Abstract
Primitive subsets of leukemic cells isolated by using fluorescence-activated cell sorting from patients with newly diagnosed Ph+/BCR–ABL+ chronic myeloid leukemia display an abnormal ability to proliferate in vitro in the absence of added growth factors. We now show from analyses of growth-factor gene expression, protein production, and antibody inhibition studies that this deregulated growth can be explained, at least in part, by a novel differentiation-controlled autocrine mechanism. This mechanism involves the consistent and selective activation of IL-3 and granulocyte colony-stimulating factor (G-CSF) production and a stimulation of STAT5 phosphorylation in CD34+ leukemic cells. When these cells differentiate into CD34− cells in vivo, IL-3 and G-CSF production declines, and the cells concomitantly lose their capacity for autonomous growth in vitro despite their continued expression of BCR–ABL. Based on previous studies of normal cells, excessive exposure of the most primitive chronic myeloid leukemia cells to IL-3 and G-CSF through an autocrine mechanism could explain their paradoxically decreased self-renewal in vitro and slow accumulation in vivo, in spite of an increased cycling activity and selective expansion of later compartments.
Keywords: stem cells, growth factors, STAT5
Chronic myeloid leukemia (CML) is a clonal multilineage myeloproliferative disorder characterized by an excessive production of granulocytes in the initial, chronic phase of the disease. In each CML patient, the clone develops from a single pluripotent hematopoietic stem cell (reviewed in refs. 1–4) that undergoes a unique rearrangement of the BCR gene on chromosome 22 and the ABL gene on chromosome 9 (5, 6). The BCR–ABL fusion gene produces a 210-kDa protein (p210BCR–ABL) (7) with increased tyrosine kinase activity (8) and an abnormal cytoplasmic location (9). Both features are important to the transforming activity of p210BCR–ABL in model systems (10–14). Expression of the BCR–ABL gene perturbs many intracellular signaling pathways, including the Ras, phosphatidylinositol-3′ kinase, and JAK/STAT pathways (15). However, the connections between these changes and the biological abnormalities characteristic of Ph+/BCR–ABL+ progenitors from CML patients (2–4) has remained unclear.
Previous studies have demonstrated similar biological and biochemical changes in primary CML cells or BCR–ABL-transfected cell lines and IL-3-stimulated cells (15), and the observation of an activation of IL-3 production in some cell lines after their transformation by BCR–ABL (16–18) has suggested a possible explanation for the ability of BCR–ABL to abrogate their factor-dependence. However, p210BCR–ABL can also activate downstream targets of IL-3, including the IL-3 receptor (19), JAK2, and STAT5 (20–23). CML progenitors have been known for many years to be growth factor-responsive (24–26), and previous attempts (including our own) to obtain evidence of an autocrine or abnormal paracrine mechanism operating in the human disease have been negative (27, 28). On the other hand, it has been shown by several groups that purified populations of primitive (CD34+) cells from CML patients (most of which are usually Ph+/BCR–ABL+) can survive and proliferate in vitro even when no exogenous growth factors (except insulin) are added, whereas analogous normal cells die rapidly under the same conditions (29–31). Using more sensitive procedures to detect growth-factor gene expression, we now demonstrate the consistent and selective activation of IL-3 and granulocyte colony-stimulating factor (G-CSF) production within the leukemic (BCR–ABL+) CD34+ cells of patients with chronic-phase CML and show that these factors serve as autocrine mediators of primitive CML cell proliferation.
Methods
Cells.
Heparinized blood was obtained from 12 chronic-phase CML patients. Seven of the CML patients were studied at diagnosis, three <1 year postdiagnosis and two >1 year postdiagnosis. All had elevated WBC counts ranging from 18 to 650 × 109 per liter, elevated circulating CD34+ cell counts ranging from 3 to 40% of viable low-density cells, elevated colony-forming cells ranging from 6 to 6,300 × 106 per liter and elevated long-term culture-initiating cells (LTC-IC) ranging from 6 to 22,000 × 103 per liter. Normal marrow was obtained from donors of allogeneic transplants or from cadaveric donors (North West Tissue Centre, Seattle). In all cases, informed consent was obtained. Low-density (<1.077 g/ml) cells were isolated by centrifugation on Ficoll-Hypaque (Amersham Pharmacia) and fractionated directly, or later, after being cryopreserved in 10% DMSO plus 90% FCS. Populations of >99% pure, viable (propidium iodide-negative, Annexin V−) CD34+ cells or subsets of CD34+ cells were isolated using a two-laser FACStarplus (Becton Dickinson) as described (31, 32). Bulk cells were collected into Eppendorf tubes containing a serum-free medium (SFM) which consisted of Iscove’s medium supplemented with BSA, insulin, and transferrin (BIT, StemCell Technologies, Vancouver), low-density lipoproteins (Sigma), and 2-mercaptoethanol (33). Single cells for culture or for reverse transcriptase (RT)-PCR analyses were sorted using the FACStar single-cell deposition unit directly into the wells of 96-well microtiter plates preloaded with 100 μl of SFM plus the additives specified, or with lysis buffer, respectively.
RT-PCR Analyses.
