Abstract
Reduction of 5,10-methylenetetrahydrofolate (methyleneTHF), a donor for methylating dUMP to dTMP in DNA synthesis, to 5-methyltetrahydrofolate (methylTHF), the primary methyl donor for methionine synthesis, is catalyzed by 5,10-methylenetetrahydrofolate reductase (MTHFR). A common 677 C → T polymorphism in the MTHFR gene results in thermolability and reduced MTHFR activity that decreases the pool of methylTHF and increases the pool of methyleneTHF. Recently, another polymorphism in MTHFR (1298 A → C) has been identified that also results in diminished enzyme activity. We tested whether carriers of these variant alleles are protected from adult acute leukemia. We analyzed DNA from a case–control study in the United Kingdom of 308 adult acute leukemia patients and 491 age- and sex-matched controls. MTHFR variant alleles were determined by a PCR-restriction fragment length polymorphism assay. The MTHFR 677TT genotype was lower among 71 acute lymphocytic leukemia (ALL) cases compared with 114 controls, conferring a 4.3-fold decrease in risk of ALL [odds ratio (OR = 0.23; 95% CI = 0.06–0.81]. We observed a 3-fold reduction in risk of ALL in individuals with the MTHFR 1298AC polymorphism (OR = 0.33; 95% CI = 0.15–0.73) and a 14-fold decreased risk of ALL in those with the MTHFR 1298CC variant allele (OR = 0.07; 95% CI = 0.00–1.77). In acute myeloid leukemia, no significant difference in MTHFR 677 and 1298 genotype frequencies was observed between 237 cases and 377 controls. Individuals with the MTHFR 677TT, 1298AC, and 1298CC genotypes have a decreased risk of adult ALL, but not acute myeloid leukemia, which suggests that folate inadequacy may play a key role in the development of ALL.
The etiology of most types of leukemia remains unknown. Established causes of leukemia such as ionizing radiation, benzene, and cancer chemotherapy account for only a small percentage of the total cases (1, 2). Moreover, leukemia is unlikely to be caused by a single genetic defect because there is only a modest familial link. Thus, leukemias are most likely the result of an adverse gene–environment interaction, with susceptibility being related to polymorphisms in multiple genes. We hypothesized that there may be a correlation between functional polymorphisms in the gene for the folate metabolizing enzyme, 5,10-methylenetetrahydrofolate reductase (MTHFR), and leukemogenesis because of the association between folate status and susceptibility to genetic damage in dividing cells (3–6).
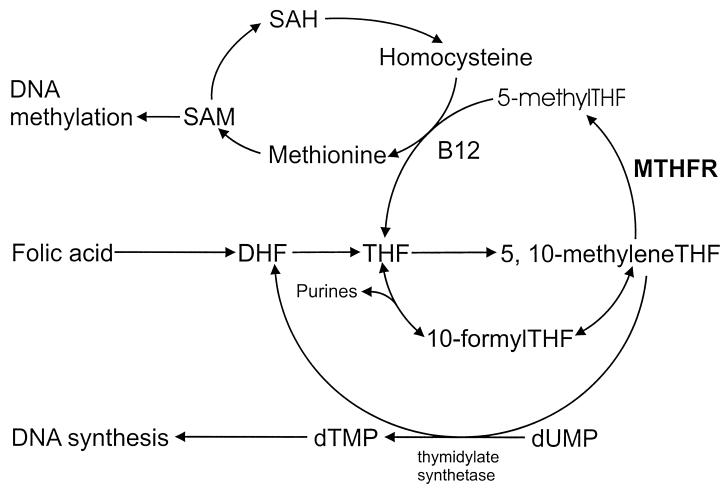
MTHFR catalyzes the reduction of 5,10-methylenetetrahydrofolate (methyleneTHF) to 5-methyltetrahydrofolate (methylTHF) (7), the predominant circulatory form of folate and carbon donor for the remethylation of homocysteine to methionine (Fig. 1). A polymorphism located at nucleotide 677 of the MTHFR gene results in an alanine-to-valine substitution. Up to 15% of individuals are homozygous for this allelic variant (8–10), which results in homocysteine accumulation (11–13) and thermolability of the enzyme with significantly reduced specific activity (8). A reduction in the activity of the MTHFR enzyme increases the pool of methyleneTHF at the expense of the pool of methylTHF. Enhanced availability of methyleneTHF in the DNA synthesis pathway reduces misincorporation of uracil into DNA, which might otherwise result in double-strand breaks during uracil excision repair processes (3, 14). Previous studies have shown that individuals with adequate folate status who are homozygous for the MTHFR 677 mutation (677TT) have a reduced incidence of colorectal cancer (15, 16). Because colorectal carcinomas and leukemias are derived from rapidly proliferating tissues that have the greatest requirement for DNA synthesis, they are likely to be affected similarly by the metabolic fate of folic acid.
Figure 1.
