Abstract
Although polyomavirus JC (JCV) is the proven pathogen of progressive multifocal leukoencephalopathy, the fatal demyelinating disease, this virus is ubiquitous as a usually harmless symbiote among human beings. JCV propagates in the adult kidney and excretes its progeny in urine, from which JCV DNA can readily be recovered. The main mode of transmission of JCV is from parents to children through long cohabitation. In this study, we collected a substantial number of urine samples from native inhabitants of 34 countries in Europe, Africa, and Asia. A 610-bp segment of JCV DNA was amplified from each urine sample, and its DNA sequence was determined. A worldwide phylogenetic tree subsequently constructed revealed the presence of nine subtypes including minor ones. Five subtypes (EU, Af2, B1, SC, and CY) occupied rather large territories that overlapped with each other at their boundaries. The entire Europe, northern Africa, and western Asia were the domain of EU, whereas the domain of Af2 included nearly all of Africa and southwestern Asia all the way to the northeastern edge of India. Partially overlapping domains in Asia were occupied by subtypes B1, SC, and CY. Of particular interest was the recovery of JCV subtypes in a pocket or pockets that were separated by great geographic distances from the main domains of those subtypes. Certain of these pockets can readily be explained by recent migrations of human populations carrying these subtypes. Overall, it appears that JCV genotyping promises to reveal previously unknown human migration routes: ancient as well as recent.
The polyomavirus JC (JCV), the proven pathogen of the fatal demyelinating disease known as progressive multifocal leukoencephalopathy (PML) (1), is ubiquitous in human beings, infecting children asymptomatically (2, 3). After primary infection, JCV moves via an unknown route to the renal tissue where it persists throughout life (4–6). In adults, the renal JCV propagates and excretes its progeny in urine (7, 8), from which JCV DNA can readily be recovered.
Although JCV transmission is categorized as horizontal transmission (9, 10), it occurs frequently from parents to children (11). The reason for this is unclear, but we assume that JCV transmission requires a prolonged cohabitation of children with a particular pair of parents shedding JCV. Inasmuch as this mode predicts that JCV is not transmitted via temporary contact with strangers, we examined to what extent the inhabitants of the Okinawa Island of Japan were invaded by JCV shed by American soldiers who have been stationed there since 1945 (12). No evidence was obtained that the Japanese children were infected by American-type JCVs. Thus, it is likely that JCV is transmitted mainly from parents to children during prolonged close contact between them.
This mode of JCV transmission appeared to link JCV with human populations. Indeed, three major genotypes (A to C) have been identified in urine samples collected from Europe, Africa, and Asia (13, 14). Type A is prevalent only in Europe, type B is primarily spread over Asia and Africa, and type C is localized to a midwestern part of Africa. However, throughout the history of mankind, human populations have frequently intermixed due to migration and/or expansion. Therefore, some modern human populations would have multiple JCV types derived from different ancient populations. These JCV types would have been conserved without undergoing recombination with other JCVs since different JCVs rarely infect the same human host (8). Thus, we considered that by elucidating the JCV genotypes prevalent in human populations located in various geographic areas, we could obtain useful information on human migrations. The worldwide distribution patterns of nine JCV subtypes presented in this study showed that typing of urinary JCV DNA offers a novel means to trace human migrations: ancient as well as recent.
MATERIALS AND METHODS
Urine Samples.
Sites of urine collection are shown in Fig. 1 and Table 1. All donors were 40 years or older and were natives of each region (exceptional ethnic groups were excluded). The donors were either volunteers or patients without PML. Urine samples were collected from about 50 donors in each geographic region. Virions were recovered from urine, and DNA was extracted as described (7).
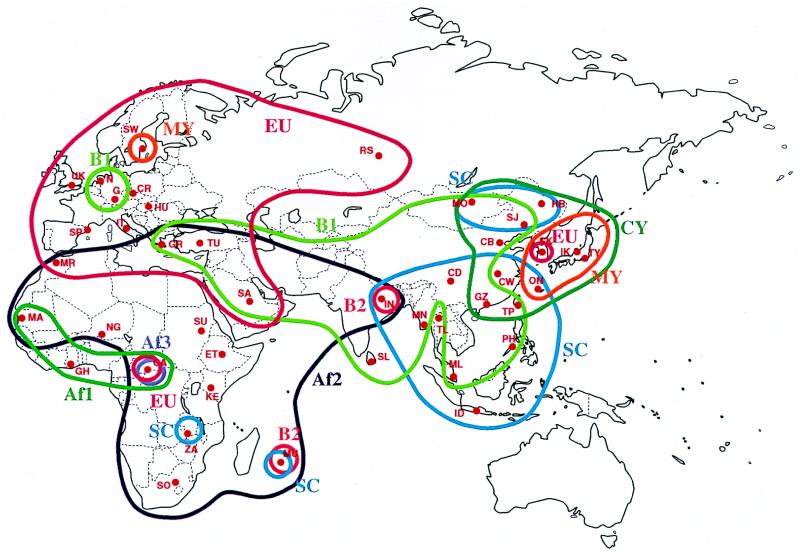
Figure 1.
