Abstract
Accurate pre-mRNA splicing requires primary splicing signals, including the splice sites, a polypyrimidine tract, and a branch site, other splicing-regulatory elements (SREs). The SREs include exonic splicing enhancers (ESEs), exonic splicing silencers (ESSs), intronic splicing enhancers (ISEs), and intronic splicing silencers (ISSs), which are typically located near the splice sites. However, it is unclear to what extent splicing-driven selective pressure constrains exonic and intronic sequences, especially those distant from the splice sites. Here, we studied the distribution of SREs in human genes in terms of DNA strand-asymmetry patterns. Under a neutral evolution model, each mononucleotide or oligonucleotide should have a symmetric (Chargaff's second parity rule), or weakly asymmetric yet uniform, distribution throughout a pre-mRNA transcript. However, we found that large sets of unbiased, experimentally determined SREs show a distinct strand-asymmetry pattern that is inconsistent with the neutral evolution model, and reflects their functional roles in splicing. ESEs are selected in exons and depleted in introns and vice versa for ESSs. Surprisingly, this trend extends into deep intronic sequences, accounting for one third of the genome. Selection is detectable even at the mononucleotide level, so that the asymmetric base compositions of exons and introns are predictive of ESEs and ESSs. We developed a method that effectively predicts SREs based on strand asymmetry, expanding the current catalog of SREs. Our results suggest that human genes have been optimized for exon and intron discrimination through an RNA landscape shaped during evolution.
Keywords: DNA strand asymmetry, exon and intron recognition, exon identity elements, intron identity elements, splicing-regulatory elements
Most mammalian genes are split, with exons (≈150 nt) separated by much longer introns (≈3,000 nt). To produce a mature transcript from a prem-RNA, introns are spliced out, and exons are ligated by a large protein/snRNA complex, the spliceosome. Extensive efforts have been made to elucidate the splicing code, i.e., the combinations of cis-regulatory elements and trans-acting factors responsible for splicing efficiency and fidelity. Besides the degenerate splice-site motifs, which are necessary but not sufficient for specific exon and intron recognition, other sequence elements are required for both constitutive and alternative splicing (1, 2). Many splicing-regulatory elements (SREs) have been identified by experimental or computational approaches (3–10). Among them, two classes of well studied SREs are exonic splicing enhancers (ESEs) recognized by SR proteins, and exonic splicing silencers (ESSs) recognized by certain hnRNP proteins (1, 2). Adding further complexity, the effect of an SRE on splicing is often context-dependent. For example, an SR-protein-dependent ESE element, when present in an intron, can act as an intronic splicing silencer (ISS) to repress splicing (11), whereas a number of ESSs, such as the GGG motif, are also potent intronic splicing enhancers (ISEs) (12). The combinatorial interactions of SR proteins and hnRNP proteins with their cognate SREs are an important aspect of splicing fidelity for most, if not all, exons and introns.
Several previous studies have focused on constitutively spliced exons and introns and revealed a nonrandom distribution of SREs, which suggests that evolution has differentiated exons from introns for the purpose of splicing (4, 8, 9). More specifically, there is a higher density of ESEs in exons than introns and vice versa for ESSs. In addition, ESEs and ESSs are preferentially located in exonic and intronic sequences near the splice sites, respectively. These observations are consistent with results from comparative-genomics studies, which demonstrated that exonic and intronic sequences near the splice sites show a higher level of sequence conservation than sequences farther from the splice sites, especially for alternatively spliced exons (13).
Despite this progress, the understanding of the extent and pattern of functional constraints for accurate splicing of mammalian genes remains incomplete. An important limitation of previous studies is the lack of “completely neutral” sequences as controls to compare with real exons and introns, which prevents a rigorous assessment of selective forces that have enriched or depleted different classes of SREs in different regions. For the same reason, it has been difficult to prove or disprove splicing-coupled selection in sequences far from the splice sites, e.g., intronic sequences beyond several hundred nucleotides, although it is commonly believed that SREs are located near the splice sites (14).
However, neutral sequence evolution is reflected in DNA strand-asymmetry patterns, which may provide a powerful tool to evaluate and characterize the signature of selection. According to Chargaff's second parity rule (PR2), the frequency of a mononucleotide or oligonucleotide should be (statistically) equal to that of its reverse complement on the same strand of a long genomic DNA (15, 16). PR2 has been validated in many organisms, from bacteria to mammals, and presumably reflects symmetric DNA mutations and repair (16). Exceptions to PR2, or DNA-strand asymmetry, do exist, reflecting different mechanisms in various organisms. In bacteria and vertebrates, strand asymmetry in gene regions is thought to arise from asymmetric but neutral transcription-coupled mutation (TCM) and transcription-coupled repair (TCR) mechanisms (17). TCM and TCR have been invoked to explain the excess of guanine (G) plus thymine (T) over adenosine (A) plus cytosine (C) in the sense strand observed in mammals (18). However, stronger strand asymmetry in intronic sequences near the splice sites was also noted and was attributed to splicing-coupled selection (19).