Overview of the human folic acid metabolic pathway and the role of MTHFR, S-adenosylmethionine (SAM), S-adenosylhomocysteine (SAH), dihydrotetrafolic acid (DHF), and tetrahydrofolic acid (THF).
Recently, a second common polymorphism in the MTHFR gene has been reported that results in a glutamate-to-alanine (A → C) change at position 1298 that also leads to diminished enzyme activity (17, 18). Allele frequencies up to 33% have been reported previously for this polymorphism (17, 18).
Our study was designed to evaluate what role, either individually or combined, the 677 and 1298 MTHFR polymorphisms play in susceptibility to leukemia because previous studies have shown evidence of interaction between them (17, 18). We hypothesized that carriers of variant alleles for MTHFR 677 and/or MTHFR 1298 may have a protective advantage against leukemia. To evaluate this hypothesis, we analyzed DNA from a population-based case–control study of adult acute leukemia.
Methods
Study Population and Sample Collection.
The study population consisted of 308 individuals diagnosed with acute leukemia that formed part of the British Leukaemia Research Fund Centre’s case–control study of acute leukemia in adults. A brief outline of the study methods are given here; they are fully described elsewhere (19). Cases were persons aged 16–70 years who were diagnosed with acute leukemia between April 1, 1991, and December 31, 1996, while resident in specific regions of north and southwest England. Patients were approached, with the physician’s permission, to participate in a face-to-face interview using a structured questionnaire. For each participating case, two controls were selected randomly from a list of all persons registered with the same local physician who were of the same sex, year of birth (±2 years), and race as the case. Cases diagnosed with acute leukemia within 6 months of being diagnosed with a prior hematological malignancy or within 2 years of any other cancer were considered ineligible. This ineligibility criterion also was applied to the corresponding controls of cases (pseudodiagnosis being taken as the date of case diagnosis). Because the prevalence of the MTHFR 677 polymorphism has been shown to vary with race (8, 9, 20–22), and the number of non-white cases included was small, the analyses here are restricted to 308 Caucasian cases and their 491 matched Caucasian controls. Blood samples were unavailable for the other 125 matched controls originally enrolled in the study.
Of the 308 cases, 71 were acute lymphocytic leukemia (ALL) with 114 individually matched controls, and 237 were acute myeloid leukemia (AML) with 377 individually matched controls. The AML cases consisted of the following subtypes: M1, 46; M2, 54; M3, 37; M4, 45; M5, 20; M6, 13; and M7, 2; a further 20 cases could not be assigned specific French–American–British subtypes. Among the ALL cases, 50 were diagnosed with ALL B, 11 were diagnosed with ALL T, and, for 10 cases, the immunophenotype could not be specified. Fifty-eight percent of the ALL patients were male, with a mean age of 41.3 years. Among AML cases, 54% were male and the average age was 47.4 years.
Analysis of the MTHFR 677 and 1298 Genotypes.
Blood samples from cases and controls were collected by venipuncture, and DNA was isolated by using a simple proteinase K treatment, followed by phenol-chloroform extraction and ethanol precipitation. The genotyping protocol for the detection of the MTHFR 677 C → T polymorphism was adapted from Frosst et al. (8). This C → T base pair substitution creates a HinfI restriction site. Briefly, 0.5–2.0 μg of human genomic DNA was amplified with 50 ng each of forward primer 5′-TGA AGG AGA AGG TGT CTG CGG GA-3′ and reverse primer 5′-AGG ACG GTG CGG TGA GAG TG-3′. PCR thermal cycling conditions were a 2-min denaturation period at 94°C and 40 cycles of the following: 94°C for 30 s, 62°C for 30 s, and 72°C for 30 s. This was followed by a 7-min extension at 72°C. The 50-μl PCR mixture contained 10 mM Tris⋅HCl (pH 8.3), 50 mM KCl, 2.5 mM MgCl2, 0.1 mg/ml gelatin, 200 μM each dNTP, and 1.25 units of Taq DNA polymerase (Sigma).
HinfI restriction digestion using 2.5 μl of buffer and 10 units of HinfI restriction enzyme (Boehringer Mannheim) added to 25 μl of PCR product was incubated at 37°C for at least 2 hr. Digestion products were visualized after electrophoresis on a 4% agarose gel with ethidium bromide. Wild types (677CC) produced a singlet band at 198 bp. Heterozygotes (677CT) produced 198-, 175-, and 23-bp fragments. Homozygous mutants (677TT) produced 175- and 23-bp fragments.