Total RNA was extracted from aliquots of 2,000 cells, and RT-PCR was performed using an oligo(dT)-based primer and poly(A) tailing procedure described previously (34). For single-cell studies, plates containing single cells were spun at 300 × g for 3 min at 4°C, RNA was extracted, and reverse transcription was performed using the same oligo(dT) primers as for the analyses of 2,000 cells. Aliquots of 1–5 μl of the first round of PCR generated from the initial amplification of total cDNA were subjected to a second round of PCR amplification in 50-μl volumes of 1× PCR buffer (GIBCO/BRL) containing 20 mM Tris⋅HCl (pH 8.4), 50 mM KCl, 2 mM MgCl2, 200 μM each dNTP (Amersham Pharmacia), 1 unit of Taq polymerase, and 10 pmol of specific primers for IL-3 (5′-GCTCCCATGACCCAGACAACGTCC-3′ and 5′-CAGATAGAACGTCAGTTTCCTCCG-3′), G-CSF (5′-CTCTGGACAGTGCAGGAAGCCACC-3′) and (5′-GCTGGGCAAGGTGGCGTAGAACGC-3′), thrombopoietin (TPO) (5′-GGCCAGAATGGAGCTGACTGAATTG-3′ and 5′-TCCTACAAGCATCAGGAAACGCACC-3′), actin (5′-GTGCGTGACATTAAGGAGAA-3′ and 5′-GGAGGGGCCGGACTCGTCA-3′), and BCR–ABL (5′- CAGGGTGCACAGCCGCAACGGCAA-3′ and 5′-GTCCAGCGAGAAGGTTTTCCTTGGA-3′) to give DNA fragments of 345 bp (IL-3), 547 bp (G-CSF), 495 bp (TPO), 471 bp (actin), and 299 or 374 bp (BCR–ABL). Thirty-five cycles (94°C for 30 sec, 62°C for 1 min, 72°C for 1 min) were then performed.
Southern Blot Analysis.
Ten-microliter aliquots of total amplified cDNA or PCR products derived by specific primers were electrophoresed in 1–1.5% agarose gel, transferred onto nylon membranes (Zeta-probe, Bio-Rad), and hybridized with [32P]dCTP (Amersham Pharmacia)-labeled probes overnight at 60°C in 4.4× saline sodium citrate, 7.5% formamide, 1.5 mM EDTA, 0.75% SDS, 0.75% skim milk, 370 mg/ml salmon sperm DNA, and 7.5% dextran sulfate. Probes were labeled by incubating the denatured fragments in the presence of hexamers and the Klenow fragment of polymerase I using a random priming kit (GIBCO/BRL) and were then purified on a Sephadex-G50 column. Membranes were finally washed twice at 60°C for 30 min in a solution of 0.1× saline sodium citrate, 0.1% SDS, and 1 mg/ml sodium pyrophosphate and then in a solution of 0.1× saline sodium citrate three times for 2 min (each time at room temperature). They were finally exposed to Kodak XAR film (Eastman Kodak) at −70°C for 6–16 h for PCR products generated from single cells using specific primers, for periods of 1–3 days for the total amplified cDNA products from positive reactions, and for up to 5 days to confirm negative reactions.
Probes.
cDNA probes for human IL-3, IL-6, G-CSF, granulocyte/macrophage-CSF, Steel factor (SF), and actin (from R. Kay and K. Humphries, Terry Fox Laboratory, Vancouver, BC), and for BCR–ABL (from J. Griffin, Dana–Farber Cancer Center, Boston) were excised from vectors with an appropriate restriction endonuclease, and the inserts were separated from the vectors by agarose gel electrophoresis. DNA fragments were recovered using the QiaEx II gel-extraction kit (Qiagen, Chatsworth, CA) and used directly as hybridization probes. For the detection of human TPO cDNA, the internal primer (5′-CCGAGTCCTCAGTAAACTGCTTCG-3′) was used as a hybridization probe.
Cultures.
Sorted populations were cultured at 37°C in the SFM described above at 105 cells per ml, with or without 20 ng/ml IL-3 (Novartis, Basel, Switzerland), 20 ng/ml IL-6 (Cangene, Mississauga, ON), 20 ng/ml G-CSF (StemCell), 100 ng/ml flt3-ligand (FL, Immunex), and 100 ng/ml SF (Amgen Biologicals). In some experiments, the following mAbs were added alone or in combination as indicated: a neutralizing anti-IL-3 antibody (R&D Systems), an anti-IL-3 receptor α-chain (anti-IL-3Rα) antibody with IL-3 antagonist activity (7G3) (35), a neutralizing anti-G-CSF antibody (R&D Systems), and a non-blocking (control) anti-IL-3Rα antibody (9F5) (35).
Western Blotting.
TF-1 cells (36) were incubated for 16 h at 37°C in RPMI medium plus 10% FCS at 2 × 105 per ml with or without 5 ng/ml human IL-3, or 10% or 30% CML conditioned medium (prepared by incubating low-density CML cells in SFM at 3 × 106 for 2–3 days and then concentrating the medium 10- to 20-fold), and 10 μg/ml indicated antibodies. Sorted CD34+ cells from normal individuals and CML patients’ samples were cultured for 16 h at 37°C in SFM at 105 cells per ml with or without antibodies as indicated. Cells were lysed in phosphorylation solubilization buffer (PSB; 50 mM Hepes buffer, 100 mM NaF, 10 mM sodium pyrophosphate, 2 mM Na3VO4, 4 mM EDTA) containing 0.5% Nonidet P-40. Western analysis was performed on lysates from 105 cells per treatment group (31) using rabbit polyclonal anti-phospho-STAT5A/B and anti-STAT5A antibodies from Upstate Biotechnology (Lake Placid, NY) and a mouse monoclonal anti-Abl antibody from PharMingen.
Results
Identification of IL-3 and G-CSF Transcripts in Primitive CML Cells.