World map showing the locations of the sites of urine collection and the territories of JCV subtypes. Red dots indicate the sites from which urine specimens were collected. One or two letters beside the dots indicate the city names by abbreviations (see Table 1). Sites where the same JCV subtype was detected are bounded by a line so that the enclosed area does not include those where this subtype was not detected. Since the German isolate GS/K (15) belonged to B1, Germany was included in the B1 domain.
Table 1.
Sites of urine collection and JCV isolates of which IG regions were identified by sequencing
| Geographic region
|
Isolates | |
|---|---|---|
| City, Country | Abbreviation | |
| Europe | ||
| London, United Kingdom | UK | UK-1, -2, -4 to -6 |
| Barcelona, Spain | SP | SP-1 to -5 |
| Rome, Italy | IT | IT-1 to -5 |
| Deventer, Netherlands | N | N1 to N7 |
| Illertissen, Germany | G | G1 to G5 |
| Stockholm, Sweden | SW | SW-1 to -4, -7, -8 |
| Prague, Czech Republic | CR | CR-1 to -7 |
| Budapest, Hungary | HU | HU-1 to -5 |
| Novosibirsk, Russia | RS | RS-1 to -5 |
| Athens, Greece | GR | GR-1 to -16 |
| Africa | ||
| Fes/Ifrane, Morocco | MR | MR-1 to -8 |
| Nouakchott, Mauritania | MA | MA-1, -2a*, -2b*, -3 to -7 |
| Accra, Ghana | GH | GH-1 to 4† |
| Bangui, Central Africa | CA | CA-1 to -11 |
| Khartoum, Sudan | SU | SU-1 to -5 |
| Tessaoua, Niger | NG | NG-1 to -5 |
| Welkom, South Africa | SO | SO-1 to -6 |
| Lusaka, Zambia | ZA | ZA-1 to -5 |
| Nairobi, Kenya | KE | KE-1 to -8 |
| Addis Ababa, Ethiopia | ET | ET-1 to -8 |
| Port Louis, Mauritius | MU | MU-1, -3 to -9 |
| Asia | ||
| Ankara, Turkey | TU | TU-1 to -15 |
| Riyadh, Saudi Arabia | SA | SA-1 to -14 |
| Varanasi, India | IN | IN-1 to -12 |
| Colombo, Sri Lanka | SL | SL-1 to -5 |
| Ulaanbaatar, Mongolia | MO | MO-1 to -12 |
| Yangon, Myanmar | MN | MN-1 to -15 |
| Chiang Mai, Thailand | TL | TL-1 to -11 |
| Masai, Malaysia | ML | ML-1 to -14 |
| Jakarta, Indonesia | ID | ID-1 to -17 |
| Pamalican Is., Philippines | PH | PH-1 to -8 |
| Harbin, China | HB | HB-1 to -6 |
| Shenyang/Jinzhou, China | SJ | SJ-1 to -7 |
| Beijing, China | CB | CB-1 to -10 |
| Wuhan, China | CW | CW-1 to -8, -10, -11 |
| Chengdu, China | CD | CD-1 to -10 |
| Guangzhou, China | GZ | GZ-1 to -13 |
| Taipei, China | TP | C1 to C9 |
| Seoul, South Korea | SK | SK-1 to -14 |
| Okinawa, Japan | ON | ON-1 to -11† |
| Ishikawa, Japan | IK | C-04 to -08, M-05 to -10† |
| Tokyo, Japan | TY | C-11 to -20, M-11 to -14† |
Amplification of the VT-Intergenic Regions (IG Regions).
From the viral DNA obtained from urine (see above), the 610-bp IG region (16) that encompasses the 3′ terminal regions of both T antigen and VP1 genes was PCR-amplified by using primers P1 and P2 (11) and KOD polymerase with proofreading activity (Toyobo, Osaka, Japan). [The IG region was previously established as a region of the JCV genome that contains abundant type-determining sites (16).] The reaction was carried out as recommended by the manufacturer. Some IG regions were amplified from established JCV DNA clones (13, 14).
