J. 1993;4:137–150. doi: 10.1046/j.1365-313x.1993.04010137.x. [DOI] [PubMed] [Google Scholar]
- 21.Saha V, Chaplin T, Gregorini A, Ayton P, Young B D. Proc Natl Acad Sci USA. 1995;92:9737–9741. doi: 10.1073/pnas.92.21.9737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu B D, Hess J L, Horning S E, Brown G A, Korsmeyer S J. Nature (London) 1995;378:505–508. doi: 10.1038/378505a0. [DOI] [PubMed] [Google Scholar]
- 23.Wydner K L, Bhattacharya S, Eckner R, Lawrence J B, Livingstone D M. Genomics. 1995;30:395–396. [PubMed] [Google Scholar]
- 24.Petrij F, Giles R H, Dauwerse H G, Saris J J, Hennekam R C M, Masuno M, Tommerup N, Ommen G B, Goodman R H, Peters D J M, Breuning M H. Nature (London) 1995;376:348–351. doi: 10.1038/376348a0. [DOI] [PubMed] [Google Scholar]
- 25.Borrow J, Stanton V P, Andresen J M, Becher R, Behm F G, Chaganti R S K, Civin C I, Disteche C, Dube I, Frischauf A M, Horsman D, Mitelman F, Volinia S, Watmore A E, Housman D E. Nat Genet. 1996;14:33–41. doi: 10.1038/ng0996-33. [DOI] [PubMed] [Google Scholar]
- 26.Kwok R P S, Lundbland J R, Chrivia J C, Richards J P, Bachinger H P, Brennan R G, Roberts S G E, Green M R, Goodman R H. Nature (London) 1994;370:223–226. doi: 10.1038/370223a0. [DOI] [PubMed] [Google Scholar]
- 27.Arias J, Alberts A S, Brindle P, Claret F X, Smeal T, Karin M, Feramisco J, Montminy M. Nature (London) 1994;370:226–229. doi: 10.1038/370226a0. [DOI] [PubMed] [Google Scholar]
- 28.Bannister A J, Oehler T, Wilhelm D, Angel P, Kouzarides T. Oncogene. 1995;11:2509–2514. [PubMed] [Google Scholar]
- 29.Dai P, Akimaru H, Tanaka Y, Hou D X, Yasukawa T, Kanei-Ishii C, Takahashi T, Ishii S. Genes Dev. 1996;10:528–540. doi: 10.1101/gad.10.5.528. [DOI] [PubMed] [Google Scholar]
- 30.Perkins N D, Felzien L K, Betts J C, Leung K, Beach D H, Nabel G J. Science. 1997;275:523–527. doi: 10.1126/science.275.5299.523. [DOI] [PubMed] [Google Scholar]
- 31.Yang X J, Ogryzko V V, Nishikawa J, Howard B H, Nakatani Y. Nature (London) 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 32.Ogryzko V V, Schiltz R L, Russanova V, Howard B H, Yoshihiro N. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 33.Bannister A J, Kouzarides T. Nature (London) 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 34.Rubinstein J H, Taybi H. Am J Dis Child. 1963;105:588–608. doi: 10.1001/archpedi.1963.02080040590010. [DOI] [PubMed] [Google Scholar]
- 35.Miller R W, Rubinstein J H. Amer J Med Genet. 1995;56:112–115. doi: 10.1002/ajmg.1320560125. [DOI] [PubMed] [Google Scholar]
- 36.Hanslip J I, Swansbury G J, Pinkerton R, Catovsky D. Leuk Lymphoma. 1992;6:479–486. [Google Scholar]
- 37.Quesnal B, Kantarjian H, Pedersen-Bjergaard J, Brault P, Estey E, Luc Lai J, Tilly H, Stoppa A, Archimbaud E, Harousseau J, Bauters F, Fenaux P. J Clin Oncol. 1993;11:2370–2379. doi: 10.1200/JCO.1993.11.12.2370. [DOI] [PubMed] [Google Scholar]