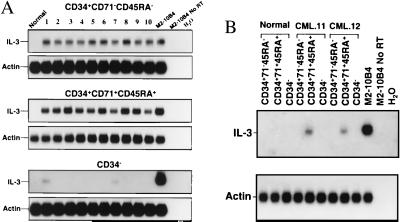
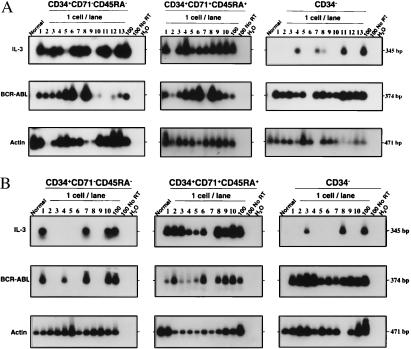
Evidence of abnormal growth-factor gene expression in CML cells was obtained from a comparative survey of highly purified CD34+CD71−CD45RA−, CD34+CD71+CD45RA+, and CD34− cells isolated from CML blood (12 chronic-phase CML patients) and normal adult marrow (4 samples). Ten of the CML samples (nos. 1–10) were from less common patients whose very early circulating progenitors (detected as LTC-IC), as well as the colony-forming cells and later hematopoietic cells, were all Ph+. These samples were included in the present analysis to ensure that growth-factor gene expression could be examined in the most primitive types of leukemic CD34+ cells. The other two samples of CML cells were from more typical chronic-phase patients whose bulk hematopoietic cells are predominantly Ph+ but whose LTC-IC, despite their increased numbers in the blood, are predominantly normal (Ph−) (32). As shown in Fig. 1, IL-3 transcripts were consistently detected in PCR-amplified cDNAs from the less primitive (CD71+CD45RA+) subset of CD34+ cells from all 12 CML patients and from the more primitive (CD71−CD45RA−) subset of CD34+ cells from the 10 CML patients’ samples in which these cells were predominantly leukemic (nos. 1–10). IL-3 transcripts were not detected in the most primitive (CD71−CD45RA−) subset of CD34+ cells from the two other CML patients studied (nos. 11 and 12) whose stem-cell reservoir was predominantly normal, or in any of the normal marrow cell populations. In these studies, IL-3 transcripts were also not detected in the mature CD34− cells from most (10/12) of the CML patients (Fig. 1). RT-PCR analysis of single FACS-sorted cells using a second, nested PCR reaction with primers specific for human IL-3 revealed an even closer concordance of IL-3 and BCR–ABL gene expression among the individual CD34+ cells present in different CML patients (Fig. 2 and Table 1). This latter comparison was strengthened by the inclusion of the two CML samples (nos. 11 and 12) that were found to contain some BCR–ABL− cells within the CD34+CD71−CD45RA− subset, as anticipated from the LTC-IC genotyping studies. Similar analysis of CD34− leukemic cells also demonstrated the presence of IL-3 transcripts in occasional cells within this population (Fig. 2). (IL-3 transcripts could also be detected in the bulk preparations by using this more sensitive procedure; data not shown).
Figure 1.
RT-PCR detection of IL-3 mRNA in CD34+CD71−CD45RA−, CD34+CD71+CD45RA+, and CD34− cells from 12 CML samples (2,000 cells per subpopulation from each patient). (A) Lane 1 contained PCR products from normal marrow cells, lanes 2–11 from the 10 CML samples with Ph+ LTC-IC, lane 12 from a mouse fibroblast cell line engineered to express human IL-3 (54) (positive control). Lane 13 is the same as lane 12 but with no RT added (negative control), and lane 14 contained water only (another negative control). The same membranes were then washed and rehybridized to a human actin cDNA probe. (B) Lanes 1–3 contained PCR products from normal marrow cells, lanes 4–9 from CML patients 11 and 12 (in which only Ph− LTC-IC were detected). Positive and negative controls are the same as in A.
Figure 2.
Southern blots showing RT-PCR detection of IL-3, BCR–ABL, and actin transcripts in single CD34+CD71−CD45RA−, CD34+CD71+CD45RA+, and CD34− cells from one of the CML patients with predominantly Ph+ LTC-IC (no. 10) (A) and one with predominantly Ph− LTC-IC (no. 11) (B). The expected sizes, in bp, are indicated for each PCR product. Negative controls are the same as in Fig. 1.
Table 1.
Frequency of CD34+ and CD34− cells in CML blood expressing IL-3 and BCR-ABL mRNA detected by single-cell RT-PCR
| Phenotype | Patient no. | Positive cells, %
|
||
|---|---|---|---|---|
| IL-3 | BCR–ABL | Actin | ||
| CD34+CD71−CD45RA− | 6 | 80 | 90 | 90 |
| 7 | 80 | 90 | 100 | |
| 10 | 84 | 76 | 93 | |
| 11 | 30 | 40 | 100 | |
| 12 | 30 | 30 | 100 | |
| CD34+CD71+CD45RA+ | 6 | 75 | 83 | 83 |
| 7 | 83 | 83 | 91 | |
| 10 | 90 | 90 | 100 | |
| 11 | 90 | 90 | 100 | |
| 12 | 90 | 90 | 90 | |
| CD34− | 6 | 30 | 84 | 92 |
| 7 | 20 | 90 | 90 | |
| 10 | 30 | 92 | 92 | |
| 11 | 20 | 100 | 90 | |
| 12 | 30 | 90 | 100 | |
A minimum of 10 individual cells per population were analyzed. Similar analysis of 1,000 cells from each population isolated from three to four different normal marrows (BM) showed no IL-3 or BCR–ABL transcripts.
Similar results to those obtained for IL-3 were seen when the same extracts (from both 2,000 cell aliquots and from single cells) were examined for G-CSF gene expression (data not shown). In addition, SF transcripts were found in both subsets of CD34+ cells isolated from 2 CML patients (nos. 7 and 10), but transcripts for IL-6, GM-CSF, and TPO were not detected in any of the populations from all 12 CML samples (data not shown). Transcripts for any of these five additional growth factors were also not detected in the normal cell samples analyzed (representative samples shown in Figs. 1 and 2).