The presence of the MTHFR 1298 A → C polymorphism was analyzed by using a modification of a method from Weisberg et al. (18). This glutamate-to-alanine substitution abolishes an MboII restriction site. Briefly, 0.5–2.0 μg of human genomic DNA was amplified with 50 ng each of the forward PCR primer 5′-CTT TGG GGA GCT GAA GGA CTA CTA C-3′ and the reverse primer 5′-CAC TTT GTG ACC ATT CCG GTT TG-3′, 10 mM Tris⋅HCl, 50 mM KCl, 1 mg/ml gelatin, 3.0 mM MgCl2, 200 μM each dNTP, and 1.25 units of DNA Taq polymerase (Sigma). PCR parameters were a 2-min denaturation cycle at 92°C and 35 cycles of the following: 92°C for 1 min, 60°C for 1 min, and 72°C for 30 s. This was followed by a 7-min extension at 72°C. The 163-bp PCR product was digested with 2.5 μl of MboII buffer and 2.5 units of MboII restriction enzyme (New England Biolabs) followed by electrophoresis on a 3% Metaphor gel (FMC). Wild types (1298AA) produced five fragments of 56, 31, 30, 28, and 18 bp, heterozygotes (1298AC) produced six fragments of 84, 56, 31, 30, 28, and 18 bp, and the homozygous mutants (1298CC) produced four fragments of 84, 31, 30, and 18 bp. The major visible bands were those of 84 and 56 bp.
Quality control samples were included in the analysis, and all laboratory personnel were blinded as to case and control status.
Statistical Analysis.
Odds ratios and 95% confidence intervals for a matched analysis were computed by using conditional logistic regression (23). Two trichotomous variables, with a level for each allelic variant, were generated for MTHFR 677 and MTHFR 1298. When cell frequencies were less than 5, exact methods were used to compute the risk estimates. Tests for independence and interaction between MTHFR 677 and MTHFR 1298 were completed by using the likelihood ratio test. All analyses were performed by using stata (24).
Results
Among the 308 adult patients diagnosed with acute leukemia, the MTHFR 677 polymorphic allele frequency was 26% compared with 29% among the 491 control subjects. Frequencies of MTHFR 677CC, 677CT, and 677TT genotypes were 57.9%, 32.5%, and 9.6% in the cases and 53.2%, 35.0%, and 11.8% in the controls, respectively. For MTHFR 1298, we observed a polymorphic allele frequency of 29% in the cases and 33% in the controls. Frequencies of MTHFR 1298AA, 1298AC, and 1298CC genotypes were 49.8%, 41.9%, and 8.3% in the cases and 45.0%, 44.3%, and 10.7% in the controls, respectively. The observed frequencies in the ALL and AML controls for MTHFR 677 and 1298 were in accordance with Hardy–Weinberg laws of equilibrium. We did not observe differences in the prevalence of either the MTHFR 677 or 1298 genotypes between the cases and controls in the total population. However, upon stratification according to leukemia type, we found significant differences in genotype frequencies among the ALL cases and controls.
Prevalence of the MTHFR 677 and 1298 Genotypes in the ALL Cases and Corresponding Controls.
After categorizing the ALL cases according to ALL-B and ALL-T immunophenotypes, no significant differences in the MTHFR 677 and 1298 mutant frequencies were observed between groups (data not shown). Therefore, ALL-B and ALL-T cases were considered as a single group for this analysis.
Listed in Table 1 are the observed frequencies of the MTHFR 677 and 1298 polymorphisms among the 71 ALL cases and 114 matched controls. We found the MTHFR 677CC allele present among 35 (50.7%) cases and 61 (53.5%) controls, the 677CT genotype among 29 (42.0%) cases and 39 (34.2%) controls, and the 677TT allele among 5 (7.2%) cases and 14 (12.3%) controls. Two cases were not genotyped successfully for MTHFR 677. For MTHFR 1298, the 1298AA genotype was observed in 45 (65.2%) of the cases and 49 (43.0%) of the controls, the 1298AC allelic variant was observed in 23 (33.3%) cases and 54 (47.4%) controls, and the rarer 1298CC variant was observed among 1 (1.5%) case and 11 (9.6%) controls. Again, two of the cases were not genotyped successfully for MTHFR 1298.
Table 1.
Number of ALL cases and controls, adjusted ORs and 95% CIs by MTHFR 677, using 677CC as a reference, and MTHFR1298, using 1298AA as a reference
| Variable | Case | Control | OR* | 95% CI |
|---|---|---|---|---|
| MTHFR 677† | ||||
| CC | 35 (50.7) | 61 (53.5) | 1 | — |
| CT | 29 (42.0) | 39 (34.2) | 0.58 | (0.27, 1.28) |
| TT | 5 (7.2) | 14 (12.3) | 0.23 | (0.06, 0.81) |
| SNA‡ | 2 | 0 | ||
| MTHFR 1298§ | ||||
| AA | 45 (65.2) | 49 (43.0) | 1 | — |
| AC | 23 (33.3) | 54 (47.4) | 0.33 | (0.15, 0.73) |
| CC | 1 (1.5) | 11 (9.6) | 0.07¶ | (0.00, 1.77)¶ |
| SNA‡ | 2 | 0 |
OR was estimated by using conditional logistic regression adjusting for other polymorphism.
