Abstract
The inv(16) is one of the most frequent chromosomal translocations associated with acute myeloid leukemia (AML). The inv(16) fusion protein acts by dominantly interfering with AML-1/core binding factor β-dependent transcriptional regulation. Here we demonstrate that the inv(16) fusion protein cooperates with AML-1B to repress transcription. This cooperativity requires the ability of the translocation fusion protein to bind to AML-1B. Mutational analysis and cell fractionation experiments indicated that the inv(16) fusion protein acts in the nucleus and that repression occurs when the complex is bound to DNA. We also found that the inv(16) fusion protein binds to AML-1B when it is associated with the mSin3A corepressor. An AML-1B mutant that fails to bind mSin3A was impaired in cooperative repression, suggesting that the inv(16) fusion protein acts through mSin3 and possibly other corepressors. Finally, we demonstrate that the C-terminal portion of the inv(16) fusion protein contains a repression domain, suggesting a molecular mechanism for AML-1-mediated repression.
The inv(16) is one of the most frequent translocations in acute myeloid leukemia (AML) (1). It fuses most of core binding factor β (CBFβ/CBFB/PEBP2B) to the C terminus of a smooth muscle myosin heavy chain (SMMHC), MYH11 (2). CBFβ is a transcription factor that does not bind DNA directly but interacts with the AML-1 DNA-binding transcription factor to increase its ability to bind DNA and regulate transcription (3, 4). AML-1 is one of the most frequently mutated genes in human leukemia (5). It is disrupted by the t(8;21), t(3;21), and t(16;21) in AML and by the t(12;21) in childhood B cell acute lymphocytic leukemia (ALL). By disrupting CBFβ, the inv(16) also disrupts AML-1 functions (6). Together, these chromosomal translocations account for nearly one-quarter of all AML cases and one-fifth of all childhood B cell ALL-containing discernible chromosomal abnormalities (7, 8).
The largest form of AML1, termed AML-1B(9), activates transcription of numerous tissue-specific genes, including genes encoding cytokines and cytokine receptors, and differentiation-specific genes such as T cell receptors, neutrophil peptide-3, and myeloperoxidase (10). CBFβ cooperates with AML-1 to activate transcription (11). Targeted deletion of the AML1 (Cbfa2/Pebp2aB2) or CBFβ genes led to identical phenotypes: embryonic lethality at day 12.5–13.5 post coitus with a complete lack of fetal liver hematopoiesis (12–15). Thus, CBFβ is required for AML-1 function.
The targeting of AML-1 by multiple chromosomal translocations in acute leukemia suggests a convergent mechanism of leukemogenesis. The t(8;21) fusion protein creates a transcriptional repressor protein by fusing the AML-1 DNA-binding domain to ETO (MTG8), a corepressor that recruits histone deacetylases to inhibit transcription (5). Likewise, the t(16;21) fuses the AML-1 DNA-binding domain to an ETO family member (16). The t(3;21) fuses the AML-1 DNA-binding domain to the Evi I transcriptional repressor (17). Alternatively, the t(12;21) fuses a repressor domain from the Translocation-Ets-Leukemia (TEL) gene to the N terminus of AML-1 to create a constitutive repressor (18–20). These proteins all repress AML-1-regulated promoters. The inv(16) fusion protein also acts as a dominant inhibitor of AML-1-dependent transactivation (21, 22). Moreover, expression of the inv(16) fusion protein during murine development resulted in embryonic lethality at day 12.5–13.5 with a phenotype similar to loss of AML-1 or CBFβ (6). The inv(16) fusion protein colocalizes with AML-1 in the nucleus and the cytoplasm, leading to the model that it blocks AML-1 function by sequestering it in inactive complexes (22, 23).
Although AML-1/CBFβ cooperates with other transcription factors to activate transcription, AML-1B is also capable of repressing transcription (11, 34). AML-1 interacts with the Groucho corepressor in yeast two-hybrid assays (24, 25) and with the mSin3 corepressors in yeast two-hybrid assays and in mammalian cells (B.L. et al., unpublished results). In this report, we demonstrate that the inv(16) fusion protein cooperates with AML-1B to repress transcription. The inv(16) fusion protein also forms a trimeric complex with AML-1B and the mSin3A corepressor. In addition, we demonstrate that CBFβ/SMMHC encodes a cryptic repression domain. Thus, we propose a mechanism of action for the inv(16) whereby it traps AML-1 in a repressive complex with mSin3A and, potentially, other corepressors.
Materials and Methods
Cell Culture.
Cos-7 cells were maintained in DMEM (BioWhittaker) containing 10% FBS, 50 units/ml penicillin, 50 μg/ml streptomycin, and 2 mM l-glutamine (BioWhittaker). NIH 3T3 cells were maintained in DMEM containing the same antibiotics, but with 10% bovine serum.
Coimmunoprecipitation, Immunoblotting, and Cell Fractionation.