Here, we systematically investigate splicing-coupled selection in human constitutive exons and introns by characterizing the patterns of DNA-strand asymmetry of mononucleotides and oligonucleotides. This approach does not require neutral sequences as controls. Instead, we examine each exonic and intronic region separately to see whether SREs can be distinguished from random elements in terms of strand asymmetry, providing a hallmark of splicing-coupled selection. We provide evidence that the distributions of many known ESEs and ESSs differ from those of random elements in both exons and introns, including deep intronic sequences. The systematic bias and the pattern of SRE distribution cannot be explained by a neutral evolution model, suggesting that human genes have been optimized during evolution for discrimination between exons and introns, among other potential functional constraints.
Results
Patterns of Mononucleotide Strand Asymmetry in Exons and Introns.
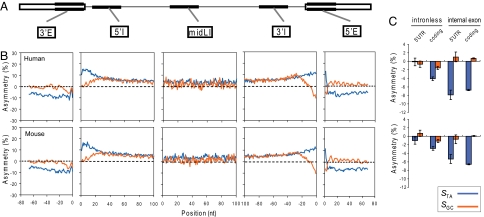
To assess the selective pressure driven by pre-mRNA splicing fidelity and/or efficiency, we initially studied the mononucleotide strand asymmetry of human and mouse genes in five regions from constitutive internal exons and introns: the first (5′E) and last (3′E) 70 nt of exons; the first (5′I) and last (3′I) 100 nt of introns; and the middle 100 nt (midLI) of long introns (≥3,000 nt) (Fig. 1A). Surprisingly, exons and introns show opposite strand asymmetry, as quantified by STA and SGC [Fig. 1B and supporting information (SI) Table S1]. T is more abundant than A, and G is more abundant than C in intronic regions, which is consistent with previous studies (18, 19). In contrast, T is less abundant than A, and there is only a slight excess of G over C in exons. The 5′ and 3′ extremities of introns generally have similar patterns, with an increased frequency of T and C (Fig. 1B). This nucleotide bias may partly reflect some longer-than-usual polypyrimidine tracts at the 3′ extremity of some introns, but the underlying reason is less apparent at the 5′ extremity.
Fig. 1.
A landscape of mononucleotide strand asymmetry in exons and introns. (A) Diagram of five types of region analyzed in this study. (B) Strand asymmetry of human (Upper) and mouse (Lower) genes. TA and GC asymmetries are shown in blue and red, respectively. For intronic regions (5′I, midLI, and 3′I), strand asymmetry was calculated for each nucleotide position. For exonic regions (5′E and 3′E), strand asymmetry was calculated in sliding 3-nt windows, to smooth out the differences among the three positions of codons. Note that the coordinates in the abscissa are not relative to the splice sites, because nucleotides that are part of the consensus motifs were removed. (C) Strand asymmetry of coding and 5′ UTR exons and coding and 5′ UTR portions of intronless genes for human (Upper) and mouse (Lower). Average strand asymmetry was calculated for each whole exon or region. Error bars represent the 95% confidence interval. The color-coding scheme is the same as in B.
The above observations indicate a more complicated landscape of strand asymmetry than can be explained by transcription-coupled mechanisms. Instead, the distinct asymmetry patterns of exons and introns could be due to protein-coding and/or splicing, whose signals are superimposed in exonic sequences. To separate these selective forces, which may have contributed to strand asymmetry in exons, we compared constitutive internal coding and 5′ UTR exons and the coding and 5′ UTR portions of intronless genes. Notably, compared with coding exons, strand asymmetry in noncoding exons is very similar (Fig. 1C), with no or only moderate differences in either TA asymmetry (P = 0.04 for human; P = 0.02 for mouse; χ2 test, df = 1; the same below, except where indicated) or GC asymmetry (P = 0.58 for human; P = 0.14 for mouse). In contrast, much weaker asymmetry, especially for STA, is observed in the coding portion of intronless genes, for which the effect of splicing is absent (P < 2.2 × 10−16 for human and mouse). Importantly, strand asymmetry in the 5′ UTR of intronless genes, in which protein-coding and splicing effects have presumably been separated, is barely detectable. For human and mouse, respectively, the TA asymmetry is estimated to be −0.1% (P = 0.8) and −1.0% (P = 0.02), and GC asymmetry is estimated to be −0.7% (P = 0.08) and 0.7% (P = 0.07) (Fig. 1C). This observation contradicts the assumption that TCR is strongest immediately downstream of the transcriptional start site (17). Although these comparisons may have overlooked other potential differences between intron-containing and intronless genes, they support the notion that the observed strand asymmetry is strongly correlated with splicing-coupled selection. Interestingly, the pattern of strand asymmetry in lower organisms differs substantially from that of mammals (Fig. S1). In particular, yeast introns have strand asymmetry in the same direction as exons (Table S1). This pattern corroborates the observation that the yeast primary splicing signals are highly conserved among different introns, which often provides sufficient discrimination between exons and introns.
Nonrandom Distribution of Known SREs.