We previously described IG sequences identified in Ghana (14) and Japan (6, 11, 12). These IG sequences were also used in this study (see Table 1).
Sequencing.
Amplified IG fragments were cloned into pUC19 and purified recombinant plasmids were sequenced as described (11). Two clones for each urine sample were sequenced.
Restriction Fragment Length Polymorphism (RFLP).
RFLP analysis was performed as described (12), with AluI, BglII, DdeI, and HinfI.
Phylogenetic Analysis.
A neighbor-joining (NJ) phylogenetic tree (17) was constructed using the clustal w program (18). Divergences were estimated by the two-parameter method (19). A phylogenetic tree was visualized using the treeview 1.4 program (20). The bootstrap test was applied to estimate the confidence of the branching patterns of the NJ tree (21). A phylogenetic tree was also constructed by the unweighted pair-group method with arithmetic averages (22) using the oden program package (23) (data not shown).
RESULTS
Identification of Nine JCV Subtypes.
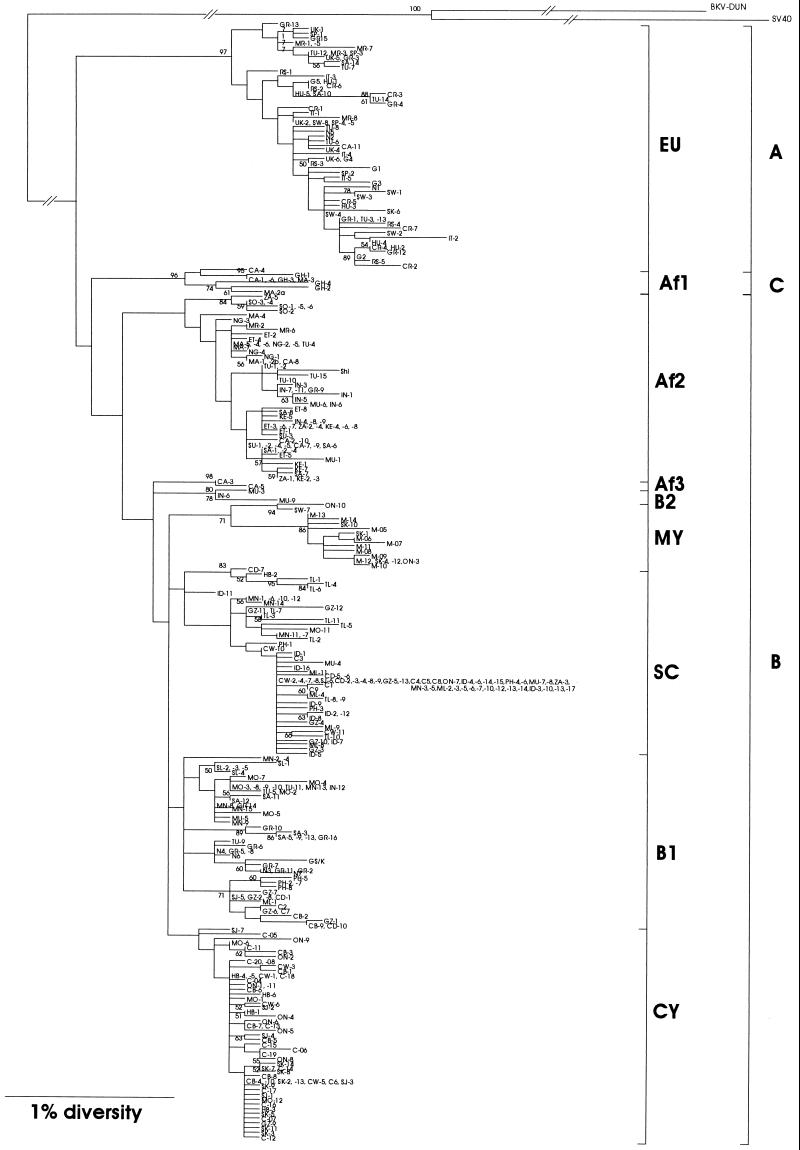
We determined the IG sequences of 379 JCV isolates (Table 1). Examination of these sequences revealed the presence of 241 different IG sequences, of which 191 were unique to one donor. The remaining 50 sequences were shared between 2 and 40 donors. A phylogenetic NJ tree (17) was constructed from the 241 IG sequences and a previously established IG sequence [GS/K (15) from the kidney of a German PML patient and JCV (Shi) (24) from the urine of Tanzanians; Fig. 2]. As the outgroup, two primate polyomaviruses, simian virus 40 (25) and BK virus (Dun) (26), were used.
Figure 2.