Cos-7 cells (3 × 106 cells in 100-mm dishes) were transfected by using lipofectamine (BRL). For coexpression experiments, 3 μg pCMV5-AML-1B and 3 μg pCMV5-inv(16) or pCMV5-CBFβ were cotransfected. Cells were extracted with PBS supplemented with 1 mM EDTA, 1.5 mg/ml iodoacetamide, 0.2 mM PMSF, and 0.1 trypsin inhibitory units per ml aprotinin and containing 0.5% Triton X-100 unless otherwise noted. For immunoprecipitations, cells were sonicated in extraction buffer. These lysates were incubated with 100 μl of formalin-fixed Staphylococcus aureus (Pansorbin; Calbiochem) for 30 min to remove proteins nonspecifically binding to protein A. After centrifugation for 5 min at 4°C, the supernatants were collected and incubated for 1 hr with affinity-purified primary antibody [K-20 anti-mSin3A (Santa Cruz Biotechnology) or anti-AML-1 N-terminal antipeptide antibody (Calbiochem)]. Fifteen microliters of a 50% slurry of protein A-Sepharose (Amersham-Pharmacia) then was added for 30 min to collect the immune complexes, and these complexes were washed three times at 4°C with lysis buffer. For immunoblot analysis, 100 μg of protein (quantitated with the Bio-Rad DC protein assay) or immune complexes were boiled in Laemmli buffer for 2 min, fractionated by SDS/PAGE, and transferred to nitrocellulose. These membranes were blocked for 1 hr with 5% nonfat dry milk dissolved in PBS and incubated with the indicated primary antibody overnight at 4°C. Proteins were visualized by enhanced chemiluminescence (Pierce).
NIH 3T3 cells were transfected by using the Superfect reagent (Qiagen) with 5 μg of the indicated plasmids. Cell pellets were resuspended in Iso-Hi buffer [10 mM Tris⋅HCl, pH 8.4/140 mM NaCl/1.5 mM MgCl2/Complete protease inhibitor (Roche)/1 mM EDTA] containing 0.5% Nonidet P-40. After 5 min on ice, the nuclei were collected by centrifugation at 2,000 rpm for 5 min. Nuclei were reextracted to ensure purity. Equal proportions of the supernatant (cytoplasmic) and nuclear fractions were analyzed by immunoblot analysis with myosin A, mSin3A, CBFβ, and AML-1 antisera.
Transcription Assays and Plasmids.
NIH 3T3 cells (35-mm dishes) were transfected by using the Superfect reagent (Qiagen) with 300 ng WWP-Luciferase (2.4 kb of the p21Waf1/Cip1 promoter) (26), 300 ng pCMV5-AML-1B (9) or deletion constructs (B.L. et al., unpublished results; ref. 27), 10–10,000 ng of pCMV5-inv(16) or pCMV5-CBFβ (3), and 200 ng of pCMV5-secreted alkaline phosphatase (SEAP) plasmids. Firefly luciferase activity was measured by using the Luciferase Assay Reagent (Promega) and normalized to SEAP activity (B.L. et al., unpublished results). The GAL4-inv(16) chimeric genes were created by subcloning the inv(16) C-terminal deletion mutants (23) (a gift from P. Paul Liu, National Institutes of Health, Bethesda, MD) in-frame with the GAL4 DNA-binding domain (residues 1–147) in the pCMV5-M1 vector (34). The AML-1B RD1,2,3 mutant was created by truncating RD1,2 (B.L. et al., unpublished results) with BssHII.
Results
The Inv(16) Protein Cooperates with AML-1B to Repress Transcription.
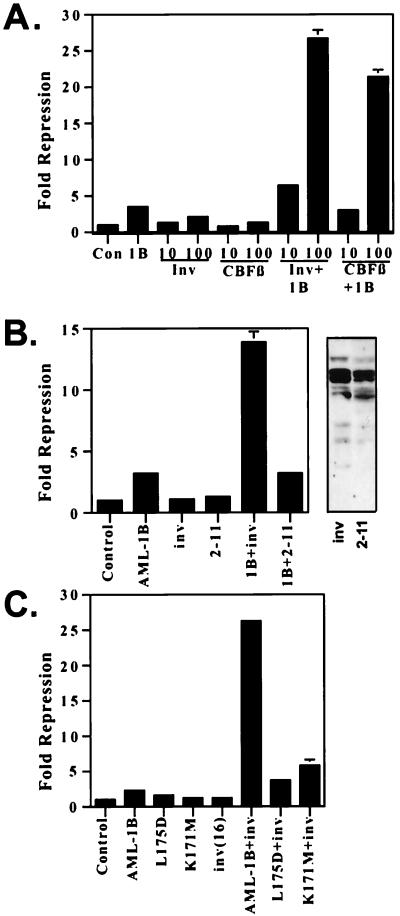
The inv(16) protein dominantly inhibits AML-1/CBFβ-dependent transactivation (21, 22). However, the effect of the inv(16) fusion protein on AML-1B-dependent repression has not been tested. We have demonstrated previously that AML-1B physically interacts with the mSin3 corepressors to repress transcription of the p21Waf1/Cip1 promoter (B.L., J. Westendorf, B. Linggi, E. Seto, and S.H., unpublished results). Therefore, we tested the inv(16) fusion protein’s ability to affect AML-1B-mediated repression. To measure potential effects of the inv(16) fusion protein, we selected a level of AML-1B that yielded 2- to 5-fold repression (Fig. 1A). On its own, the inv(16) fusion protein had little effect on transcription from the p21Waf1/Cip1 promoter (Fig. 1A). However, with as little as 10 ng of inv(16) plasmid there was an enhancement of AML-1B-mediated repression, and at 100 ng of input plasmid the inv(16) fusion protein synergized with AML-1B to repress the p21Waf1/Cip1 promoter (Fig. 1A). Importantly, the internal control promoter was not affected by either AML-1B or the inv(16) (data not shown). Because CBFβ stimulates AML-1’s ability to bind DNA and activate transcription, we tested the ability of wild-type CBFβ to cooperate with AML-1B in this assay. CBFβ did not repress on its own nor did it cooperate with AML-1B at low levels of plasmid. However, at 100 ng of input plasmid, CBFβ also cooperated with AML-1B to repress transcription (Fig. 1A). Thus, although CBFβ can cooperate with AML-1B to both activate (11) and repress (Fig. 1A) transcription, the inv(16) fusion protein instead blocks AML-1-dependent activation and it cooperates with AML-1B to repress transcription (Fig. 1A).