MTHFR 677CC as reference.
Sample not available or did not amplify.
MTHFR 1298AA as reference.
OR and 95% CI computed by using exact methods with adjustment for sex, age, and the other polymorphism.
Because each individual has both a MTHFR 677 and 1298 genotype, the associations between disease and the allelic variants of one polymorphism are first reported so that their effects are described independently of the allelic variants of the other polymorphism (Table 1). Irrespective of the allelic variants of MTHFR 1298, the effect of the MTHFR 677CT allele relative to 677CC shows a nonsignificant decrease in the risk of ALL [odds ratio (OR) = 0.58; 95% CI = 0.27–1.28]. However, we observed a significant 4.3-fold decreased risk of ALL among subjects who have the 677TT variant of MTHFR compared with those who are 677CC (OR = 0.23; 95% CI = 0.06–0.81). We also found that individuals with the MTHFR 1298 variant C allele may be protected similarly from ALL. Individuals with the 1298AC genotype had a 3.0-fold decrease in the risk of ALL (OR = 0.33; 95% CI = 0.15–0.73) and those with the 1298CC genotype had a 14-fold reduction in risk of ALL (OR = 0.07; 95% CI = 0.00–1.77) when using 1298AA as a reference.
We next investigated the joint effects of the two polymorphisms as shown in Table 2. Here we found that 677CT/1298AC individuals were at a 5.6-fold decreased risk of developing ALL when using 677CC/1298AA as the reference group (OR = 0.18; 95% CI = 0.04–0.75). Although there was no evidence of statistical interaction between MTHFR 677 and MTHFR 1298 (χ2 = 0.42, P = 0.81), we did observe linkage between the two; namely, all cases and controls with the 677TT genotype had the1298AA genotype, and those with the 1298CC genotype had the 677CC genotype. Previous population studies have reported the rare existence of these mutations in cis and the total absence of the double-homozygous mutants (18, 20, 25). Here we also present evidence of their existence in cis because we found two ALL controls with the 677TT/1298AC-genotype combination. We observed similar trends in our AML cases and controls as described below.
Table 2.
Number of ALL cases and controls, adjusted ORs and 95% CIs by MTHFR 677 and MTHFR 1298, using MTHFR 677CC and MTHFR 1298AA as a reference
| MTHFR 677 | MTHFR 1298 | Case (%) | Control (%) | OR* | 95% CI |
|---|---|---|---|---|---|
| CC | AA | 16 (23.5) | 15 (13.2) | 1 | — |
| CC | AC | 18 (26.5) | 35 (30.7) | 0.39 | (0.14, 1.04) |
| CC | CC | 1 (1.5) | 11 (9.6) | 0.07† | (0.00, 1.77)† |
| CT | AA | 23 (33.8) | 22 (19.3) | 0.68 | (0.26, 1.82) |
| CT | AC | 5 (7.4) | 17 (14.9) | 0.18 | (0.04, 0.75) |
| CT | CC | 0 | 0 | — | — |
| TT | AA | 5 (7.4) | 12 (10.5) | 0.27 | (0.07, 1.07) |
| TT | AC | 0 | 2 (1.8) | 0† | — |
| TT | CC | 0 | 0 | — | — |
| SNA‡ | 3 | 0 | |||
Test for interaction: χ2 = 0.42 (P = 0.81).
OR was estimated by using conditional logistic regression.
OR computed by using exact methods, with adjustment for sex and age.
Sample not available or did not amplify.
Prevalence of the MTHFR 677 and 1298 Genotypes in the AML Cases and Corresponding Controls.
Frequencies of the MTHFR 677 and 1298 polymorphisms in the 237 AML cases and 377 controls are listed in Table 3. For 677CC, 677CT, and 677TT, we observed 134 (60.1%), 66 (29.6%), and 23 (10.3%) AML cases, respectively. Similar proportions were found among their matched controls; that is, 196 (53.1%) were 677CC, 130 (35.2%) were 677CT, and 43 (11.7%) were 677TT. DNA of 14 cases and 8 controls was not amplified or was not available. No significant differences were observed for 677CT or 677TT compared with 677CC, even after adjusting for MTHFR 1298 (677CT: OR = 0.73, 95% CI = 0.48–1.11; 677TT: OR = 0.84, 95% CI = 0.43–1.62). Allele frequencies of 1298AA, 1298AC, and 1298CC were 99 (45.0%), 98 (44.5%), and 23 (10.5%) among cases and 165 (45.6%), 157 (43.4%), and 40 (11.0%) among controls, respectively. DNA for 17 cases and 15 controls was not amplified or was unavailable. Again, there was no evidence of a protective effect of MTHFR 1298 against AML. Adjusting for MTHFR 677 made little difference to the risk estimates (1298AC: OR = 0.99, 95% CI = 0.66–1.50; 1298CC: OR = 0.95, 95% CI = 0.51–1.76). Stratifying by French–American–British subtype led to small numbers, and so the power to detect differences was substantially reduced. In an attempt to increase power, MTHFR 677CT and 677TT and MTHFR 1298AC and 1298CC were combined. No odds ratios were significantly less than 1 (data not shown).