Production of Bioactive IL-3 and G-CSF by CML Cells.
FACS analyses of fixed cell preparations showed specific positive intracellular staining by 10–25% of the low-density cells analyzed from 10 of 10 CML samples (using a phycoerythrin-labeled anti-human IL-3 antibody from BioSource International, Camarillo, CA), whereas cells from all 3 normal marrow samples similarly analyzed were negative (data not shown). Using sensitive ELISA kits for IL-3 (Biosource), G-CSF, SF, and TPO (R&D Systems), immunoreactive IL-3 and G-CSF were both shown to be present in medium conditioned for 2–3 days by low-density cells (at 3 × 106 per ml SFM) from all CML patients tested and then concentrated 10- to 20-fold (0.5 ± 0.3 ng/ml, n = 7 for IL-3 and 0.1 ± 0.01 ng/ml, n = 6 for G-CSF). Consistent with the RT-PCR findings, neither TPO (n = 7) nor SF (n = 6) could be detected in any of these media except in one case where a low level of SF (0.01 ng/ml) was found. This was one of the two patients whose CD34+ cells had been found to contain detectable SF transcripts by RT-PCR. Concentrated media similarly conditioned by normal marrow cells (n = 3) contained <0.02 ng/ml of IL-3, <0.005 ng/ml G-CSF, <0.005 ng/ml SF, and <0.02 ng/ml TPO.
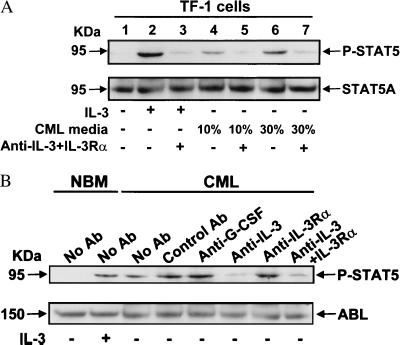
The CML cell-conditioned media were also able to stimulate the proliferation of human TF-1 cells, a human hematopoietic cell line that can respond to a variety of growth factors including IL-3 but not G-CSF (36). Moreover, up to 80% of the stimulation observed could be specifically blocked by the addition to the TF-1 cell cultures of neutralizing anti-human IL-3 antibody, or anti-human IL-3Rα antibody with IL-3 antagonist activity (35), or both in combination (data not shown). Similarly, CML cell-conditioned medium was able to mimic the ability of 5 ng/ml IL-3 to induce the phosphorylation of STAT5 in TF-1 cells, and this effect could be blocked by the addition of a combination of 10 μg/ml anti-IL-3 plus 10 μg/ml antagonistic anti-IL-3Rα antibodies (Fig. 3A).
Figure 3.
Inhibition of STAT5 phosphorylation in TF-1 cells stimulated by conditioned medium from CML cells (A) and in CD34+ cells from CML patient no. 9 (B), by antibodies to IL-3 and IL-3Rα. In each experiment, a Western blot was performed first with an anti-phospho-STAT5 A/B antibody (P-STAT5, Upper). The membranes were then stripped and reprobed with antibodies to STAT5A (A) and ABL (B) as indicated. Control antibody was the non-blocking anti-IL-3Rα antibody (9F5) (35). For further experimental details, see Methods.
IL-3 and G-CSF Produced by CML Cells Are Required for Their Autonomous Growth.
CD34+ and CD34− cells from both CML and normal samples were cultured in SFM in the presence or absence of a mixture of hematopoietic growth factors (i.e., FL, SF, IL-3, IL-6, and G-CSF) previously found to stimulate the proliferation and amplification in vitro of very primitive normal marrow cells (33, 37). In all cultures containing the five growth factors, there was a marked and similar cell expansion (≈100-fold) over a period of 3 weeks. In the absence of growth factor addition, all subpopulations of normal cells declined to undetectable numbers (a >1,000-fold decrease) within 10 days (Fig. 4). The CD34− cells from the CML samples, regardless of the relative proportion or absolute frequency of Ph+/BCR–ABL+ LTC-IC present in their CD34+ populations, also declined to undetectable levels within 3 weeks, although less rapidly than the CD34− cells from normal marrow. In contrast, the number of viable cells was maintained at input levels in cultures initiated with CD34+CD71+CD45A+ CML cells. In cultures initiated with CD34+CD71−CD45RA− cells that were predominantly BCR–ABL+ (Table 1), there was almost as much cell growth in the absence of growth factors as when growth factors were added (Fig. 4A); whereas, in cultures initiated with CD34+CD71−CD45RA− cells that contained a substantial proportion of BCR–ABL− cells (and Ph− LTC-IC), there was an early transient decline in cell numbers followed by a later regrowth of viable cells (Fig. 4B). Analysis of the frequency of factor-independent cells within the total CD34+ CML population in single-cell cultures has shown this to be a feature of 30–50% of these cells (see Table 2 and ref. 38).
Figure 4.
Factor-independent growth of CML cells in culture decreases with their differentiated state. Highly purified CD34+CD71−CD45RA− (diamonds), CD34+CD71+CD45RA+ (squares), and CD34− (triangle) CML cells (n = 5) or normal marrow cells (circles, n = 3) were cultured at 105 cells per ml in SFM with (solid symbols) or without (open symbols) growth factors (IL-3, IL-6, G-CSF, FL, or SF). Viable cell numbers were determined by hematocytometer counts of trypan blue-excluding cells. Results are expressed as the mean fold-change compared with input values ± SEM. (A) CML patients with predominantly Ph+ LTC-IC (n = 3). (B) CML patients with predominantly Ph− LTC-IC (n = 2).