Figure 1.
The inv(16) fusion protein cooperates with AML-1B to repress transcription. (A) CBFβ and CBFβ/SMMHC cooperate with AML-1 to repress. Increasing amounts of the inv(16)-expressing plasmid or CBFβ-expressing plasmid were cotransfected with 300 ng of p21(WWP)-firefly luciferase reporter plasmid in the presence or absence of 300 ng of AML-1B-expressing plasmid. (B) Comparison of the ability of wild-type inv(16) fusion protein and the 2–11 deletion mutant. The inv(16) or inv(16) 2–11 plasmid (150 ng) was cotransfected with 300 ng of AML-1B and 300 ng of p21-reporter plasmid. (Right) Immunoblot analysis of the inv(16) and 2–11 deletion mutant by using anti-CBFβ serum. Note that the inv(16) lane is the same as in Fig. 2. (C) AML-1B DNA-binding activity is required for AML-1B/inv(16) cooperative repression. An inv(16)-expressing plasmid was cotransfected with 300 ng of p21(WWP)-firefly luciferase reporter plasmid and 300 ng of plasmids expressing AML-1B or the AML-1B mutant proteins. Fold repression was calculated after correcting for transfection efficiency by using a plasmid expressing a secreted form of alkaline phosphatase (SEAP). The levels shown are the average of duplicate experiments. In those samples lacking error bars, the error was too small to graph. Control, empty expression vector.
To confirm the specificity of the role of inv(16) in repression, we used a mutant of the inv(16), which lacks residues 2–11 and which fails to bind AML-1 (23). Detection of the inv(16) fusion protein and this mutant was difficult to confirm at the low levels of input plasmid used in these experiments. However, previous work has established that the 2–11 mutant is stably expressed (23), and at higher levels we confirmed this result (Fig. 1B). Titration experiments indicated that at 30 ng of input, inv(16)-expressing plasmid repression reached maximal levels (data not shown). Therefore, to judge the effectiveness of the 2–11 deletion protein, we used levels of input plasmid five times higher than that needed for maximal repression by wild-type inv(16)-expressing plasmid. Even at 150 ng of input plasmid, this mutant failed to cooperate with AML-1B (Fig. 1B).
AML-1B DNA Binding Is Required for Cooperation with the Inv(16) Fusion Protein.
Random mutagenesis identified two point mutations in AML-1B that affect DNA binding and association with CBFβ (28). To confirm that the cooperative repression observed between AML-1B and the inv(16) fusion protein occurred at the level of DNA binding, we used the AML-1B L175D mutant (leucine 175 changed to aspartic acid) that fails to bind DNA and CBFβ and the K171M (lysine 171 changed to methionine) mutant that weakly binds DNA, but binds CBFβ at wild-type levels (K171M) (28). Both of these mutants failed to repress the p21Waf1/Cip1 promoter on their own (Fig. 1C). The L175D mutant also failed to cooperate with either CBFβ or the inv(16) protein to repress transcription. The K171M mutant was impaired in its ability to cooperate with the inv(16) fusion protein, but retained some activity (Fig. 1C). This is probably because the K171M mutant retains approximately 10% of wild-type DNA-binding function (28). Therefore, AML-1B DNA binding is required for AML-1/inv(16) cooperative repression.
The Inv(16) Fusion Protein Is Found in Both the Cytoplasm and the Nucleus.