We reasoned that if the landscape of strand asymmetry in exons and introns is associated with splicing-coupled selection, the bias of mononucleotides per se may not have a direct functional meaning. Rather, splicing factors, such as SR proteins and hnRNPs, could have preferences for certain sequence motifs, whose nature and frequency would determine the overall strand asymmetry in exons and introns. To evaluate splicing-coupled selection more directly, we analyzed the distribution of known and putative hexameric SREs in human exons and introns, in comparison with random hexamers. For each type of sequence (exon, 5′I, 3′I, and midLI), we divided all unique hexamers, including SREs, into three groups: those with positive (S>0), negative (S<0), or no (S = 0) asymmetry, in which a hexamer is more, less, or equally frequent, respectively, in the sense strand than in the antisense strand. Because all hexamers are part of reverse-complementary pairs (except self-complementary or palindromic ones), the number of hexamers with positive asymmetry has to be equal to the number with negative asymmetry independently of the sequences under consideration. Our null hypothesis is the neutral-evolution model, under which SREs should be subject to the same selective pressure as random elements, so that the strand asymmetry of SREs should not differ from that of random elements. Alternatively, if the sequences are not neutral and certain elements are enriched (depleted), more than half of those asymmetric elements should have positive (negative) asymmetry. Therefore, a systematic bias in the direction of strand asymmetry of SREs would provide direct evidence of splicing-coupled selection.
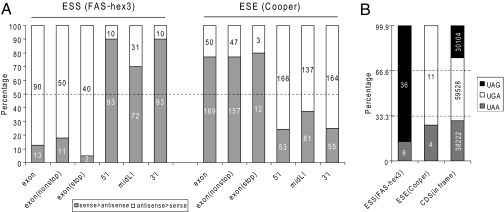
We first tested this hypothesis by examining the distribution of experimentally determined ESSs and ESEs. A panel of 103 ESS hexamers, dubbed FAS-hex3, was derived by cell-based selection from a library of random decamers engineered into an alternative exon in a fluorescent splicing reporter (8). These ESS hexamers do have a lower frequency in exons compared with flanking intronic sequences (8), but it was unclear whether the distribution deviates from neutral evolution in exons or introns, or both. We found that the ESS hexamers show very biased strand asymmetries in both exons and introns, yet opposite in direction. As shown in Fig. 2A, 90 ESS hexamers (87%) have negative asymmetry in exons, implying that ESSs tend to be depleted in exons (P = 3.2 × 10−14). In contrast, in introns, especially in the 5′I and 3′I regions, most ESS hexamers have positive asymmetry (93 of 103 or 90% in both regions), implying that ESSs tend to be enriched in introns (P = 2.9 × 10−16). Even in the midLI region, 70% (72 of 103) of ESS hexamers have positive asymmetry (P = 5.3 × 10−5), suggesting a role in repression of exon-like sequences (pseudoexons) in introns. Therefore, the distribution of ESSs deviates from the prediction of the neutral-evolution model in both exons and introns, including deep intronic sequences, and is consistent with the role of ESSs in exon silencing.
Fig. 2.
Distinct strand-asymmetry patterns of known SREs that discriminate between exons and introns. (A) The percentage of SREs with positive asymmetry (filled areas) and SREs with negative asymmetry (blank areas). In exons, SREs were also subdivided into two groups, depending on whether the hexamer comprises a stop codon, and the percentages were calculated separately for each group. Actual counts are shown inside each box. (B) The frequencies of the three stop codons in ESS hexamers, ESE hexamers, and coding sequences (CDS). Actual counts are shown inside each box.
We similarly studied a panel of 220 ESE hexamers, dubbed “Cooper ESEs,” identified by in vivo functional SELEX experiments (3). The distribution of these ESEs is also significantly nonrandom and has an opposite pattern compared with ESSs (Fig. 2A). Among the 219 nonpalindromic hexamers, 169 (77%) ESE hexamers have positive asymmetry in exons (P = 8.9 × 10−16), whereas in the 5′I and 3′I regions, most (166 or 76%, P = 2.2 × 10−14 for 5′I; 164 or 75%, P = 1.8 × 10−13 for 3′I) have negative asymmetry. Again, even in the midLI region, 63% (137 of 218; one is absent in the midLI region, P = 1.5 × 10−4) have negative asymmetry. Similar results were also observed from two additional panels of experimentally determined ESEs: “Kole-ESEs” identified by in vitro functional SELEX experiments (7) and “literature ESEs” compiled in a survey of multiple studies (10) (Fig. S2 and Fig. S3). Therefore, ESEs tend to be enriched in exons and depleted in introns, including deep intronic sequences, which is consistent with their functional roles in exon recognition.