(On the opposite page.) NJ tree relating JCVs detected in Europe, Africa, and Asia. Alignment of IG sequences (379 in total) determined in this and other studies (see Table 1) generated 241 unique sequences. A NJ tree (17) was constructed from the 241 IG sequences and previously established IG sequence [GS/K (15) from the kidney of a German PML patient and JCV (Shi) (24) from the urine of Tanzanians] by using the clustal w program (18). The tree was rooted by using two primate polyomaviruses, simian virus 40 (25) and BK virus (Dun) (26), as the outgroup. The phylogenetic tree was visualized by the treeview 1.4 program (20). The symbol to each sequence is shown in Table 1. The numbers at the nodes indicate bootstrap confidence levels obtained by 100 replicates (only those 50% are shown). Subtypes and types are indicated to the right of the tree.
In the phylogenetic tree (Fig. 2), these JCV isolates formed nine discrete clusters that were designated as subtypes as shown in Fig. 2. We also made an independent analysis of the same data by using the unweighted pair-group method with arithmetic averages (22). This method also yielded nine subtypes that corresponded well to the above noted nine subtypes identified by the NJ method.
JCV isolates in Europe, Africa, and Asia were previously classified into types A-C according to RFLP (types A and B) or phylogenetic analysis (type C) (13, 14). The phylogentic tree (Fig. 2) revealed that types A and C of the previous studies corresponded exactly to subtypes EU and Af1 determined by the present sequencing study. On the other hand, RFLP type B revealed itself to be an amalgam that encompassed all the remaining seven subtypes: Af2, Af3, B2, MY, SC, B1, and CY. Another previously described type was JCV (Shi) (or type 3) found in Tanzania and the United States (24, 27). This type belonged to Af2 subtype.
According to the phylogenetic tree (Fig. 2) that was constructed using both simian virus 40 and BK virus as the outgroup, EU was the first to diverge from the common root for all JCVs, followed by Af1, Af2, and the other subtypes. However, the above noted order of divergence of various subtypes should be verified by an independent study using other regions of the JCV genome for comparison.
From the phylogenetic tree (Fig. 2), we roughly estimated the evolutionary rate for the JCV IG region, on the assumption that the first divergence of JCV subtypes occurred 100,000 years ago, and we obtained a value of about 1–3 × 10−7 per site per year.
JCV Subtypes Prevalent in the Old World.
We determined the subtypes of 379 isolates by sequencing IG regions, as described above. In addition, 106 isolates in Europe, Africa, and western Asia were identified as EU or Af2 by RFLP analysis. The incidence of JCV subtypes thus determined is shown in Table 2 for each region of the Old World. The data can be summarized as follows.
Table 2.
Geographic distribution of JCV subtypes
| Region* | No. of isolates | Incidence (%) of JCV
subtype
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EU | Af1 | Af2 | Af3 | B1 | B2 | SC | CY | MY | ||||
| Europe | ||||||||||||
| UK | 6 | 100 | —† | — | — | — | — | — | — | — | ||
| SP | 13 | 100 | — | — | — | — | — | — | — | — | ||
| IT | 12 | 100 | — | — | — | — | — | — | — | — | ||
| N | 12 | 67 | — | — | — | 33 | — | — | — | — | ||
| G | 8 | 100 | — | — | — | — | — | — | — | — | ||
| SW | 18 | 94 | — | — | — | — | — | — | — | 6 | ||
| CR | 18 | 100 | — | — | — | — | — | — | — | — | ||
| HU | 17 | 100 | — | — | — | — | — | — | — | — | ||
| RS | 14 | 100 | — | — | — | — | — | — | — | — | ||
| GR | 20 | 50 | — | 5 | — | 45 | — | — | — | — | ||
| Africa | ||||||||||||
| MR | 21 | 57 | — | 43 | — | — | — | — | — | — | ||
| MA | 10 | — | 20 | 80 | — | — | — | — | — | — | ||
| GH‡ | 4 | — | 100 | — | — | — | — | — | — | — | ||
| CA | 11 | 9 | 27 | 45 | 18 | — | — | — | — | — | ||
| SU | 9 | — | — | 100 | — | — | — | — | — | — | ||