Table 2.
Inhibition of proliferation of CD34+ CML cells by antibodies to IL-3, IL-3Rα, and G-CSF
| Patient no. | Bulk cultures
|
Single-cell cultures
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Control Ab | Anti- IL-3 | Anti- IL-3Rα | Anti- G-CSF | Anti-IL-3 + anti-IL-3Rα | Anti-IL-3 + anti-G-CSF | Control Ab | Anti- IL-3 | Anti- IL-3Rα | Anti- G-CSF | Anti-IL-3 + anti-IL-3Rα | Anti-IL-3 + anti-G-CSF | |
| 1 | 3 | 55 (61) | 39 (42) | 25 | 63 (68) | 57 | 0 | 66 | 52 | 52 | 72 | 72 |
| 3 | 3 | 37 | 20 | 16 | 46 | 42 | — | — | — | — | — | — |
| 8 | 6 | 62 | 38 | 28 | 69 | 62 | — | — | — | — | — | — |
| 9 | 8 | 68 (71) | 41 (43) | 30 | 72 (75) | 69 | 6 | 72 | 62 | 63 | 84 | 88 |
| 10 | 5 | 29 | 27 | 25 | 37 | 30 | — | — | — | — | — | — |
| 11 | — | — | — | — | — | — | 8 | 75 | 58 | 59 | 82 | 85 |
| 12 | — | — | — | — | — | — | 10 | 70 | 60 | 57 | 78 | 78 |
Bulk cultures were initiated (three replicates per group) with 104 CD34+ cells per 100 μL of SFM, and antibodies added at 50 μg/ml on day 0 and again 24 hours later. After a total of 5 days at 37°C, [3H]thymidine was added for 4 hours, and the cells were then harvested onto glass fiber filters for determination of isotope incorporation or for hemacytometer counts of viable (trypan blue-excluding) cells (data shown in parentheses). Single-cell cultures were maintained in SFM for the same period, but the antibodies were added only on day 0 and at a lower concentration, i.e., 10 μg/ml for single antibodies and 5 μg/ml each when two antibodies were added together. Proliferation was scored as the observation of ≥2 viable (refractile) cells detectable under the microscope at the end of the 5 days in culture. In both bulk and single-cell experiments, the control Ab was the non-blocking anti-IL-3R antibody (9F5) (35). Values shown are the percent inhibition of proliferation (cpm or cell yield in bulk cultures and frequency of wells with ≥2 viable cells in single-cell cultures) relative to the proliferation measured in cultures to which no antibodies were added (maximum cpm in these bulk cultures = 23,000; frequency of clones = 30–50%, 72–96 wells assessed per group).
We then repeated these experiments incubating purified CD34+ cells in the presence (or absence) of the same neutralizing anti-ligand and anti-receptor or control antibodies that had been used to demonstrate the presence of bioactive IL-3 and G-CSF in CML cell-conditioned media. Addition of these antibodies consistently inhibited the autonomous proliferation seen in cultures to which no antibody or a control antibody was added, both as measured by [3H]thymidine incorporation assays and by counts of viable cells. In bulk cultures, relatively high concentrations of antibodies (50 μg/ml) were required to consistently inhibit the autonomous proliferation of CD34+ CML cells by >40% (Table 2). The apparent insensitivity of these cells to anti-ligand or anti-receptor antibodies is similar to previous findings with BCR–ABL-transduced cell lines in which evidence of an autocrine IL-3 mechanism was demonstrated (16–18). Such a result could be attributable to a high local concentration of growth factor around individual autocrine cells, intracellular receptor activation (39), or paracrine effects potentially operative in bulk cultures. To address this latter possibility, the effect of the same antibodies on the proliferation of single CD34+ CML cells was examined. These experiments showed >70% inhibition of all four CML samples tested in the presence of 5 μg/ml anti-IL-3 and 5 μg/ml either anti-G-CSF or anti-IL-3Rα (7G3) antibody (Table 2).
IL-3 but Not G-CSF Contributes to the Constitutive Activation of STAT5 in CD34+ CML Cells.
To investigate further the role of autocrine IL-3/G-CSF on CD34+ CML cell growth, we assessed STAT5 phosphorylation in these cells after they had been cultured overnight in the presence or absence of the same antibodies described above. As shown for a representative experiment in Fig. 3B, CD34+ CML cells cultured in SFM alone or with control antibody resembled normal CD34+ cells stimulated with 5 ng/ml of IL-3 but, in the presence of 10 μg/ml anti-IL-3 (±anti-IL-3Rα) antibody, the level of phosphorylated STAT5 was reduced 4.4 ± 2.2-fold (n = 3). No reduction of phosphorylated STAT5 was seen in the presence of anti-G-CSF antibody.