The skewed asymmetry of ESSs and ESEs in exons is not due to the depletion of in-frame stop codons. To demonstrate this, we separately examined the strand asymmetry of stop-codon-containing SREs and non-stop-codon-containing SREs and found the same pattern of strand asymmetry for both groups (Fig. 2A). Interestingly, the frequencies of the three stop codons in ESSs, ESEs, and the termini of coding sequences (actual stop codons) are very different (Fig. 2B): UAG is much more frequent in ESSs (86%, P < 2.2 × 10−16) but almost absent in ESEs (P = 0.06, moderate significance due to limited sample size; more significant results observed in Fig. S2B and Fig. S3B) compared with its use as a stop codon (24%). This bias likely reflects the similarity of UAG with the consensus motif (UAGGGA/U) of hnRNP A1 (20), which represses exon recognition and splicing when bound to exons. Taken together, the analyses of both ESSs and ESEs provide strong evidence that the distribution of SREs is selected to maximize splicing fidelity in both exons and introns, even for deep intronic sequences, which were assumed to be neutral (14).
Prediction of New SREs, Using Strand Asymmetry.
The distinct landscape of strand asymmetry of known SREs also suggests a method for de novo SRE prediction. Instead of the four conventional categories of SREs (ESE, ESS, ISE, and ISS), we define two categories: exon-identity elements (EIEs), which are enriched in exons and important for exon recognition, and intron-identity elements (IIEs), which are enriched in introns and important for intron recognition. This definition reflects the functional overlap between ESEs and ISSs, which together approximately correspond to EIEs, and between ESSs and ISEs, which together approximately correspond to IIEs. In addition, this dual classification of elements may have a more natural correspondence with the two main categories of ubiquitous splicing-regulatory proteins, i.e., SR proteins and hnRNPs.
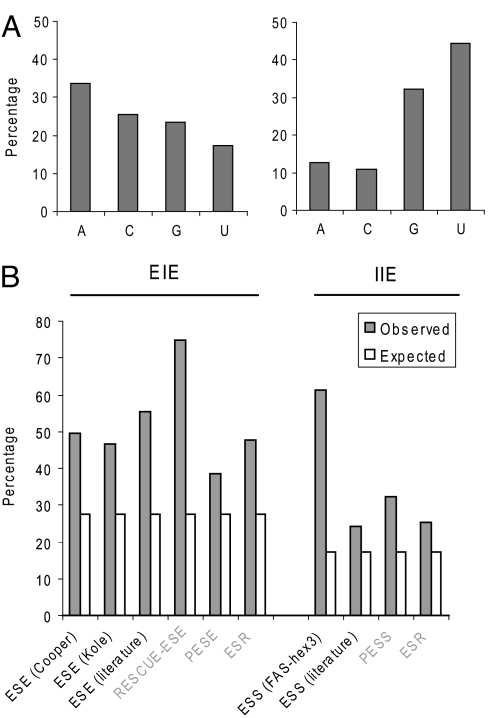
Overall, we predicted 1,131 hexamers with the strongest positive asymmetry in constitutive exons as EIEs (z = 5, P = 0.001, after Bonferroni correction for multiple testing) (Fig. 3 and Dataset S1). At the same significance level, we similarly predicted 569 and 568 hexamers with the strongest positive asymmetry in 5′I and 3′I sequences as IIEs, respectively. The 5′ and 3′ IIEs largely overlap, and their union gives 708 IIEs (Fig. 3 and Dataset S2). Among the EIEs, the hexamer GAAGAA, which is recognized by SF2/ASF and enhances exon recognition (21), is ranked third from the top (S = 44%, z = 40). AC-rich elements are also abundant among EIEs (3). In contrast, a number of top IIEs are U-rich elements, which can be recognized by several hnRNP proteins, such as hnRNP C (20).
Fig. 3.
Predicted EIEs and IIEs, using strand asymmetry. (A) Asymmetric base composition of EIEs and IIEs. (B) Overlap of predicted EIEs or IIEs with known SREs. The height of each bar represents the observed (filled bars) or expected (open bars) percentages of previously reported SREs predicted here as EIEs or IIEs. In all of the comparisons, the overlaps are statistically significant (P < 0.007 in the worst case). Computationally derived SREs, which might have implicit biases for this comparison, are labeled in gray letters.
To evaluate the method more quantitatively, we performed extensive comparisons of the predicted EIEs and IIEs with known SREs (3–5, 7–10), especially those determined by unbiased experimental approaches (3, 7, 8, 10). We found significant overlaps between EIEs and ESEs and between IIEs and ESSs, respectively (Fig. 3B). In particular, 61% (63 of 103) of FAS-hex3 ESSs are predicted as IIEs, 3.5-fold greater than expected by chance (P < 2.2 × 10−16). Among them, five of the six (83%) representative ESS hexamers derived from clustering analysis (8) are predicted as IIEs. For Cooper ESEs (3), 50% (109 of 220) of the derived hexamers are predicted as EIEs (1.8-fold enrichment compared with random hexamers, P < 1.3 × 10−13). We note that comparisons with previous computationally defined elements are likely biased, because such methods explicitly used the enrichment or depletion in exons (introns) to derive the elements. Nevertheless, the overlap between EIEs and Cooper ESEs, and that between IIEs and FAS-hex3 ESSs, which are unbiased, are among the largest in all of the comparisons. In contrast, the overlaps between ESEs and IIEs and between ESSs and EIEs are significantly smaller than expected by chance (data not shown).