| NG | 8 | — | — | 100 | — | — | — | — | — | — | ||
| SO | 6 | — | — | 100 | — | — | — | — | — | |||
| ZA | 5 | — | — | 80 | — | — | — | 20 | — | — | ||
| KE | 8 | — | — | 100 | — | — | — | — | — | — | ||
| ET | 8 | — | — | 100 | — | — | — | — | — | — | ||
| MU | 8 | — | — | 25 | — | 13 | 25 | 38 | — | — | ||
| Asia | ||||||||||||
| TU | 15 | 47 | — | 33 | — | 20 | — | — | — | — | ||
| SA | 20 | 10 | — | 60 | — | 30 | — | — | — | — | ||
| IN | 17 | — | — | 82 | — | 6 | 6 | 6 | — | — | ||
| SL | 5 | — | — | — | — | 100 | — | — | — | — | ||
| MO | 12 | — | — | — | — | 67 | — | 8 | 25 | — | ||
| MN | 15 | — | — | — | — | 46 | — | 53 | — | — | ||
| TL | 11 | — | — | — | — | — | — | 100 | — | — | ||
| ML | 14 | — | — | — | — | 7 | — | 93 | — | — | ||
| ID | 17 | — | — | — | — | — | — | 100 | — | — | ||
| PH | 8 | — | — | — | — | 50 | — | 50 | — | — | ||
| HB | 6 | — | — | — | — | — | — | 17 | 83 | — | ||
| SJ | 7 | — | — | — | — | 14 | — | 14 | 71 | — | ||
| CB | 10 | — | — | — | — | 20 | — | — | 80 | — | ||
| CW | 10 | — | — | — | — | — | — | 60 | 40 | — | ||
| CD | 10 | — | — | — | — | 20 | — | 80 | — | — | ||
| GZ | 13 | — | — | — | — | 38 | — | 54 | 8 | — | ||
| TP | 9 | — | — | — | — | 22 | — | 67 | 11 | — | ||
| SK | 14 | 7 | — | — | — | — | — | — | 64 | 29 | ||
| ON‡ | 11 | — | — | — | — | — | — | 9 | 73 | 18 | ||
| IK‡ | 11 | — | — | — | — | — | — | — | 45 | 55 | ||
| TY‡ | 14 | — | — | — | — | — | — | — | 71 | 29 | ||
JCV Subtypes in Europe.
A single subtype (EU) was predominant in all areas of Europe. However, one (B1) and two additional subtypes (B1 and Af2) were identified in Deventer (the Netherlands) and Athens (Greece), respectively. Furthermore, one minor subtype (MY) was found in Stockholm (Sweden).
JCV Subtypes in Africa.
A single subtype (Af2) was predominant in all areas of Africa with the following exceptions. Both EU and Af2 were prevalent in Fes/Ifrane (Morocco), and a single subtype (Af1) was predominant in Accra (Ghana). In addition, the following minor subtypes were found in indicated regions: Af1 in Nouakchott (Mauritania) and Bangui (Central Africa), Af3 in Bangui, EU in Bangui and SC in Lusaka (Zambia). Furthermore, four subtypes (Af2, B1, B2, and SC) were prevalent in Port Louis (Mauritius).
JCV Subtypes in Asia.
Prevalent subtypes markedly varied between regions. Three subtype (EU, Af2, and B1) were prevalent in Ankara (Turkey). One major (Af2) and two minor subtypes (EU and B1) were prevalent in Riyadh (Saudi Arabia). One major (Af2) and three minor subtypes (B1, B2 and SC) were prevalent in Varanasi (India). A single subtype (B1) was predominant in Colombo (Sri Lanka). One major (B1) and two minor subtypes (SC and CY) were prevalent in Ulaanbaatar (Mongolia). Two subtypes (B1 and SC) were prevalent in Yangon (Myanmar). A single subtype (SC) was predominant in southeast Asia including Chiang Mai (Thailand), Masai (Malaysia), and Jakarta (Indonesia) with a minor subtype of B1 (Masai). Two subtypes (SC and B1) were equally prevalent in Pamalican Island (the Philippines). Three major (B1, SC, and CY) were prevalent in China. Two subtypes (CY and MY) were prevalent in Far-East Asia including Seoul (South Korea), Okinawa, Ishikawa, and Tokyo (Japan) with minor subtypes of EU (Seoul) and SC (Okinawa).
Territories of JCV Subtypes.
To illustrate the territories of JCV subtypes, sites where the same JCV subtype was detected is enclosed by a line that excludes areas where this subtype was not detected (Fig. 1).
Five subtypes (EU, Af2, B1, SC, and CY) occupied rather large domains that overlapped with each other at their boundaries. Thus, the entire Europe, northern Africa, and western Asia was the domain of EU, whereas the domain of Af2 included nearly all of Africa and southwestern Asia all the way to the northeastern edge of India. A wide region extending from western to eastern Asia was the domain of B1; southern China and southeastern Asia was that of SC; and northeastern Asia was that of CY.