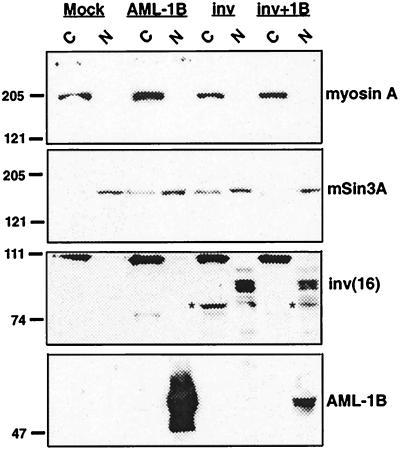
Direct and indirect fluorescence experiments have indicated that the inv(16) fusion protein is either cytoplasmic, cytoplasmic and nuclear, or nuclear (22, 23, 29). However, in inv(16)-containing leukemic blasts, as determined by both cell fractionation and by immunofluorescence assays, the protein is primarily nuclear (29). Moreover, our transcriptional analysis implies that the inv(16) fusion protein acts in the nucleus, at the promoter. To determine the cellular localization of the inv(16) fusion protein in our cells, we fractionated NIH 3T3 cells expressing AML-1B, the inv(16) fusion protein, or both of these proteins into cytoplasmic and nuclear fractions to assess the relative amounts of these proteins in both cellular compartments. To extend these results, we used a detergent extraction and isotonic buffer to avoid possible multimerization of CBFβ/SMMHC. As a control, we used the cytoplasmic protein nonmuscle myosin A, and it indeed was found in the cytoplasmic fractions (Fig. 2, top image). As a nuclear marker, we used mSin3A and found that it was mostly nuclear under these conditions, although some mSin3A did leak into the cytoplasmic fractions (Fig. 2, second image). When expressed alone, the inv(16) fusion protein was found in both cytoplasmic and nuclear fractions, although the predominant form found in the cytoplasm migrated faster (denoted by the asterisk) than the CBFβ/SMMHC found in the nuclear factions (Fig. 2, third image). The identity of these bands as inv(16) fusion proteins was confirmed by using two mAbs that recognize CBFβ (ref. 13 and data not shown; note that CBFβ is an approximately 25-kDa protein and was electrophoresed off this gel). The prominent approximately 110-kDa protein in the cytoplasmic fractions is not related to CBFβ/SMMHC as it is also in the untransfected sample and, thus, serves as a further control for the fractionation. Finally, AML-1B is a component of the nuclear matrix (27), and, even under the stringent conditions used here where mSin3A was partially extracted, it was found in the nuclear fractions (Fig. 2 bottom image). When AML-1B was coexpressed with the inv(16) fusion protein, AML-1B and CBFβ/SMMHC were mostly nuclear. Upon longer exposure CBFβ/SMMHC also was found in cytoplasmic fractions, consistent with previous results (22, 23). Thus, the inv(16) fusion protein likely stimulates AML-1’s repressive functions in the nucleus.
Figure 2.
The inv(16) fusion protein is nuclear and cytoplasmic. NIH 3T3 cells were transfected with pCVM5-AML-1B, pCMV5-inv(16), or pCVM5-inv(16) + pCMV5-AML-1B. Cells were disrupted by using isotonic buffer containing 0.5% Nonidet P-40 on ice for 5′ and nuclei were collected by low-speed centrifugation as described (27). Equal proportions of each fraction were analyzed by immunoblot with anti-myosin A (top), anti-mSin3A (second), anti-CBFβ (third), and anti-AML-1 (bottom). Asterisk denotes a faster-migrating form of CBFβ/SMMHC.
The Inv(16) Associates with AML-1B in a Complex with the mSin3A Corepressor.
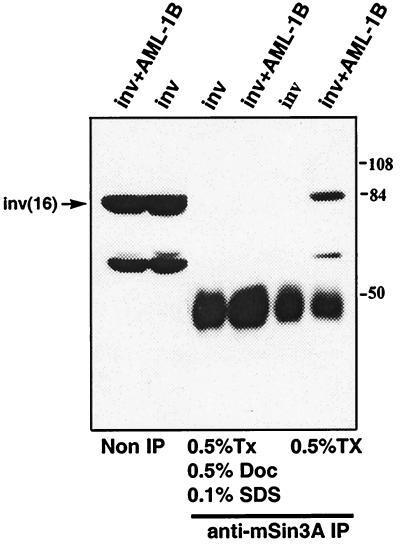
AML-1B repressed the p21Waf1/Cip1 promoter, in part, through association with the mSin3A corepressor (B.L., J. Westendorf, B. Linggi, E. Seto, and S.H., unpublished results). Therefore, we asked whether the inv(16) fusion protein could associate with mSin3A directly or could interact with the AML-1B/mSin3A complex. The inv(16) fusion protein was expressed in Cos-7 cells in the absence or the presence of AML-1B. Cell lysates were immunoprecipitated by using anti-mSin3A IgG and the inv(16) fusion protein detected by immunoblot analysis by using anti-CBFβ IgG (Fig. 3). Under these conditions, the inv(16) fusion protein did not associate with mSin3A directly. However, in the absence of SDS, the fusion protein was present in mSin3A immune complexes when AML-1B was coexpressed (Fig. 3). Thus, we infer that the inv(16) fusion protein forms a ternary complex with AML-1B and mSin3A.
Figure 3.
The inv(16) fusion protein forms a trimeric complex with AML-1B and mSin3A. Cos-7 cells were transfected with pCVM5-AML-1B, pCMV5-inv(16), or pCVM5-inv(16) + pCMV5-AML-1B. Cell lysates were prepared with the indicated detergents and analyzed by immunoblot either before or after immunoprecipitation with anti-mSin3A IgG. The mobility of molecular weight markers are shown at the right. Tx, Triton X-100; Doc, sodium deoxycholate.