Next, we examined the strand-asymmetry patterns of predicted EIEs and IIEs to evaluate functional selection. Because we did not use introns for predicting EIEs, our prediction method should not bias the strand asymmetry of EIEs in introns; a similar argument holds for IIEs in exons and midLI regions. However, we found significantly biased strand asymmetries for both EIEs and IIEs (Fig. S4) qualitatively similar to what we observed from known ESEs and ESSs, respectively (Fig. 2A). Therefore, these results provide an independent line of evidence that exons and introns—even intronic sequences distant from the splice sites—are under splicing-coupled selection.
Correlation Between Strand Asymmetry of Oligonucleotides and Mononucleotides.
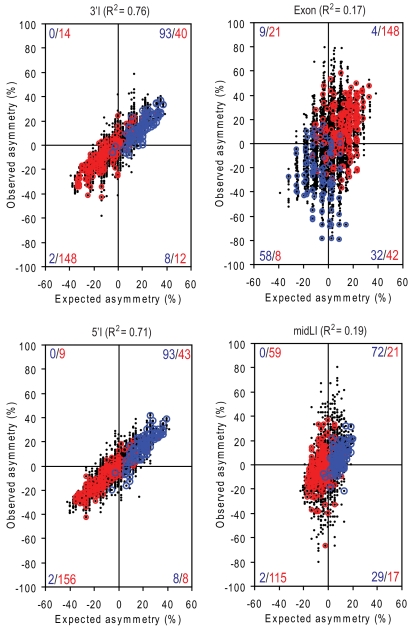
We noticed that EIEs and IIEs have a strongly nonuniform base composition, with T > A and G > C in IIEs and the opposite pattern in EIEs (Fig. 3A). This pattern is consistent with the compositional bias of overall exonic and intronic sequences (22) and with that of known ESSs (8). To understand the relationship between mononucleotide and oligonucleotide strand asymmetries, we asked whether the base composition reflects only neutral evolution by examining the relationship between the observed strand asymmetry of hexamers and that expected from their base composition. Strikingly, ESEs and ESSs can be largely separated based on the strand asymmetry predicted from the base composition in exons and all three types of intronic regions (Fig. 4). This again suggests that the skewed base composition may be also constrained by splicing-coupled selection, probably because many SREs are degenerate and ubiquitous, and have nucleotide compositional biases.
Fig. 4.
Correlation between the strand asymmetry of hexamers and their base composition. Black dots represent hexamers. The ordinate shows the strand asymmetry of each hexamer calculated from its observed occurrences (high-order asymmetry). The abscissa shows the strand asymmetry of each hexamer expected from mononucleotide composition (low-order asymmetry). The squared Pearson correlation of the two values is indicated at the top. The FAS-hex3 ESSs and Cooper ESEs are overlaid and highlighted by blue and red circles, respectively. The number of ESSs or ESEs in each quadrant is also given in blue and red, respectively.
However, we cannot exclude other selective pressures that might also cause mononucleotide asymmetry, especially in exons. Indeed, for coding exons, the three positions of codons have very different patterns of strand asymmetry, suggesting that the bias is in part related to protein-coding (Fig. S5). Importantly, at the fourfold degenerate (synonymous) sites (14), which are under the weakest selective pressure from the protein-coding perspective, we found that C > G and T > A (Fig. S6). This pattern is distinct from the overall pattern of coding exons and that of noncoding exons (Fig. 1 and Table S1). The excess of C over G is consistent with our model of splicing-coupled selection, although other interpretations have been proposed to relate this bias to RNA secondary structure (23). The excess of T over A cannot be readily explained by our model. However, we noticed that the abundance of A increases near the splice sites, where ESEs are more abundant (24). A similar position-dependent skewness has been recently found for certain amino acids, and is related to the enrichment of ESEs near splice sites (25).
Discussion
Detecting noncoding sequences under functional selection is an important step to decode the genetic information in the genome. In this study, we provide evidence for splicing-coupled selective forces and characterize the resulting sequence patterns in human exons and introns, including deep intronic sequences. The widespread evidence of selection in multiple-exon genes, accounting for one-third of the human genome, is surprising. Previous studies estimated that 5% of nucleotides in the genome are under evolutionary constraints, as deduced from multiple-species sequence alignments (26). In most cases, deep intronic sequences were assumed to be nearly neutral, unless significant cross-species conservation was detected. However, these alignment-based methods may fail to detect sequences under weak selection, because of the difficulties in precise alignment. In addition, these studies used fourfold degenerate sites or ancient repeats as a practical proxy for neutral sequences, which may also result in an underestimate of constrained sequences.