The other four subtypes (Af1, Af3, B2, and MY) had rather small domains, located in unique geographic regions. These minor domains were usually existed within the larger ones. Thus, Af1, Af3, and B2 were within that of Af2; and MY was within the CY domain.
Several subtypes had a pocket or pockets separated by geographic distances from the main domains. We identified the following pockets: MY pocket in Stockholm (Sweden), EU pockets in Seoul (South Korea) and Bangui (Central Africa), and SC pockets in Lusaka (Zambia) and Port Louis (Mauritius).
DISCUSSION
We have developed a new method using urinary JCV DNA to trace human migrations. This method is based on the fact that each of the nine JCV subtypes has a unique territory in the Old World (Fig. 1). Although these distribution patterns of JCV subtypes themselves have implications for the differentiations and migrations of human beings (this point will be discussed below), they should also offer the basis for further studies using the current method. For example, if a group of individuals from a previously unstudied area are found to be infected with a particular known JCV subtype, these are migrants from the domain of that subtype. Conversely, if a certain number of individuals in a region show a previously unknown JCV subtype distinctly different from prevalent subtypes of that area, these are migrants whose origin must be reevaluated. This analysis may be applied to any human populations, including Amerindians in the New World and original inhabitants of polar regions, since JCV is prevalent in essentially all human populations (3).
Although other viruses have also been employed to trace human migrations, JCV genotyping is the most advantageous. For example, human T cell lymphotropic virus type I, which has most frequently been used for the purpose described above (28–32), is limited in that it offers only knowledge on migrations of populations carrying this virus since it is not endemic in many countries (33). Another example is human papillomavirus type 16 where although there exist clusters of variants characteristic for different geographic regions (34), the original correlation between these clusters and human races appears to be markedly obscured by human migrations that have occurred since the 16th century.
Mitochondrial DNA is now widely used as a genetic marker to infer the ethnic backgrounds of human subjects. However, mitochondrial DNA is hypervariable, and it is generally observed that mitochondrial lineages are markedly intermingled between human populations (35–37). Therefore, population trees are frequently constructed on the basis of the intra- and interpopulational genetic distances (38) that can be calculated from mitochondrial DNA sequence diversities (37, 39, 40). Furthermore, analysis of mitochondrial DNA RFLP haplotypes are sometimes used to infer human migrations (41–43).
We emphasize that various human migrations on the earth can readily and clearly be indicated by the current method. It is now widely accepted that our own species, Homo sapiens sapiens, originated in Africa less than a million years ago, and subsequently a small band of the original population migrated out of Africa to the near East and further spread to West, East, and North, in the process differentiating into Caucasoids and Mongoloids (44). The overall distribution of JCV subtypes (Fig. 1) is compatible with the above notion. The domain of Af2 extends from most of Africa through the near East to the eastern edge of India, whereas EU could have originated in the near East and accompanied the migration of protoCaucasoids to Europe. B1 too could have originated in the near East and its extension eastward might have accompanied the migration of protoMongoloids.
As to more recent past, several routes of human migrations in East Asia were inferred on the basis of the domains of JCV subtypes shown in Fig. 1: a route from mainland China to Southeast Asia was inferred from the SC domain; two routes from mainland China to mainland Japan, one via Korea and the other via Okinawa, were inferred from the CY domain; a route from mainland China to Taiwan was inferred from the CY, SC, and B1 domains; and a route from Korea to mainland Japan then to Okinawa was inferred from the MY domain.
The power of this method was further suggested by the identification of pockets that were separated by great geographic distances from the main domains. For example, MY that was detected as one of the two major subtypes in South Korea and Japan was also seen in one of the 18 samples from Sweden. Similarly, EU of the territory composed of Europe and western Asia was also found in one sample each from Central Africa and South Korea. Although at this time, no rational explanation can be given to the presence of these pockets, the presence of B2 subtype both in the northeastern extreme of India and in Mauritius off the east coast of Africa can readily be explained by recent migration of a population from the former to the latter. Overall, it appears that JCV genotyping promises to reveal previously unknown human migration routes: ancient as well as recent.
Acknowledgments
We thank Y. Nagai and S. Sekiguchi for helpful suggestions, A. Kato and H. Ebihara for help in sequencing, and N. Shimokawa for excellent technical assistance. This study was supported in part by grants from the Ministry of Education, Sciences, Sports and Culture and from the Ministry of Welfare, Japan.