Definition of AML-1B Domains Required for Inv(16) Cooperation.
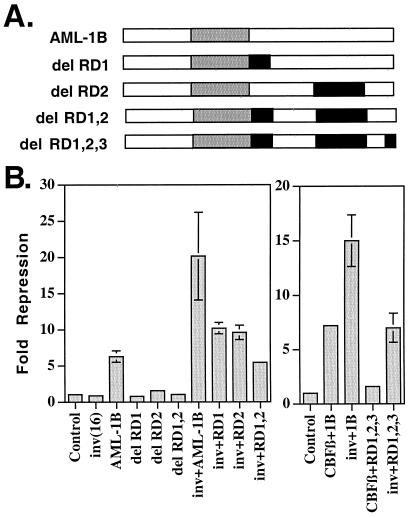
The mSin3A interaction domain is necessary, but not sufficient, for AML-1B-mediated repression of the p21 promoter (34). To define the regions of AML-1B that are required for cooperation with the inv(16) fusion protein, we used deletion mutants of AML-1B (Fig. 4A) that remove the mSin3A interaction domain (residues 208–237, RD1) and a second repression domain (RD2, residues 290–432) (B.L., J. Westendorf, B. Linggi, E. Seto, and S.H., unpublished results). Deletion of the mSin3A interaction domain reduced, but did not eliminate, the cooperative repression observed upon coexpression with CBFβ/SMMHC (Fig. 4B). Deletion of residues 290–432, which ablated the function of AML-1B in repressing the p21Waf1/Cip-1 promoter, had only a 2-fold effect on AML-1/inv(16)-cooperative repression (Fig. 4B). When the deletion of the mSin3A-binding domain was coupled with the 290–432 mutation, cooperative repression was impaired further; however, even this double mutant was still capable of cooperating with the inv(16) protein to some degree (Fig. 4B). Therefore, we coupled a deletion of the Groucho-binding motif (RD3) with the deletion of RD1 and RD2 and directly compared the ability of CBFβ and CBFβ/SMMHC to repress transcription (Fig. 4B Right). CBFβ stimulated AML-1B-dependent repression, but had no effect in the context of the triple mutant. By contrast, the inv(16) protein retained the ability to act through this AML-1B mutant. The ability of the inv(16) protein to repress transcription by interacting with this mutant AML-1 suggests that simply recruiting the fusion protein to a promoter is sufficient for repression.
Figure 4.
Definition of AML-1B C-terminal domains required for cooperation with the inv(16) fusion protein. (A) Schematic representation of AML-1B and the C-terminal deletion mutants used. RD, repression domain. (B) Transcriptional analysis of the AML-1B mutants alone or when coexpressed with the Inv(16) fusion protein or CBFβ. AML-1B (300 ng) was cotransfected with 300 ng of p21-reporter plasmid in the absence or presence of 150 ng of the inv(16) or CBFβ plasmids. Fold repression was calculated after correcting for transfection efficiency by using a plasmid expressing a secreted form of alkaline phosphatase (SEAP). Control, empty expression vector.
The Inv(16) Fusion Protein Contains a Repression Domain.
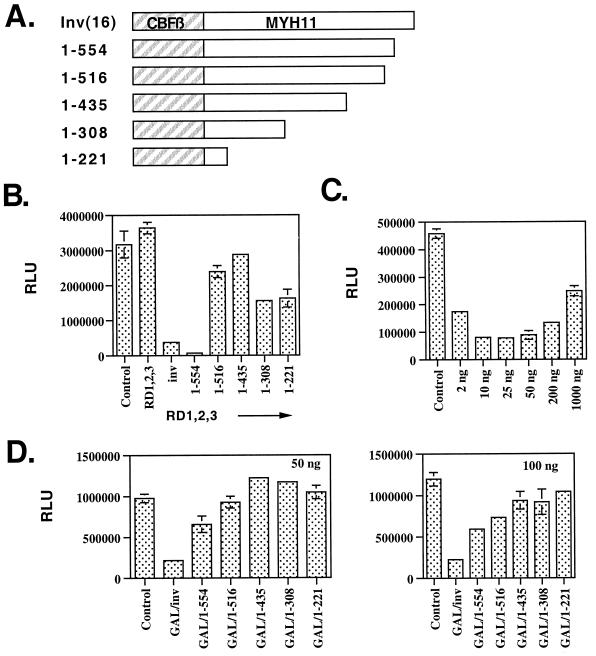
Given that the inv(16) fusion protein, but not CBFβ, acted through the AML-1B RD1,2,3 mutant, we tested whether the SMMHC sequences of the inv(16) protein contain a repression domain. A series of C-terminal inv(16) deletion mutants was used for this analysis (Fig. 5A). Once again, wild-type CBFβ/SMMHC cooperated with the AML-1B mutant to repress transcription approximately 6-fold (Fig. 5B; note that in this figure we show the corrected relative light units). Deletion of the C-terminal 57 aa did not impair CBFβ/SMMHC function; rather, this mutant was consistently a better repressor in this assay (1–554, Fig. 5B). However, deletion of as little as 95 aa from the C terminus (1–516, Fig. 5B) ablated CBFβ/SMMHC’s effects.