The widespread selection is consistent with and provides further insight into the current understanding of mechanisms that confer splicing fidelity. We have recently shown that alternative splicing events that represent evolutionary precursors or errors are prevalent in mammals and weakly deleterious, so that a purifying selective force is discernible (27). Indeed, the distribution pattern of ESEs and ESSs compared with that of random elements cannot be explained by neutral evolution. The enrichment of ESEs (ISSs) in exons and their depletion in introns, together with the opposite pattern for ESSs (ISEs), maximizes the distinction between exon and intron identity and therefore maximizes splicing fidelity. The same trend—albeit weaker in magnitude—in deep intronic sequences suggests selective pressure to suppress pseudoexons. Therefore, the present genome has evolved into an optimal landscape to discriminate between exons and introns. Although the different densities of SREs in exons and introns were noted, earlier studies could not identify the exact pattern of selective constraints because of the lack of a neutral model (4, 8, 9). In contrast, we used the baseline from the strand-asymmetry pattern of random elements to gauge whether SREs are more enriched or depleted than expected by chance. We note that the SREs we used for this purpose were originally derived from screens of random-sequence libraries inserted into the alternative exon of a splicing reporter. As far as we can tell, there is no apparent bias among these SREs due to the base composition, protein-coding, or other characteristics of human genes. Therefore, the pattern of strand asymmetry of SREs we observed is unlikely to be artifactual.
An application of the characteristic strand-asymmetry landscape is to predict new SREs. We predicted elements with the strongest strand asymmetry in exons and introns as EIEs and IIEs, respectively. The number of hexamers showing significant asymmetry is considerably larger than the sets of SREs identified in several previous studies (4, 5, 8). According to comparisons with known ESEs and ESSs, our method is very effective in recovering many known elements. Therefore, many unknown elements are expected to be functional SREs as well, although further experimental validation will be required. However, the predictions could also include elements involved in other steps of posttranscriptional regulation. For example, elements with strongest asymmetry in 3′ UTRs were recently used to predict microRNA targets (28). However, lack of asymmetry, e.g., for palindromic sequences, does not exclude a possible function in splicing regulation. Another potential caveat in this method is the assumption of symmetric neutral sequences to assign a significance value of strand asymmetry for each hexamer. This may represent an over-simplification, because background asymmetry might exist because of asymmetric, yet neutral mutation or repair processes. A solution to this problem is to control for low-order strand asymmetry (i.e., asymmetric base composition), using a Markov model. However, useful information might be lost in the process, because we observed that strand asymmetry estimated by using merely base composition can largely distinguish between known ESEs and ESSs. As a proof of principle, here we used the simplest approach, before this issue can be addressed more rigorously in future studies. Although the significance level assigned to each hexamer might be biased, this does not affect the conclusion that the hexamers with the strongest asymmetry are more likely to be functional SREs, as observed in practice.
The correlation between higher-order strand asymmetry (e.g., hexamers) and that of low order (e.g., mononucleotides) can be at least partly explained by the degeneracy in the binding specificity of SR proteins and hnRNPs. As a general mechanism for splicing fidelity, the splicing machinery needs to have sufficient flexibility and robustness so that it can recognize signals embedded in various sequence contexts. This is especially important in coding exons, where splicing signals are superimposed on the more restrictive protein code. A direct consequence of the degeneracy of the binding motifs is that SREs are highly ubiquitous. Therefore, the higher-order constraints are also reflected in the base composition, because exonic (intronic) nucleotide substitutions toward EIEs (IIEs) are favored for the discrimination between exons and introns (29). However, we could not distinguish whether exonic and intronic sequences adapted to the specificity of the splicing machinery during early evolution or vice versa. Given the considerable differences in both the exonic and intronic strand-asymmetry patterns and in splicing-regulatory proteins across eukaryotic species, a coevolution scheme appears more likely, such that multiple selective forces and mutational processes can be reconciled to be compatible with the nearly optimal genetic code (30).
Materials and Methods
Data Compilation.
Constitutive internal exons and introns for six species (human, mouse, rat, zebrafish, Drosophila melanogaster, and Caenorhabditis elegans) were compiled from our database dbCASE (http://rulai.cshl.edu/dbCASE), which was based on high-quality transcripts (mRNA/EST) and genome alignment. The data were filtered to include only exons and introns flanked by AG/GT splice sites and supported by ≥4 transcripts. To exclude primary splicing signals, the first 1 nt and last 3 nt of exons, and the first 10 nt and last 30 nt of introns were removed. To avoid overlap of sequences, only exons ≥144 nt were used for 5′E and 3′E regions; similarly, only introns ≥240 nt were used for 5′I and 3′I regions (Fig. 1A). Repeat-masked sequences in different regions were then extracted. Alignments of yeast protein-coding genes were downloaded from the UCSC genome browser (assembly October 2003, the SGD gene track), from which exons and introns were extracted. Introns that overlap with other genes were excluded. Nucleotides that overlap with primary splicing signals were also removed similarly.
The coding information of dbCASE constitutive exons was based on CDS annotations of RefSeq transcripts to identify coding and 5′ UTR exons. To minimize contamination of 5′ UTR exons by coding sequences, we further filtered the data by checking each putative noncoding exon against coding exons of all RefSeq and UCSC Known Gene exons. A putative noncoding exon was removed if there was any overlap with coding exons. Similarly, intronless genes were extracted according to the aligned RefSeq transcripts, followed by the exclusion of those overlapping with any other genes (e.g., embedded in the intronic region of another gene). Stop-codon usage was obtained from the codon usage database (31).