ABBREVIATIONS
- JCV
JC virus
- PML
progressive multifocal leukoencephalopathy
- IG region (sequence)
VT-intergenic region (sequence)
- RFLP
restriction fragment length polymorphism
- NJ tree
neighbor joining tree
Footnotes
References
- 1.Padgett B L, Walker D L, ZuRhein G M, Eckroade R J, Dessel B H. Lancet. 1971;i:1257–1260. doi: 10.1016/s0140-6736(71)91777-6. [DOI] [PubMed] [Google Scholar]
- 2.Padgett B L, Walker D L. J Infect Dis. 1973;127:467–470. doi: 10.1093/infdis/127.4.467. [DOI] [PubMed] [Google Scholar]
- 3.Padgett B L, Walker D L. Prog Med Virol. 1976;22:1–35. [PubMed] [Google Scholar]
- 4.Chesters P M, Heritage J, McCance D J. J Infect Dis. 1983;147:676–684. doi: 10.1093/infdis/147.4.676. [DOI] [PubMed] [Google Scholar]
- 5.Tominaga T, Yogo Y, Kitamura T, Aso Y. Virology. 1992;186:736–741. doi: 10.1016/0042-6822(92)90040-v. [DOI] [PubMed] [Google Scholar]
- 6.Kitamura T, Sugimoto C, Kato A, Ebihara H, Suzuki M, Taguchi F, Kawabe K, Yogo Y. J Clin Microbiol. 1997;35:1255–1257. doi: 10.1128/jcm.35.5.1255-1257.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kitamura T, Aso Y, Kuniyoshi N, Hara K, Yogo Y. J Infect Dis. 1990;161:1128–1133. doi: 10.1093/infdis/161.6.1128. [DOI] [PubMed] [Google Scholar]
- 8.Kitamura T, Kunitake T, Guo J, Tominaga T, Kawabe K, Yogo Y. J Clin Microbiol. 1994;32:2359–2363. doi: 10.1128/jcm.32.10.2359-2363.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Daniel R, Shah K, Madden D, Stagno S. Infect Immun. 1981;33:319–321. doi: 10.1128/iai.33.1.319-321.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coleman D V, Wolfendale M R, Daniel R A, Dhanjal N K, Gardner S D, Gibson P E, Field A M. J Infect Dis. 1980;142:1–8. doi: 10.1093/infdis/142.1.1. [DOI] [PubMed] [Google Scholar]
- 11.Kunitake T, Kitamura T, Guo J, Taguchi F, Kawabe K, Yogo Y. J Clin Microbiol. 1995;33:1448–1451. doi: 10.1128/jcm.33.6.1448-1451.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kato A, Kitamura T, Sugimoto C, Ogawa Y, Nakazato K, Nagashima K, Hall W W, Kawabe K, Yogo Y. Arch Virol. 1997;142:875–882. doi: 10.1007/s007050050125. [DOI] [PubMed] [Google Scholar]
- 13.Yogo Y, Iida T, Taguchi F, Kitamura T, Aso Y. J Clin Microbiol. 1991;29:2130–2138. doi: 10.1128/jcm.29.10.2130-2138.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo J, Kitamura T, Ebihara H, Sugimoto C, Kunitake T, Takehisa J, Na Y Q, Al-Ahdal M N, Hallin A, Kawabe K, Taguchi F, Yogo Y. J Gen Virol. 1996;77:919–927. doi: 10.1099/0022-1317-77-5-919. [DOI] [PubMed] [Google Scholar]
- 15.Loeber G, Dörries K. J Virol. 1988;62:1730–1735. doi: 10.1128/jvi.62.5.1730-1735.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ault G S, Stoner G L. J Gen Virol. 1992;73:2669–2678. doi: 10.1099/0022-1317-73-10-2669. [DOI] [PubMed] [Google Scholar]
- 17.Saitou N, Nei M. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 18.Thompson J D, Higgins D G, Gibson T J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kimura M. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 20.Page R D M. Comput Appl Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- 21.Felsenstein J. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 22.Sneath P H A, Sokal R R. Numerical Taxonomy. San Francisco: Freeman; 1973. [Google Scholar]
- 23.Ina Y. Comput Appl Biosci. 1994;10:11–12. doi: 10.1093/bioinformatics/10.1.11. [DOI] [PubMed] [Google Scholar]
- 24.Agostini H T, Brubaker G R, Shao J, Levin A, Ryschkewitsch C F, Blattner W A, Stoner G L. Arch Virol. 1995;140:1919–1934. doi: 10.1007/BF01322682. [DOI] [PubMed] [Google Scholar]