Table 3.
Number of AML cases and controls, adjusted ORs and 95% CIs by MTHFR 677, using 677CC as a reference, and MTHFR 1298, using 1298AA as a reference
| Variable | Case | Control | OR* | 95% CI |
|---|---|---|---|---|
| MTHFR 677† | ||||
| CC | 134 (60.1) | 196 (53.1) | 1 | — |
| CT | 66 (29.6) | 130 (35.2) | 0.73 | (0.48, 1.11) |
| TT | 23 (10.3) | 43 (11.7) | 0.84 | (0.43, 1.62) |
| SNA‡ | 14 | 8 | ||
| MTHFR 1298§ | ||||
| AA | 99 (45.0) | 165 (45.6) | 1 | — |
| AC | 98 (44.5) | 157 (43.4) | 0.99 | (0.66, 1.50) |
| CC | 23 (10.5) | 40 (11.0) | 0.95 | (0.51, 1.76) |
| SNA‡ | 17 | 15 |
OR was estimated by using conditional logistic regression adjusting for other polymorphism.
MTHFR 677CC as reference.
Sample not available or did not amplify.
MTHFR 1298AA as reference.
We similarly found no evidence of protection against AML when looking at the joint effects of the two polymorphisms (Table 4). However, we did detect tight linkage of the mutations because we observed no 677TT/1298CC genotypes and only three each of the 677CT/1298CC and 677TT/1298AC variants. We next calculated the expected frequencies of the MTHFR 677/1298 combinations in our control group by multiplying those observed for 677 and 1298 (Table 4). We found that the frequencies in the AML controls differed markedly from those that would be expected (χ2 = 94.3; P < 0.001); namely, the total frequency of the 677CT/1298CC, 677TT/1298AC, and 677TT/1298CC combinations was a mere 1.4%, whereas they should have comprised 10.3% of the population. The absence of linkage could be quite disadvantageous because suboptimal genotypes such as 677TT/1298CC could lead to total inactivity of the enzyme, which may be incompatible with life.
Table 4.
Number of AML cases and controls, expected MTHFR 677/1298 allele and frequencies, adjusted ORs and 95% CIs by MTHFR 677 and MTHFR 1298 using MTHFR 677CC and MTHFR 1298AA as a reference
| MTHFR 677 | MTHFR 1298 | Cases, n = 211 | Controls n = 356 | OR* | 95% CI | Expected frequency of MTHFR 677/1298 in controls† |
|---|---|---|---|---|---|---|
| CC | AA | 39 (18.5) | 58 (16.3) | 1 | — | (24.2) |
| CC | AC | 66 (31.3) | 95 (26.7) | 0.90 | (0.52, 1.56) | (23.0) |
| CC | CC | 20 (9.5) | 36 (10.1) | 0.82 | (0.41, 1.64) | (5.8) |
| CT | AA | 33 (15.6) | 65 (18.3) | 0.66 | (0.35, 1.23) | (16.1) |
| CT | AC | 27 (12.8) | 60 (16.9) | 0.60 | (0.31, 1.18) | (15.3) |
| CT | CC | 3 (1.4) | 2 (0.6) | 1.95 | (0.30, 12.7) | (3.9) |
| TT | AA | 20 (9.5) | 37 (10.4) | 0.72 | (0.35, 1.47) | (5.3) |
| TT | AC | 3 (1.4) | 3 (0.8) | 2.29 | (0.34, 15.3) | (5.1) |
| TT | CC | 0 | 0 | — | — | (1.3) |
| SNA‡ | 26 | 21 | ||||
Test for interaction: χ2 = 4.25 (P = 0.37).
OR of combined MTHFR 677/1298 genotypes with 677CC/1298AA as a reference was estimated using conditional logistic regression.
Expected MTHFR 677/1298 allele frequencies calculated by multiplying observed MTHFR 677 and 1298 control frequencies.
Sample not available or did not amplify.
Discussion
In the present study, we report an association between susceptibility to adult ALL and polymorphisms in the gene encoding the enzyme MTHFR. Specifically, we found that individuals with at least one MTHFR mutation at 677 (C → T) or 1298 (A → C) were less likely to contract ALL. Statistically significant reductions were demonstrated for those who were either 677TT (OR = 0.23; 95% CI = 0.06–0.81; equivalent to a decreased risk of 4.3-fold with 95% CI = 1.2–16.7) or 1298AC (OR = 0.33; 95% CI = 0.15–0.73; equivalent to a decreased risk of 3.0-fold with 95% CI = 1.4–6.7). We observed that individuals with the 1298CC mutation were most highly protected because only 1 among the 71 cases had this genotype (OR = 0.07). However, because of the relatively small numbers of individuals with the 1298CC genotype (1 case and 11 controls) and the fact that this was a matched case–control study, the 95% CI around this odds ratio exceeded 1. When the matching was ignored, the odds ratio generated was similar (OR = 0.09), and the CI was smaller (95% CI = 0.00–0.60) and highly significant. We also examined joint effects between the two polymorphisms. We found that the double heterozygotes (677CT/1298AC) were approximately five times less likely than 677CC/1298AA individuals to develop ALL. We detected no significant difference in associated risk of AML between the cases and controls for either polymorphism.