Figure 5.
The inv(16) encodes a repression domain. (A) Schematic diagram of the inv(16) mutants. (B) Transcriptional analysis of the mutants shown in A in conjunction with the AML-1B RD1,2,3 mutant. (C) Transcriptional repression by a GAL4-inv(16) chimeric protein. Increasing amounts of GAL4-inv(16) were transfected with 500 ng of GAL4-TK-Luc plasmid. (D) Mapping the CBFβ/SMMHC repression domain. GAL4 fusion proteins containing the deletion mutants shown in A were assayed at 50 ng (Left) or 100 ng (Right) by using the GAL4-responsive promoter. RLU, relative light units. Control, empty expression vector.
To further test whether tethering the inv(16) fusion protein to a promoter is sufficient for repression, we fused the wild-type and C-terminally truncated CBFβ/SMMHC proteins to the GAL4 DNA-binding domain. The GAL4 DNA-binding domain did not significantly affect the GAL4 reporter gene (data not shown). However, at only 2 ng of input plasmid, the GAL-4/inv(16) fusion protein was able to repress transcription more than 2.5-fold from a reporter plasmid containing four GAL4 DNA-binding sites upstream of the thymidine kinase promoter (Fig. 5C). At 10 ng of input plasmid, maximal levels of more than 5-fold repression were obtained. At higher levels of effector plasmid, repression diminished, perhaps because of “squelching.” Cell fractionation experiments confirmed that the GAL4/inv(16) chimeric protein was nuclear (data not shown). The expression of the C-terminal inv(16) deletion mutants was confirmed in Cos7 cells (data not shown), but at the levels of plasmid used in these transcription assays, we were unable to confirm expression of the GAL-4-inv(16) fusion mutants. Therefore, we tested the activity of the C-terminal truncation mutants at 50 and 100 ng of plasmid, which is 5- to 10-fold more plasmid than required for the wild-type plasmid to repress transcription. In this assay, truncation of as little as 57 aa from the C terminus impaired repression, and deletion of 95 aa nearly ablated the repression (Fig. 5D). Thus, in two different contexts, the MYH11 portion of the fusion protein appears to encode a cryptic transcriptional repression domain.
Discussion
The inv(16) fusion protein dominantly interferes with AML-1-dependent transactivation of transcription (22). We have demonstrated that CBFβ/SMMHC stimulates the ability of AML-1B to repress transcription (Figs. 1, 4, and 5). Repression was not limited to the p21 promoter because we also observed repression when using the Rous sarcoma virus long terminal repeat (data not shown). Cooperative repression required an intact AML-1 DNA-binding domain, implying that it occurs at the promoter. Moreover, we showed that the inv(16) fusion protein associates with AML-1 in a complex with the mSin3A corepressor (Fig. 3). CBFβ/SMMHC, but not CBFβ, repressed transcription through an AML-1B mutant lacking three repression domains and when linked to the GAL4 DNA-binding domain. We conclude that the inv(16) fusion protein functions as an AML-1 corepressor.
Our cell fractionation studies (Fig. 2) indicated that the inv(16) fusion protein is both cytoplasmic and nuclear, which is consistent with other overexpression studies (22, 23). However, we found that the majority of the protein is nuclear, which is more consistent with previous studies of leukemic blasts containing the inv(16) (29). Although our results do not address a possible role for the fusion protein in the cytoplasm, our work does define a nuclear function for CBFβ/SMMHC. CBFβ stimulates AML-1-dependent activation (22) and repression (Figs. 1, 4, and 5). However, the inv(16) fusion protein only represses transcription [that is, it blocks AML-1-dependent activation (22) or cooperates with AML-1 to repress]. The use of AML-1B mutants lacking the repression domains and the C-terminal truncations of the inv(16) fusion protein uncovered a repression function unique to the inv(16). This observation may explain why the inv(16) protein only stimulates AML-1’s repressive functions. We hypothesize that the fusion of the cryptic repression domain in MYH11 to CBFβ converts CBFβ from a protein that can cooperate with AML-1 to both activate (22) and repress transcription (Fig. 1) to one that can only repress transcription. Thus, it is possible that the inv(16) fusion protein traps AML-1 in a complex with corepressors to constitutively repress AML-1-regulated genes.