- 38.Doggett, N. A., Coogwin, L. A., Tesmer, J. G., Meincke, L. J., Bruce, D. C., Clark, L. M., Altherr, M. R., Ford, A. A., Chi, H. C. & Murrone, B. L. (1995) Nature (London) Suppl., 377, 335–365. [DOI] [PubMed]
- 39.Rowley J D, Diaz M O, Espinosa R, III, Patel Y D, van Melle E, Ziemin S, Taillon-Miller P, Lichter P, Evans G A, Kersey J H, Ward D C, Domer P H, Le Beau M M. Proc Natl Acad Sci USA. 1990;87:9358–9362. doi: 10.1073/pnas.87.23.9358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roulston D, Anastasi J, Rudinsky R, Nucifora G, Zeleznik-Le J, Rowley J D. Blood. 1995;86:3613–3614. [PubMed] [Google Scholar]
- 41.Rowley J D, Vignon C, Rosenberg C L, Gollin S M, Wyandt H E, Mulinsky A. N Engl J Med. 1996;334:601–603. doi: 10.1056/NEJM199602293340916. [DOI] [PubMed] [Google Scholar]
- 42.Mbangkollo D, Burnett R, McCabe N, Thirman M, Gill H, Yu H, Rowley J D, Diaz M O. DNA Cell Biol. 1995;14:475–483. doi: 10.1089/dna.1995.14.475. [DOI] [PubMed] [Google Scholar]
- 43.Chrivia J C, Kwok R P S, Lamb N, Hagiwara M, Montminy M R, Goodman R H. Nature (London) 1993;365:855–859. doi: 10.1038/365855a0. [DOI] [PubMed] [Google Scholar]
- 44.McCabe N R, Burnett R C, Gill H J, Thirman M J, Mbangkollo D, Kipiniak M, van Melle E, Ziemin-van der Poel S, Rowley J D, Diaz M O. Proc Natl Acad Sci USA. 1992;89:11794–11798. doi: 10.1073/pnas.89.24.11794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aplan P D, Chervinsky D S, Stanulla M, Burhans W C. Blood. 1996;87:2649–2658. [PubMed] [Google Scholar]
- 46.Strissel, P. L., Hart, C., Harden, A., Rowley, J. D. & Zeleznik-Le, N. J. (1996) Blood Suppl. 1, 250 (abstr.).
- 47.Brownell J E, Zhou J, Ranali T, Kobayashi R, Edmonston D G, Roth S Y, Allis C D. Cell. 1996;84:843–851. doi: 10.1016/s0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- 48.Haynes S R, Dollard C, Winston F, Beck S, Trowsdale J, Dawid I B. Nucleic Acids Res. 1992;20:2603. doi: 10.1093/nar/20.10.2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tamkun J W, Deuring R, Scott M P, Kissinger M, Pattatucci A M, Kaufman T C, Kennison J A. Cell. 1992;68:561–572. doi: 10.1016/0092-8674(92)90191-e. [DOI] [PubMed] [Google Scholar]
- 50.Corral J, Lavenir I, Impey H, Warren A J, Forster A, Larson T A, Bell S, McKenzie A N J, King C, Rabbitts T H. Cell. 1996;85:853–861. doi: 10.1016/s0092-8674(00)81269-6. [DOI] [PubMed] [Google Scholar]
- 51.Shilatifard A, Lane W S, Jackson K W, Conaway R C, Conaway J W. Science. 1996;271:1873–1876. doi: 10.1126/science.271.5257.1873. [DOI] [PubMed] [Google Scholar]
- 52.Taki T, Sako M, Tsuchida M, Hayashi Y. Blood. 1997;89:3945–3950. [PubMed] [Google Scholar]
- 53.Satake, N., Ishida, Y., Otoh, Y., Hinohara, S., Kobayashi, H., Sakashita, A., Maseki, N. & Kaneko, Y. (1997) Genes Chromosomes Cancer, in press. [PubMed]
- 54.Giles R H, Petrij F, Dauwerse H G, Hollander A I, Lushnikova T, van Ommen G-J B, Goodman R H, Deaven L L, Doggett N A, Peters D J M, Breuning M H. Genomics. 1997;42:96–114. doi: 10.1006/geno.1997.4699. [DOI] [PubMed] [Google Scholar]