Because of the relatively small sample size of ALL cases in the present study, we determined the prevalence of the MTHFR 677 and 1298 polymorphisms of another 44 ALL cases from the United Kingdom that were not part of the case–control study to further validate our findings. For MTHFR 677, we observed that 20 (48%) cases were 677CC, 19 (45%) were 677CT, and 3 (7%) were 677TT. Two samples did not amplify for MTHFR 677. For MTHFR 1298, 27 (61%) cases were 1298AA, 16 (36%) were 1298AC, and only 1 (2%) ALL case had the 1298CC variant. These frequencies were consistent with those found for ALL cases in the present case–control study with low percentages of the 677TT and 1298CC genotypes. Again, we did not observe any individuals with the 677TT/1298CC genotype.
Although it is not clear whether genetic make-up is related to the controls’ participation in the study, the polymorphic allele frequencies are similar to those reported in previous studies (16, 18, 22). Among our controls, the prevalence of the T variant for MTHFR 677 was 29%, lying within the published range of 23–41% for persons of European descent (26–28). Although the MTHFR 1298 polymorphism has not been studied as extensively, our reported frequency of 32% for the C variant among controls is consistent with the previously reported population frequencies of 28% and 33% (17, 18). It is also reassuring that the distributions of the polymorphic alleles within our two control groups are similar, suggesting that the decreased risks observed for ALL are not a consequence of a peculiar polymorphic distribution among that specific control group. To duplicate these findings in highly multiethnic populations such as in the United States, ethnically matched study groups should be formed to avoid disparities because of racial genetic differences.
This study provides further insight into the mechanisms of leukemogenesis and distinctions in folate metabolism between acute myeloid and lymphoid leukemias resulting from the observed protective effects of these two polymorphisms in ALL and not AML. ALL and AML are clinically distinct diseases that arise from disparate cell lineages and differ in response to treatment. Methotrexate, an antifolate, is a common and effective chemotherapeutic agent for ALL but not for AML. Folypolyglutamate synthetase (FPGS) catalyzes the formation of long-chain folate and antifolate polyglutamate derivatives (29), which determines their residency time within cells. It is the enhanced expression of FPGS in lymphoid compared with myeloid cells that contributes to the high efficacy of methotrexate in ALL (30). This increased FPGS activity also is seen in normal lymphoid compared with myeloid progenitor cells (31), the target of malignant transformations in acute leukemia. These differences suggest that lymphoid cells may have higher folate requirements and be more susceptible to folate deficiency and resultant DNA damage than myeloid cells. This may partly explain why the MTHFR 677 and 1298 mutations were found to be protective for ALL and not for AML.
The marked effects of the MTHFR 677TT polymorphism in reducing the specific activity and thermolability of the enzyme and increasing plasma homocysteine levels have been well documented (8, 20, 32). More recent studies have shown the joint effects of the MTHFR 677 and 1298 polymorphisms and their resultant disturbance in folic acid metabolism (17, 18). Although the effects of the 1298AC or 1298CC mutations are not as profound in impairing MTHFR enzyme activity as that of 677CT or 677TT, it has been shown that heterozygotes for both mutations have lower MTHFR enzyme activity (comparable to that seen in the 677TT mutation), elevated homocysteine, and decreased plasma folate levels compared with those heterozygous for one mutation alone (17, 18).
Reduced MTHFR enzyme activity inhibits the methylTHF pathway and leads to increased levels of cytosolic methyleneTHF available for thymidylate, an essential precursor of de novo biosynthesis of DNA (Fig. 1). Uracil is a normal RNA base, but is misincorporated in DNA because of the deficient methylation of uridylate to thymidylate (dUMP to dTMP) involved in DNA synthesis (3). Studies have shown that folate/methyl-deficient diets lead to elevated dUMP/dTMP ratios and DNA double-strand breaks in humans (3), genomewide hypomethylation (33), increased micronucleus frequency in lymphocytes (34), diminished DNA repair capacity (35), and premutagenic apyrimidinic lesions (5). Transient nicks formed during the removal of DNA uracil and subsequent repair may cause DNA strand breaks and alterations that lead to chromosomal damage and possible carcinogenesis.