Expression of the inv(16) fusion protein during embryogenesis produced the same phenotype as AML-1 deficiency (6). Coupled with the ability of the inv(16) fusion protein to block AML-1-dependent transactivation, these observations lead to the hypothesis that the inv(16) creates a dominant negative protein to block AML-1’s functions. However, our work suggests that the inv(16) creates a dominant transcriptional repressor, thus blocking only the transactivation function of AML-1. Thus, we speculate that the developmental phenotype of AML-1 deficiency is due to the lack of AML-1-dependent transactivation. AML-1 is expressed early in the developing embryo, but the mice lacking AML-1 develop normally until embryonic day 12.5–13.5. It is possible that, like its Drosophila homologue Runt (25), AML-1 is a repressor early in development and acts as a molecular switch to turn genes on at specific times in development. Our results would suggest that, when expressed during development, the inv(16) fusion protein traps AML-1 in the repressive state and inhibits the expression of AML-1 target genes, which leads to embryonic lethality. This is consistent with the observation that the t(8;21) also creates a dominant repressor of AML-1 target genes (5) and expression of the t(8;21) fusion protein during development leads to a phenotype similar to that observed with embryonic expression of CBFβ/SMMHC (30, 31). Therefore, our proposed model of the inv(16) fusion protein acting as an active repressor explains the functional data to date: the inv(16) protein blocks AML-1-dependent activation by adding a repression domain to CBFβ, it yields a phenotype in mice similar to AML-1/ETO (a repressor), and it cooperates with AML-1 to repress transcription.
AML-1 is the target of multiple translocations in acute leukemia. The t(8;21) and the t(12;21) fusion proteins interact with mSin3A and other corepressors to form AML-1-dominant repressor proteins (5, 34). Our current work indicates that the inv(16) also creates a dominant repressor that can use the mSin3A corepressor (Figs. 3 and 4). Thus, these translocations may act through a common mechanism. Likewise, the t(15;17) and t(11;17), which are characteristic of acute promyelocytic leukemia, create fusion proteins that interact with N-CoR, SMRT, the mSin3 corepressors, and histone deacetylases to repress transcription (32). The t(8;21) also interacts with N-CoR and histone deacetylases 1 and 2 to repress transcription. Given that mSin3A can recruit histone deacetylases to repress transcription (33), it is possible that targeting mSin3 or histone deacetylases may provide therapeutic benefit to a large number of leukemia patients, including those containing the inv(16).
Acknowledgments
We thank Robert Adelstein for the nonmuscle myosin A antibody. We especially thank Paul Liu and Neeraj Adya for the unpublished C-terminal inv(16) mutants and for critical evaluation of the manuscript and helpful comments. This work was supported by the VICC DNA-sequencing core, by National Institutes of Health/National Cancer Institute Grants RO1-CA64140 (S.W.H.) and RO1-CA77274 (S.W.H.), by a Center Grant CA68485 from the National Cancer Institute, and by the Vanderbilt-Ingram Cancer Center. B.L. is a Fellow of the Leukemia Society of America.
Abbreviations
- AML
acute myeloid leukemia
- CBFβ
core binding factor β
- SMMHC
smooth muscle myosin heavy chain
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Shurtleff S A, Meyers S, Hiebert S W, Raimondi S C, Head D R, Willman C L, Wolman S, Slovak M L, Carroll A J, Behm F, et al. Blood. 1995;85:3695–3703. [PubMed] [Google Scholar]
- 2.Liu P, Tarle S A, Hajra A, Claxton D F, Marlton P, Freedman M, Siciliano M J, Collins F S. Science. 1993;261:1041–1044. doi: 10.1126/science.8351518. [DOI] [PubMed] [Google Scholar]
- 3.Wang S, Wang Q, Crute B E, Melnikova I N, Keller S R, Speck N A. Mol Cell Biol. 1993;13:3324–3339. doi: 10.1128/mcb.13.6.3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ogawa E, Inuzuka M, Maruyama M, Satake M, Naito-Fujimoto M, Ito Y, Shigesada K. Virol. 1993;194:314–331. doi: 10.1006/viro.1993.1262. [DOI] [PubMed] [Google Scholar]
- 5.Fenrick R, Hiebert S W. J Cell Biochem. 1998;31,Suppl.:194–202. doi: 10.1002/(SICI)1097-4644(1998)72:30/31+<194::AID-JCB24>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 6.Castilla L H, Wijmenga C, Wang Q, Stacy T, Speck N A, Eckhaus M, Marin-Padilla M, Collins F S, Wynshaw-Boris A, Liu P P. Cell. 1996;87:687–696. doi: 10.1016/s0092-8674(00)81388-4. [DOI] [PubMed] [Google Scholar]
- 7.Look A T. Science. 1997;278:1059–1064. doi: 10.1126/science.278.5340.1059. [DOI] [PubMed] [Google Scholar]