Discussion
These studies provide definitive evidence of an autocrine mechanism in naturally occurring chronic-phase CML. Since completion of the studies described here, we have detected IL-3 and G-CSF transcripts in the CD34+ cells isolated from an additional 8 (of 8) CML patients (total = 20 of 20), indicating a strong association of a deregulated expression of IL-3 and G-CSF with this disease. Of particular note is the observation of IL-3/G-CSF transcripts in individual leukemic stem-cell candidates (identified by a CD34+CD71−CD45RA− phenotype and coexpression of BCR–ABL) that correlated with the high autonomous growth capacity of this population. In contrast, later (CD34−) cell types continue to express BCR–ABL but stop expressing IL-3 and G-CSF and become factor-dependent. Thus, with differentiation, the growth of CML cells is ultimately extinguished, regardless of the factors present, and the clone must be sustained by the proliferation of more primitive leukemic cells. The restricted presence of IL-3 and G-CSF transcripts in CD34+ CML cells raises many interesting questions about the nature of a mechanism that would link differentiated state to the deregulated expression of these two growth-factor genes. Although future studies will be required to delineate how this may occur, the resultant short-lived autonomous growth potential of most CD34+ cells helps to explain historical experience that optimal colony formation by CML progenitors in vitro requires the addition of growth factors to the cultures (26, 29). It should be noted that an examination of bulk CD34+ populations from most CML patients would typically also not detect the more extensive autonomous growth potential of the very primitive CD34+CD71−CD45RA− subset because of their minor representation in the total CD34+ population. Moreover, for many patients, isolation of the CD34+CD71−CD45RA− cells might still be uninformative because of a predominance of residual normal (Ph−/BCR–ABL−) cells in this population (4). In the present study, this problem was circumvented by selection of a number of CML patients in whom leukemic cells were known to be prevalent in this stem cell-enriched subpopulation of CD34+ cells.
The autocrine IL-3/G-CSF growth loop demonstrated here for primitive CML cells recapitulates in the primary disease a phenomenon previously observed only in certain BCR–ABL-transduced cell lines (16–18) and now suggested by emerging data from mice transplanted with mouse bone-marrow cells transduced with BCR–ABL retroviral vectors (40, 41). However, the significance to CML of these observations has not been generally accepted because of their lack of confirmation in patients’ cells. In addition, only recently has it been appreciated that IL-3 alone is sufficient to stimulate very primitive normal cells to enter S phase (42) and that exposure of very primitive normal cells to excess IL-3 promotes their differentiation with an enhanced amplification occurring only among their more mature progeny (37, 43). Thus, the cell population dynamics exhibited by primitive CML cells growing in vitro in the absence of exogenous growth factor addition can now be seen to match closely the behavior of analogous primitive normal cells cultured in the presence of excess IL-3 (Fig. 5). Interestingly, these findings also have strong parallels with the pattern of CML cell expansion in vivo. During the initial chronic phase of CML, the most primitive types of CML progenitors do not increase their numbers at a rate predicted by their increased cycling activity (2, 4), and clonal dominance is achieved only at later stages of differentiation by their progeny. In vivo, the ability of intermediate types of CML progenitors to produce and respond in a lineage-unrestricted fashion to IL-3 and G-CSF would be expected to give the CML colony-forming cells a significant, albeit short-lived, selective advantage over any coexisting normal colony-forming cells, for example, by counteracting the effects of endogenous levels of inhibitors that constrain the proliferation and amplification of their normal counterparts (44, 45). Interestingly, an expanded population of leukemic colony-forming cells is a well established feature of CML despite the relatively slow accumulation of more primitive leukemic cells (4).
Figure 5.
Schematic diagram comparing the predicted effect on different CML cell subpopulations of an autocrine IL-3/G-CSF mechanism based on documented effects of excess exposure to these two growth factors on their phenotypically normal counterparts (37). Dotted arrows indicate the different conditions of growth-factor stimulation compared (supplied exogenously, in the case of the normal cells, or by an autocrine mechanism, in the case of the CML cells). Solid arrows indicate the self-renewal (arched arrows) vs. differentiation (vertical arrows) responses elicited. In each case, the width of the arrow reflects the magnitude of the stimulus or response indicated. The pathology of CML cell expansion in vivo (slow at the level of the leukemic stem-cell compartment and then more rapidly with their progressive differentiation) is consistent with what is observed in vitro when analogous types of very primitive normal hematopoietic cells are exposed to excessive concentrations of IL-3 (±G-CSF).
The present findings also provide an explanation for the intriguing similarities noted between human CML and murine models of myeloproliferative disease resulting from the transplantation of mouse bone-marrow cells retrovirally transduced or transgenically engineered to produce IL-3 or G-CSF (46–50). Such associations alone do not establish BCR–ABL as the cause of the autocrine IL-3 and G-CSF seen in human CML; however, this seems likely in view of the inactivation of BCR–ABL- induced growth factor production by mutation of BCR–ABL (18) and the close correlation observed here in growth factor and BCR–ABL gene expression in individual CD34+ cells from CML patients. Interestingly, however, we have recently identified a very rare and unusual subset of quiescent leukemic CD34+ cells in chronic-phase CML patients that express BCR–ABL (51) but not IL-3 and G-CSF (38). Thus, other events must also be involved in deregulating IL-3 and G-CSF production in primitive CML cells. In addition, although an autocrine mechanism may contribute to the disease process in vivo, it is unlikely to explain all of the biological abnormalities produced by BCR–ABL. Nevertheless, the strong dependence of STAT5 phosphorylation in CD34+ CML cells on autocrine IL-3 (but not G-CSF) demonstrated here identifies one important pathway involved and underscores the role of an autocrine mechanism in primary CML progenitors from patients with chronic-phase disease. The present observations also have potentially important implications for therapy. For example, either enhancement of the differentiation-promoting effect of intrinsically produced IL-3 on very primitive CML cells, or in vivo blockade of IL-3 and G-CSF action using antibody, ligand–toxin, or other strategies for interfering with this mechanism of autocrine stimulation may have clinically useful effects. With the recent introduction of xenotransplant models that support the engraftment of primary CML cells (52, 53), it should now be possible to assess the potential of these new approaches in a preclinical setting.