Our data suggest that the enhanced availability of 5,10-methyleneTHF may play a protective role in the onset of ALL in adults. These findings are consistent with those of Ma et al. (16), who have shown that individuals with the 677TT variant have half the risk of colorectal cancer compared with those with the 677CC genotype. Although impaired MTHFR activity, because of polymorphic variation, reduces the amount of methylTHF available for the methylation of homocysteine to methionine, our findings provide support that uracil misincorporation during DNA synthesis may be more important in the development of adult ALL. However, folate deficiencies in individuals with these MTHFR polymorphisms may negate some of the protective effects against ALL that we have observed. Because we were unable to ascertain the folate status of the populations studied herein, it may be that the presence of the MTHFR 677TT, 1298AC, or 1298CC mutation in individuals with adequate folate levels confers an even greater level of protection against ALL than presently reported.
In conclusion, our data suggest that folic acid metabolism plays an important role in adult ALL and that the MTHFR gene is central in this. We currently are investigating the effect of the MTHFR polymorphism in childhood ALL, which is the most common form of leukemia in children. Adult ALL is a relatively rare disease, making it difficult to perform a large epidemiological study, but studies in other ALL populations should be undertaken to confirm the findings presented here.
Acknowledgments
We are grateful to Dr. Yafei Liu for his contribution to the setup of the PCR-based MTHFR 677 assay and to Professors Barry Shane, Bruce Ames, and Patricia Buffler for their critical reading of the manuscript. This research was supported by the National Foundation for Cancer Research and Center Grant P30ES01896 from the National Institute of Environmental Health Sciences (to M.T.S.) and by the British Leukemia Research Fund (to G.M. and R.C.).
Abbreviations
- methyleneTHF
5,10-methylenetetrahydrofolate
- methylTHF
5-methyltetrahydrofolate
- MTHFR
methylenetetrahydrofolate reductase
- ALL
acute lymphocytic leukemia
- AML
acute myeloid leukemia
- OR
odds ratio
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
See commentary on page 12216.
References
- 1.Greaves M F. Lancet. 1997;349:344–349. doi: 10.1016/s0140-6736(96)09412-3. [DOI] [PubMed] [Google Scholar]
- 2.Smith M T, Zhang L. Environ Health Perspect. 1998;106, Suppl. 4:937–946. doi: 10.1289/ehp.98106s4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blount B C, Mack M M, Wehr C M, MacGregor J T, Hiatt R A, Wang G, Wickramasinghe S N, Everson R B, Ames B N. Proc Natl Acad Sci USA. 1997;94:3290–3295. doi: 10.1073/pnas.94.7.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim Y I, Pogribny I P, Basnakian A G, Miller J W, Selhub J, James S J, Mason J B. Am J Clin Nutr. 1997;65:46–52. doi: 10.1093/ajcn/65.1.46. [DOI] [PubMed] [Google Scholar]
- 5.Pogribny I P, Muskhelishvili L, Miller B J, James S J. Carcinogenesis. 1997;18:2071–2076. doi: 10.1093/carcin/18.11.2071. [DOI] [PubMed] [Google Scholar]
- 6.Trentin G A, Moody J, Heddle J A. Mutat Res. 1998;405:81–87. doi: 10.1016/s0027-5107(98)00147-x. [DOI] [PubMed] [Google Scholar]
- 7.Shane B. In: Folic Acid Metabolism in Health and Disease. Picciano M F, Stokstad E L R, Gregory J F, editors. New York: Wiley; 1990. pp. 65–78. [Google Scholar]
- 8.Frosst P, Blom H J, Milos R, Goyette P, Sheppard C A, Matthews R G, Boers G J, den Heijer M, Kluijtmans L A, van den Heuvel L P, et al. Nat Genet. 1995;10:111–113. doi: 10.1038/ng0595-111. [DOI] [PubMed] [Google Scholar]
- 9.de Franchis R, Sebastio G, Mandato C, Andria G, Mastroiacovo P. Lancet. 1995;346:1703. doi: 10.1016/s0140-6736(95)92865-0. [DOI] [PubMed] [Google Scholar]
- 10.Nishio H, Lee M J, Fujii M, Kario K, Kayaba K, Shimada K, Matsuo M, Sumino K. Jpn J Hum Genet. 1996;41:247–251. doi: 10.1007/BF01875985. [DOI] [PubMed] [Google Scholar]