Experimentally Determined ESEs and ESSs.
Several studies identified ESEs or ESSs by screening a library of random sequences inserted into an alternative exon of a minigene as a splicing reporter, although technical details varied (3, 7, 8). The SREs identified by these studies represent a relatively unbiased sample of SREs, which are not restricted to a few specific splicing factors and are therefore appropriate to characterize general distribution patterns of SREs. Another compilation of ESEs identified in separate experimental studies was also examined (10). Because the original SRE sequences are relatively long, they had to be converted into hexamers to calculate strand asymmetry. For the ESSs, 103 hexamers that appear at least three times among ESS decamers, dubbed FAS-hex3, were derived in the original study (8) and were used here. For the other three ESE datasets, the original sequences were converted into overlapping hexamers, resulting in 220 (Cooper ESEs), 386 (Kole ESEs), and 279 (literature ESEs) hexamers, respectively.
Calculation of Strand Asymmetry.
For each type of sequence from exons, 5′I, 3′I or midLI regions, the strand asymmetry (skewness) of a mononucleotide or oligonucleotide (hexamer in particular) was calculated by
where Ns and Na denote its total count in the sense and antisense strands of sequences, respectively (32). In particular, the mononucleotide TA asymmetry and GC asymmetry were denoted as STA and SGC, respectively. At the mononucleotide level, we also calculated strand asymmetry for each nucleotide position, in the five types of regions (5′E, 3′E, 5′I, 3′I, and midLI), to study the dependence on the distance of the position to the splice sites.
The standard deviation of strand asymmetry was estimated by 2 using the binomial distribution, where r = (Ns + 1)/(N + 2) and N = Ns + Na.
The expected strand asymmetry of a hexamer was also predicted from the base composition (fA, fC, fG, fT) of the sequences under consideration:
where Bi,s and Bi,a represent the base at position i of the hexamer in the sense and antisense strands, respectively.
Predicting EIEs and IIEs, using strand asymmetry.
To test the significance of the strand asymmetry, we made the simplifying assumption that, under the neutral model, the sequences are symmetric, i.e., r = 0.5, although strand asymmetry of base composition was observed. The reason for this assumption is that we found a correlation between SREs and base composition and suspected that the base composition might have been skewed by selection (see Discussion). We tested the null hypothesis by a normal approximation, z = (r − 0.5)/. A hexamer is predicted as an EIE if the z score calculated by using exon sequences is ≥5, which corresponds to the significance level P = 0.001 after Bonferroni correction. Similarly, a hexamer is predicted as a 5′IIE or 3′IIE if the z score calculated in 5′I or 3′I is ≥5. The two sets of IIEs largely overlap and were pooled together.
Statistical Analysis.
The difference in strand asymmetry between two groups was tested by a χ2 test, using R software (www.R-project.org).
Supplementary Material
Acknowledgments.
We thank Drs. Phillip Green, Xavier Roca, and Xiaowo Wang for valuable comments on the manuscript. This work was supported by National Institutes of Health Grant GM74688 (to M.Q.Z. and A.R.K.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0801692105/DCSupplemental.
References
- 1.Cartegni L, Chew SL, Krainer AR. Listening to silence and understanding nonsense: Exonic mutations that affect splicing. Nat Rev Genet. 2002;3:285–298. doi: 10.1038/nrg775. [DOI] [PubMed] [Google Scholar]
- 2.Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- 3.Coulter LR, Landree MA, Cooper TA. Identification of a new class of exonic splicing enhancers by in vivo selection. Mol Cell Biol. 1997;17:2143–2150. doi: 10.1128/mcb.17.4.2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fairbrother WG, Yeh R-F, Sharp PA, Burge CB. Predictive identification of exonic splicing enhancers in human genes. Science. 2002;297:1007–1013. doi: 10.1126/science.1073774. [DOI] [PubMed] [Google Scholar]
- 5.Goren A, et al. Comparative analysis identifies exonic splicing regulatory sequences–the complex definition of enhancers and silencers. Mol Cell. 2006;22:769–781. doi: 10.1016/j.molcel.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 6.Smith PJ, et al. An increased specificity score matrix for the prediction of SF2/ASF-specific exonic splicing enhancers. Hum Mol Genet. 2006;15:2490–2508. doi: 10.1093/hmg/ddl171. [DOI] [PubMed] [Google Scholar]