- 8.Rowley J D. Annu Rev Genet. 1998;32:495–519. doi: 10.1146/annurev.genet.32.1.495. [DOI] [PubMed] [Google Scholar]
- 9.Meyers S, Lenny N, Hiebert S W. Mol Cell Biol. 1995;15:1974–1982. doi: 10.1128/mcb.15.4.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lenny N, Westendorf J J, Hiebert S W. Mol Biol Rep. 1997;24:157–168. doi: 10.1023/a:1006859700409. [DOI] [PubMed] [Google Scholar]
- 11.Kanno T, Kanno Y, Chen L, Ogawa E, Kim W, Ito Y. Mol Cell Biol. 1998;18:2444–2454. doi: 10.1128/mcb.18.5.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Q, Stacy T, Binder M, Marin-Padilla M, Sharpe A H, Speck N A. Proc Natl Acad Sci USA. 1996;93:3444–3449. doi: 10.1073/pnas.93.8.3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Q, Stacy T, Miller J D, Lewis A F, Gu T L, Huang X, Bushweller J H, Bories J C, Alt F W, Ryan G, et al. Cell. 1996;87:697–708. doi: 10.1016/s0092-8674(00)81389-6. [DOI] [PubMed] [Google Scholar]
- 14.Okuda T, van Deursen J, Hiebert S W, Grosveld G, Downing J R. Cell. 1996;84:321–330. doi: 10.1016/s0092-8674(00)80986-1. [DOI] [PubMed] [Google Scholar]
- 15.Niki M, Okada H, Takano H, Kuno J, Tani K, Hibino H, Asano S, Ito Y, Satake M, Noda T. Proc Natl Acad Sci USA. 1997;94:5697–5702. doi: 10.1073/pnas.94.11.5697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kitamura E, Hosoda F, Shimizu K, Shinohara K, Hayashi Y, Nagase T, Yokoyama Y, Ohki M. Blood. 1998;91:4028–4037. [PubMed] [Google Scholar]
- 17.Nucifora G, Begy C R, Erickson P, Drabkin H A, Rowley J D. Proc Natl Acad Sci USA. 1993;90:7784–7788. doi: 10.1073/pnas.90.16.7784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Golub T R, Barker G F, Bohlander S K, Hiebert S W, Ward D C, Bray-Ward P, Morgan E, Raimondi S C, Rowley J D, Gilliland D G. Proc Natl Acad Sci USA. 1995;92:4917–4921. doi: 10.1073/pnas.92.11.4917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Romana S P, Mauchauffe M, Le Coniat M, Chumakov I, Le Paslier D, Berger R, Bernard O A. Blood. 1995;85:3662–3670. [PubMed] [Google Scholar]
- 20.Hiebert S W, Sun W, Davis J N, Golub T, Shurtleff S, Buijs A, Downing J R, Grosveld G, Roussell M F, Gilliland D G, et al. Mol Cell Biol. 1996;16:1349–1355. doi: 10.1128/mcb.16.4.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cao W, Britos-Bray M, Claxton D F, Kelley C A, Speck N A, Liu P P, Friedman A D. Oncogene. 1997;15:1315–1327. doi: 10.1038/sj.onc.1201305. [DOI] [PubMed] [Google Scholar]
- 22.Kanno Y, Kanno T, Sakakura C, Bae S C, Ito Y. Mol Cell Biol. 1998;18:4252–4261. doi: 10.1128/mcb.18.7.4252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Adya N, Stacy T, Speck N A, Liu P P. Mol Cell Biol. 1998;18:7432–7443. doi: 10.1128/mcb.18.12.7432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aronson B D, Fisher A L, Blechman K, Caudy M, Gergen J P. Mol Cell Biol. 1997;17:5581–5587. doi: 10.1128/mcb.17.9.5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Levanon D, Goldstein R E, Bernstein Y, Tang H, Goldenberg D, Stifani S, Paroush Z, Groner Y. Proc Natl Acad Sci USA. 1998;95:11590–11595. doi: 10.1073/pnas.95.20.11590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.el-Deiry W S, Tokino T, Velculescu V E, Levy D B, Parsons R, Trent J M, Lin D, Mercer W E, Kinzler K W, Vogelstein B. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 27.Zeng C, van Wijnen A J, Stein J L, Meyers S, Sun W, Shopland L, Lawrence J B, Penman S, Lian J B, Stein G S, Hiebert S W. Proc Natl Acad Sci USA. 1997;94:6746–6751. doi: 10.1073/pnas.94.13.6746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lenny N, Meyers S, Hiebert S W. Oncogene. 1995;11:1761–1769. [PubMed] [Google Scholar]
- 29.Liu P P, Wijmenga C, Hajra A, Blake T B, Kelley C A, Adelstein R S, Bagg A, Rector J, Cotelingam J, Willman C L, Collins F S. Genes Chromosomes Cancer. 1996;16:77–87. doi: 10.1002/(SICI)1098-2264(199606)16:2<77::AID-GCC1>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 30.Yergeau D A, Hetherington C J, Wang Q, Zhang P, Sharpe A H, Binder M, Marin-Padilla M, Tenen D G, Speck N A, Zhang D E. Nat Genet. 1997;15:303–306. doi: 10.1038/ng0397-303. [DOI] [PubMed] [Google Scholar]
- 31.Okuda T, Cai Z, Yang S, Lenny N, Lyu C, van Deursen J, Harada H, Downing J. Blood. 1998;91:3134–3143. [PubMed] [Google Scholar]
- 32.Melnick A, Licht J D. Blood. 1999;93:3167–3215. [PubMed] [Google Scholar]
- 33.Torchia J, Glass C, Rosenfeld M G. Curr Opin Cell Biol. 1998;10:373–383. doi: 10.1016/s0955-0674(98)80014-8. [DOI] [PubMed] [Google Scholar]
- 34.Fenrick R, Amann J M, Lutterbach B, Wang L, Westendorf J J, Downing J R, Hiebert S W. Mol Cell Biol. 1999;19:6566–6574. doi: 10.1128/mcb.19.10.6566. [DOI] [PMC free article] [PubMed] [Google Scholar]