Acknowledgments
We thank the Division of Hematology for assistance in procuring patient samples and clinical data, the Stem Cell Assay Service for initial cell processing, Gayle Thornbury and Giovanna Cameron for operating the FACS, Margaret Hale for technical assistance, and Bernadine Fox for manuscript preparation. We also thank O. Witte (Howard Hughes Medical Institute, Los Angeles, CA) and J. Griffin (Dana–Farber Cancer Center, Boston) for helpful comments; J. Griffin for the BCR–ABL plasmid; R. Kay, K. Humphries, and P. Lansdorp (Terry Fox Laboratory, Vancouver, BC) for various growth-factor cDNA probes and antibodies; and Amgen, Cangene, Immunex, Novartis, and StemCell for gifts of other reagents. This work was supported by grants from the National Cancer Institute of Canada (NCIC) with funds from the Terry Fox Run and from Novartis Canada. T.H. holds a United Kingdom Leukemia Research Fund Senior Lectureship. C.E. is a Terry Fox Cancer Research Scientist of the NCIC.
Abbreviations
- CML
chronic myeloid leukemia
- FL
flt3–ligand
- G-CSF
granulocyte colony-stimulating factor
- LTC-IC
longterm culture-initiating cells
- RT
reverse transcriptase
- SF
Steel factor
- SFM
serum-free medium
- TPO
thrombopoietin
References
- 1.Raskind W H, Fialkow P J. Adv Cancer Res. 1987;49:127–167. doi: 10.1016/s0065-230x(08)60796-4. [DOI] [PubMed] [Google Scholar]
- 2.Eaves A C, Barnett M J, Ponchio L, Cashman J D, Petzer A, Eaves C J. Stem Cells (Dayton) 1998;16:77–83. doi: 10.1002/stem.5530160809. [DOI] [PubMed] [Google Scholar]
- 3.Verfaillie C M. In: Stem Cells in Chronic Myelogenous Leukemia. Zon L I, editor. Vol. 4. Philadelphia: W. B. Saunders; 1998. pp. 1079–1114. [DOI] [PubMed] [Google Scholar]
- 4.Eaves C, Cashman J, Eaves A. Leuk Res. 1998;22:1085–1096. doi: 10.1016/s0145-2126(98)00113-1. [DOI] [PubMed] [Google Scholar]
- 5.Groffen J, Stephenson J R, Heisterkamp N, De Klein A, Bartram C R, Grosveld G. Cell. 1984;36:93–99. doi: 10.1016/0092-8674(84)90077-1. [DOI] [PubMed] [Google Scholar]
- 6.Shtivelman E, Lifshitz B, Gale R P, Canaani E. Nature (London) 1985;315:550–554. doi: 10.1038/315550a0. [DOI] [PubMed] [Google Scholar]
- 7.Ben-Neriah Y, Daley G Q, Mes-Masson A M, Witte O N, Baltimore D. Science. 1986;233:212–214. doi: 10.1126/science.3460176. [DOI] [PubMed] [Google Scholar]
- 8.Konopka J B, Witte O N. Mol Cell Biol. 1985;5:3116–3123. doi: 10.1128/mcb.5.11.3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Van Etten R A, Jackson P, Baltimore D. Cell. 1989;58:669–678. doi: 10.1016/0092-8674(89)90102-5. [DOI] [PubMed] [Google Scholar]
- 10.Daley G Q, Van Etten R A, Baltimore D. Science. 1990;247:824–830. doi: 10.1126/science.2406902. [DOI] [PubMed] [Google Scholar]
- 11.Heisterkamp N, Jenster G, ten Hoeve J, Zovich D, Pattengale P K, Groffen J. Nature (London) 1990;344:251–253. doi: 10.1038/344251a0. [DOI] [PubMed] [Google Scholar]
- 12.Lugo T G, Pendergast A M, Muller A J, Witte O N. Science. 1990;247:1079–1082. doi: 10.1126/science.2408149. [DOI] [PubMed] [Google Scholar]
- 13.Evans C A, Owen-Lynch J, Whetton A D, Dive C. Cancer Res. 1993;53:1735–1738. [PubMed] [Google Scholar]
- 14.Druker B J, Tamura S, Buchdunger E, Ohno S, Segal G M, Fanning S, Zimmermann J, Lydon N B. Nat Med. 1996;2:561–566. doi: 10.1038/nm0596-561. [DOI] [PubMed] [Google Scholar]
- 15.Sattler M, Salgia R. Cytokine Growth Factor Rev. 1997;8:63–79. doi: 10.1016/s1359-6101(96)00047-0. [DOI] [PubMed] [Google Scholar]
- 16.Hariharan I K, Adams J M, Cory S. Oncogene Res. 1988;3:387–399. [PubMed] [Google Scholar]
- 17.Sirard C, Laneuville P, Dick J. Blood. 1994;83:1575–1585. [PubMed] [Google Scholar]
- 18.Anderson S M, Mladenovic J. Blood. 1996;87:238–244. [PubMed] [Google Scholar]
- 19.Wilson-Rawls J, Xie S, Liu J, Laneuville P, Arlinghaus R B. Cancer Res. 1996;56:3426–3430. [PubMed] [Google Scholar]
- 20.Shuai K, Halpern J, ten Hoeve J, Rao X, Sawyers C L. Oncogene. 1996;13:247–254. [PubMed] [Google Scholar]
- 21.Carlesso N, Frank D, Griffin J. J Exp Med. 1996;183:811–820. doi: 10.1084/jem.183.3.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chai S K, Nichols G L, Rothman P. J Immunol. 1997;159:4720–4728. [PubMed] [Google Scholar]