- 7.Tian H, Kole R. Selection of novel exon recognition elements from a pool of random sequences. Mol Cell Biol. 1995;15:6291–6298. doi: 10.1128/mcb.15.11.6291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang ZF, et al. Systematic identification and analysis of exonic splicing silencers. Cell. 2004;119:831–845. doi: 10.1016/j.cell.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 9.Zhang XH-F Chasin LA. Computational definition of sequence motifs governing constitutive exon splicing. Genes Dev. 2004;18:1241–1250. doi: 10.1101/gad.1195304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zheng Z-M. Regulation of alternative RNA splicing by exon definition and exon sequences in viral and mammalian gene expression. J Biomed Sci. 2004;11:278–294. doi: 10.1159/000077096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ibrahim EC, Schaal TD, Hertel KJ, Reed R, Maniatis T. Serine/arginine-rich protein-dependent suppression of exon skipping by exonic splicing enhancers. Proc Natl Acad Sci USA. 2005;102:5002–5007. doi: 10.1073/pnas.0500543102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McCullough AJ, Berget SM. G triplets located throughout a class of small vertebrate introns enforce intron borders and regulate splice site selection. Mol Cell Biol. 1997;17:4562–4571. doi: 10.1128/mcb.17.8.4562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sorek R, Ast G. Intronic sequences flanking alternatively spliced exons are conserved between human and mouse. Genome Res. 2003;13:1631–1637. doi: 10.1101/gr.1208803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chamary JV, Parmley JL, Hurst LD. Hearing silence: Non-neutral evolution at synonymous sites in mammals. Nat Rev Genet. 2006;7:98–108. doi: 10.1038/nrg1770. [DOI] [PubMed] [Google Scholar]
- 15.Lin HJ, Chargaff E. On denaturation of deoxyribonucleic acid. 2. Effects of concentration. Biochim Biophys Acta. 1967;145:398–409. doi: 10.1016/0005-2787(67)90058-5. [DOI] [PubMed] [Google Scholar]
- 16.Albrecht-Buehler G. Asymptotically increasing compliance of genomes with Chargaff's second parity rules through inversions and inverted transpositions. Proc Natl Acad Sci USA. 2006;103:17828–17833. doi: 10.1073/pnas.0605553103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Svejstrup JQ. Mechanisms of transcription-coupled DNA repair. Nature Rev Mol Cell Biol. 2002;3:21–29. doi: 10.1038/nrm703. [DOI] [PubMed] [Google Scholar]
- 18.Green P, et al. Transcription-associated mutational asymmetry in mammalian evolution. Nat Genet. 2003;33:514–517. doi: 10.1038/ng1103. [DOI] [PubMed] [Google Scholar]
- 19.Touchon M, Arneodo A, d'Aubenton -Carafa Y, Thermes C. Transcription-coupled and splicing-coupled strand asymmetries in eukaryotic genomes. Nucleic Acids Res. 2004;32:4969–4978. doi: 10.1093/nar/gkh823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Swanson MS, Dreyfuss G. RNA-binding specificity of hnRNA proteins—a subset bind to the 3′ end of introns. EMBO J. 1988;7:3519–3529. doi: 10.1002/j.1460-2075.1988.tb03228.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tacke R, Manley JL. The human splicing factors ASF/SF2 and SC35 possess distinct, functionally significant RNA binding specificities. EMBO J. 1995;14:3540–3551. doi: 10.1002/j.1460-2075.1995.tb07360.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.International Human Genome Sequencing Consortium. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 23.Chamary JV, Hurst L. Evidence for selection on synonymous mutations affecting stability of mRNA secondary structure in mammals. Genome Biol. 2005;6:R75. doi: 10.1186/gb-2005-6-9-r75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fairbrother WG, Holste D, Burge CB, Sharp PA. Single nucleotide polymorphism - based validation of exonic splicing enhancers. PLoS Biol. 2004;2:e268. doi: 10.1371/journal.pbio.0020268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parmley JL, Urrutia AO, Potrzebowski L, Kaessmann H, Hurst LD. Splicing and the evolution of proteins in mammals. PLoS Biol. 2007;5:e14. doi: 10.1371/journal.pbio.0050014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.The ENCODE Project Consortium. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang C, Krainer AR, Zhang MQ. Evolutionary impact of limited splicing fidelity in mammalian genes. Trends Genet. 2007;23:484–488. doi: 10.1016/j.tig.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 28.Cora D, Di Cunto F, Caselle M, Provero P. Identification of candidate regulatory sequences in mammalian 3′ UTRs by statistical analysis of oligonucleotide distributions. BMC Bioinformatics. 2007;8:174. doi: 10.1186/1471-2105-8-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ke S, Zhang XHF, Chasin LA. Positive selection acting on splicing motifs reflects compensatory evolution. Genome Res. 2008 doi: 10.1101/gr.070268.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Itzkovitz S, Alon U. The genetic code is nearly optimal for allowing additional information within protein-coding sequences. Genome Res. 2007;17:405–412. doi: 10.1101/gr.5987307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakamura Y, Gojobori T, Ikemura T. Codon usage tabulated from international DNA sequence databases: Status for the year 2000. Nucleic Acids Res. 2000;28:292. doi: 10.1093/nar/28.1.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baisnee P-F, Hampson S, Baldi P. Why are complementary DNA strands symmetric? Bioinformatics. 2002;18:1021–1033. doi: 10.1093/bioinformatics/18.8.1021. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.