- 25.Fiers W, Contreras R, Haegeman G, Rogiers R, van de Voorde A, van Heuverswyn H, van Herreweghe J, Volckaert G, Ysebaert M. Nature (London) 1978;273:113–120. doi: 10.1038/273113a0. [DOI] [PubMed] [Google Scholar]
- 26.Seif I, Khoury G, Dhar R. Cell. 1979;18:963–977. doi: 10.1016/0092-8674(79)90209-5. [DOI] [PubMed] [Google Scholar]
- 27.Agostini H T, Ryschkewitsch C F, Brubaker G R, Shao J, Stoner G L. Arch Virol. 1997;142:637–655. doi: 10.1007/s007050050108. [DOI] [PubMed] [Google Scholar]
- 28.Ishida T, Hinuma Y. Nature (London) 1986;322:504. doi: 10.1038/322504a0. [DOI] [PubMed] [Google Scholar]
- 29.Miura T, Fukunaga T, Igarashi T, Yamashita M, Ido E, Funahashi S, Ishida T, Washio K, Ueda S, Hashimoto K, Yoshida M, Osame M, Singhal B S, Zaninovic V, Cartier L, Sonoda S, Tajima K, Ina Y, Gojobori T, Hayami M. Proc Natl Acad Sci USA. 1994;91:1124–1127. doi: 10.1073/pnas.91.3.1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yanagihara R. Adv Virus Res. 1994;43:147–186. doi: 10.1016/s0065-3527(08)60048-2. [DOI] [PubMed] [Google Scholar]
- 31.Yanagihara, R., Saitou, N., Nerurkar, V. R., Song, K.-J., Bastian, I., Franchini, G. & Gajdusek, D. C. (1995) Cell. Mol. Biol. 41, Suppl. 1, S145–S161. [PubMed]
- 32.Mahieux R, Ibrahim F, Mauclere P, Herve V, Michel P, Tekaia F, Chappey C, Garin B, van der Ryst E, Guillemain B, Ledru E, Delaporte E, de The G, Gessain A. J Virol. 1997;71:1317–1333. doi: 10.1128/jvi.71.2.1317-1333.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tajima K, Hinuma Y. Gann Monogr. 1992;39:129–149. [Google Scholar]
- 34.Ho L, Chan S-Y, Burk R D, Das B C, Fujinaga K, Icenogle J P, Kahn T, Kiviat N, Lancaster W, Mavromara-Nazos P, Labropoulou V, Mitrani-Rosenbaum S, Norrild B, Pillai M R, Stoerker J, Syrjaenen K, Syrjaenen S, Tay S-K, Villa L L, Wheeler C M, Williamson A-L, Bernard H-U. J Virol. 1993;67:6413–6423. doi: 10.1128/jvi.67.11.6413-6423.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cann R L, Stoneking M, Wilson A C. Nature (London) 1987;325:31–36. doi: 10.1038/325031a0. [DOI] [PubMed] [Google Scholar]
- 36.Horai S, Kondo R, Murayama K, Hayashi S, Koike H, Nakai N. Philos Trans R Soc London B. 1991;333:409–417. doi: 10.1098/rstb.1991.0091. [DOI] [PubMed] [Google Scholar]
- 37.Horai S, Murayama K, Hayasaka K, Matsubayashi S, Hattori Y, Fucharoen G, Harihara S, Park K S, Omoto K, Pan I-H. Am J Hum Genet. 1996;59:579–590. [PMC free article] [PubMed] [Google Scholar]
- 38.Nei M, Jin L. Mol Biol Evol. 1989;6:290–300. doi: 10.1093/oxfordjournals.molbev.a040547. [DOI] [PubMed] [Google Scholar]
- 39.Watson E, Bauer K, Aman R, Weiss G, von Haeseler A, Pääbo S. Am J Hum Genet. 1996;59:437–444. [PMC free article] [PubMed] [Google Scholar]
- 40.Bonatto S L, Salzano F M. Proc Natl Acad Sci USA. 1997;94:1866–1871. doi: 10.1073/pnas.94.5.1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wallace D C. Proc Natl Acad Sci USA. 1994;91:8739–8746. doi: 10.1073/pnas.91.19.8739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Merriwether D A, Hall W W, Vahlne A, Ferrell R E. Am J Hum Genet. 1996;59:204–212. [PMC free article] [PubMed] [Google Scholar]
- 43.Easton R D, Merriwether D A, Crews D E, Ferrell R E. Am J Hum Genet. 1996;59:213–225. [PMC free article] [PubMed] [Google Scholar]
- 44.Vigilant L, Pennington R, Harpending H, Kocher T D, Wilson A C. Proc Natl Acad Sci USA. 1989;86:9350–9354. doi: 10.1073/pnas.86.23.9350. [DOI] [PMC free article] [PubMed] [Google Scholar]