- 11.Engbersen A M, Franken D G, Boers G H, Stevens E M, Trijbels F J, Blom H J. Am J Hum Genet. 1995;56:142–150. [PMC free article] [PubMed] [Google Scholar]
- 12.Goyette P, Frosst P, Rosenblatt D S, Rozen R. Am J Hum Genet. 1995;56:1052–1059. [PMC free article] [PubMed] [Google Scholar]
- 13.Harmon D L, Woodside J V, Yarnell J W, McMaster D, Young I S, McCrum E E, Gey K F, Whitehead A S, Evans A E. Q J Med. 1996;89:571–577. doi: 10.1093/qjmed/89.8.571. [DOI] [PubMed] [Google Scholar]
- 14.Dianov G L, Timchenko T V, Sinitsina O I, Kuzminov A V, Medvedev O A, Salganik R I. Mol Gen Genet. 1991;225:448–452. doi: 10.1007/BF00261686. [DOI] [PubMed] [Google Scholar]
- 15.Chen J, Giovannucci E, Kelsey K, Rimm E B, Stampfer M J, Colditz G A, Spiegelman D, Willett W C, Hunter D J. Cancer Res. 1996;56:4862–4864. [PubMed] [Google Scholar]
- 16.Ma J, Stampfer M J, Giovannucci E, Artigas C, Hunter D J, Fuchs C, Willett W C, Selhub J, Hennekens C H, Rozen R. Cancer Res. 1997;57:1098–1102. [PubMed] [Google Scholar]
- 17.van der Put N M, Gabreëls F, Stevens E M, Smeitink J A, Trijbels F J, Eskes T K, van den Heuvel L P, Blom H J. Am J Hum Genet. 1998;62:1044–1051. doi: 10.1086/301825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weisberg I, Tran P, Christensen B, Sibani S, Rozen R. Mol Genet Metab. 1998;64:169–172. doi: 10.1006/mgme.1998.2714. [DOI] [PubMed] [Google Scholar]
- 19.Kane, E. V., Roman, E., Carwright, R. A., Parker, J. & Morgan, G. (1999) Br. J. Cancer, in press. [DOI] [PMC free article] [PubMed]
- 20.van der Put N M, Steegers-Theunissen R P, Frosst P, Trijbels F J, Eskes T K, van den Heuvel L P, Mariman E C, den Heyer M, Rozen R, Blom H J. Lancet. 1995;346:1070–1071. doi: 10.1016/s0140-6736(95)91743-8. [DOI] [PubMed] [Google Scholar]
- 21.Ou C Y, Stevenson R E, Brown V K, Schwartz C E, Allen W P, Khoury M J, Rozen R, Oakley G P, Jr, Adams M J., Jr Am J Med Genet. 1996;63:610–614. doi: 10.1002/(SICI)1096-8628(19960628)63:4<610::AID-AJMG15>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 22.Whitehead A S, Gallagher P, Mills J L, Kirke P N, Burke H, Molloy A M, Weir D G, Shields D C, Scott J M. Q J Med. 1995;88:763–766. [PubMed] [Google Scholar]
- 23.Breslow N E, Day N E. Statistical Methods in Cancer Research: The Analysis of Case-Control Studies. Vol. 1. Lyon, France: International Agency for Research on Cancer; 1980. pp. 162–186. [PubMed] [Google Scholar]
- 24.stata (1997) (Stata Corporation, College Station, TX).
- 25.Donnelly J C, Isotalo P A. Clin Chem. 1999;45:A39. [PubMed] [Google Scholar]
- 26.Gudnason V, Stansbie D, Scott J, Bowron A, Nicaud V, Humphries S. Atherosclerosis. 1998;136:347–354. doi: 10.1016/s0021-9150(97)00237-2. [DOI] [PubMed] [Google Scholar]
- 27.Bowen D J, Bowley S, John M, Collins P W. Thromb Haemostasis. 1998;79:949–954. [PubMed] [Google Scholar]
- 28.Franco R F, Araújo A G, Guerreiro J F, Elion J, Zago M A. Thromb Haemostasis. 1998;79:119–121. [PubMed] [Google Scholar]
- 29.Shane B. Vitam Horm. 1989;45:263–335. doi: 10.1016/s0083-6729(08)60397-0. [DOI] [PubMed] [Google Scholar]
- 30.Longo G S, Gorlick R, Tong W P, Ercikan E, Bertino J R. Blood. 1997;90:1241–1245. [PubMed] [Google Scholar]
- 31.Barredo J C, Synold T W, Laver J, Relling M V, Pui C H, Priest D G, Evans W E. Blood. 1994;84:564–569. [PubMed] [Google Scholar]
- 32.Goyette P, Christensen B, Rosenblatt D S, Rozen R. Am J Hum Genet. 1996;59:1268–1275. [PMC free article] [PubMed] [Google Scholar]
- 33.Jacob R A, Gretz D M, Taylor P C, James S J, Pogribny I P, Miller B J, Henning S M, Swendseid M E. J Nutr. 1998;128:1204–1212. doi: 10.1093/jn/128.7.1204. [DOI] [PubMed] [Google Scholar]
- 34.Fenech M F, Dreosti I E, Rinaldi J R. Carcinogenesis. 1997;18:1329–1336. doi: 10.1093/carcin/18.7.1329. [DOI] [PubMed] [Google Scholar]
- 35.Choi S W, Mason J B. Am J Gastroenterol. 1998;93:2013–2016. doi: 10.1111/j.1572-0241.1998.02013.x. [DOI] [PubMed] [Google Scholar]