- 23.Nieborowska-Skorska M, Wasik M A, Slupianek A, Salomoni P, Kitamura T, Calabretta B, Skorski T. J Exp Med. 1999;189:1229–1242. doi: 10.1084/jem.189.8.1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moore M A S, Williams N, Metcalf D. J Natl Cancer Inst. 1973;50:591–602. doi: 10.1093/jnci/50.3.591. [DOI] [PubMed] [Google Scholar]
- 25.Moberg C, Olofsson T, Olsson I. Scand J Haematol. 1974;12:381–390. doi: 10.1111/j.1600-0609.1974.tb00225.x. [DOI] [PubMed] [Google Scholar]
- 26.Metcalf D. Hemopoietic Colonies: In Vitro Cloning of Normal and Leukemic Cells. Berlin: Springer; 1977. [PubMed] [Google Scholar]
- 27.Duncombe A S, Heslop H E, Turner M, Meager A, Priest R, Exley T, Brenner M K. J Immunol. 1989;143:3828–3834. [PubMed] [Google Scholar]
- 28.Otsuka T, Eaves C J, Humphries R K, Hogge D E, Eaves A C. Leukemia. 1991;5:861–868. [PubMed] [Google Scholar]
- 29.Strife A, Lambek C, Wisniewski D, Wachter M, Gulati S C, Clarkson B D. Cancer Res. 1988;48:1035–1041. [PubMed] [Google Scholar]
- 30.Bedi A, Zehnbauer B A, Barber J, Sharkis S, Jones R. Blood. 1994;83:2038–2044. [PubMed] [Google Scholar]
- 31.Maguer-Satta V, Burl S, Liu L, Damen J, Chahine H, Krystal G, Eaves A, Eaves C. Oncogene. 1998;16:237–248. doi: 10.1038/sj.onc.1201533. [DOI] [PubMed] [Google Scholar]
- 32.Petzer A L, Eaves C J, Lansdorp P M, Ponchio L, Barnett M J, Eaves A C. Blood. 1996;88:2162–2171. [PubMed] [Google Scholar]
- 33.Petzer A L, Zandstra P W, Piret J M, Eaves C J. J Exp Med. 1996;183:2551–2558. doi: 10.1084/jem.183.6.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maguer-Satta V, Petzer A L, Eaves A C, Eaves C J. Blood. 1996;88:1796–1804. [PubMed] [Google Scholar]
- 35.Sun Q, Woodcock J M, Rapoport A, Stomski F C, Korpelainen E I, Bagley C J, Goodall G J, Smith W B, Gamble J R, Vadas M A, Lopez A F. Blood. 1996;87:83–92. [PubMed] [Google Scholar]
- 36.Drexler H G, Zaborski M, Quentmeier H. Leukemia. 1997;11:701–708. doi: 10.1038/sj.leu.2400633. [DOI] [PubMed] [Google Scholar]
- 37.Zandstra P W, Conneally E, Petzer A L, Piret J M, Eaves C J. Proc Natl Acad Sci USA. 1997;94:4698–4703. doi: 10.1073/pnas.94.9.4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Holyoake T, Jiang X, Eaves C J, Eaves A C. Blood. 1998;92, Suppl. 1:317. [Google Scholar]
- 39.Keating M T, Williams L T. Science. 1988;239:914–916. doi: 10.1126/science.2829358. [DOI] [PubMed] [Google Scholar]
- 40.Zhang X, Ren R. Blood. 1998;92:3829–3840. [PubMed] [Google Scholar]
- 41.Li S, Ilaria R L, Million R P, Daley G Q, Van Etten R A. J Exp Med. 1999;189:1399–1412. doi: 10.1084/jem.189.9.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ponchio L, Eaves C. J Hematother. 1995;3:217. [Google Scholar]
- 43.Yonemura Y, Ku H, Hirayama F, Souza L M, Ogawa M. Proc Natl Acad Sci USA. 1996;93:4040–4044. doi: 10.1073/pnas.93.9.4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Eaves C J, Cashman J D, Wolpe SD, Eaves A C. Proc Natl Acad Sci USA. 1993;90:12015–12019. doi: 10.1073/pnas.90.24.12015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cashman J D, Eaves C J, Sarris A H, Eaves A C. Blood. 1998;92:2338–2344. [PubMed] [Google Scholar]
- 46.Chang J M, Metcalf D, Lang R A, Gonda T J, Johnson G R. Blood. 1989;73:1487–1497. [PubMed] [Google Scholar]
- 47.Wong P M C, Chung S, Dunbar C E, Bodine D M, Ruscetti S, Nienhuis A W. Mol Cell Biol. 1989;9:798–808. doi: 10.1128/mcb.9.2.798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chang J M, Metcalf D, Gonda T J, Johnson G R. J Clin Invest. 1989;84:1488–1496. doi: 10.1172/JCI114324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Just U, Katsuno M, Stocking C, Spooncer E, Dexter M. Growth Factors. 1993;9:41–55. doi: 10.3109/08977199308991581. [DOI] [PubMed] [Google Scholar]
- 50.Yamada T, Kaneko H, Iizuka K, Matsubayashi Y, Kokai Y, Fujimoto J. Lab Invest. 1996;74:384–394. [PubMed] [Google Scholar]
- 51.Holyoake T, Jiang X, Eaves C, Eaves A. Blood. 1999;94:2056–2064. [PubMed] [Google Scholar]
- 52.Lewis I D, McDiarmid L A, Samels L M, Bik To L, Hughes T P. Blood. 1998;91:630–640. [PubMed] [Google Scholar]
- 53.Wang J C Y, Lapidot T, Cashman J D, Doedens M, Addy L, Sutherland D R, Nayar R, Laraya P, Minden M, Keating A, et al. Blood. 1998;91:2406–2414. [PubMed] [Google Scholar]
- 54.Hogge D E, Lansdorp P M, Reid D, Gerhard B, Eaves C J. Blood. 1996;88:3765–3773. [PubMed] [Google Scholar]