Abstract
The Cuatro Ciénegas Basin (CCB) in the central part of the Chihuahan desert (Coahuila, Mexico) hosts a wide diversity of microorganisms contained within springs thought to be geomorphological relics of an ancient sea. A major question remaining to be answered is whether bacteria from CCB are ancient marine bacteria that adapted to an oligotrophic system poor in NaCl, rich in sulfates, and with extremely low phosphorus levels (<0.3 μM). Here, we report the complete genome sequence of Bacillus coahuilensis, a sporulating bacterium isolated from the water column of a desiccation lagoon in CCB. At 3.35 Megabases this is the smallest genome sequenced to date of a Bacillus species and provides insights into the origin, evolution, and adaptation of B. coahuilensis to the CCB environment. We propose that the size and complexity of the B. coahuilensis genome reflects the adaptation of an ancient marine bacterium to a novel environment, providing support to a “marine isolation origin hypothesis” that is consistent with the geology of CCB. This genomic adaptation includes the acquisition through horizontal gene transfer of genes involved in phosphorous utilization efficiency and adaptation to high-light environments. The B. coahuilensis genome sequence also revealed important ecological features of the bacterial community in CCB and offers opportunities for a unique glimpse of a microbe-dominated world last seen in the Precambrian.
Keywords: evolution, genomic adaptation, horizontal gene transfer, phosphorus stress, sulfolipids
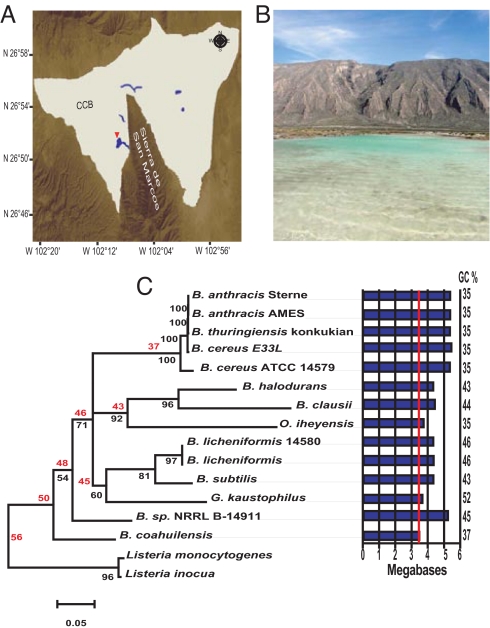
The Cuatro Ciénegas Basin (CCB) is located in a valley ≈740 m above sea level in the state of Coahuila, Mexico, that measures ≈30 km by 40 km and is surrounded by high mountains (>3,000 m) (Fig. 1). CCB is an enclosed evaporitic basin that receives ≈150 mm of annual precipitation. Despite the dry climate of the valley, the CCB harbors an extensive system of springs, streams, and pools (1). The CCB ecosystem is not only characterized by a high endemism of macrooganisms and biodiversity of microorganisms (1, 2), but also by extremely oligotrophic waters that are unable to sustain algal growth, making microbial mats the base of the food web (3). In particular, phosphorus (P) levels in CCB appear to be rather low, because they were below the level of detection of several methods used (0.3 μM) and the extremely high biomass C:P and N:P ratios (>100 by moles) previously reported for CCB stromatolites (3, 4). Unlike the present sea, the Churince spring water is poor in NaCl and carbonates, but it is rich in sulfates, magnesium, and calcium (4). Characterization of the microbiological diversity by sequencing 16S rRNA genes revealed that nearly half of the phylotypes from the CCB were closely related to bacteria from marine environments (2). Bacillus coahuilensis is a free-living, spore-forming bacteria isolated from the water column of a shallow desiccation lagoon in the Churince system at CCB (4) (Fig. 1 A and B). A molecular phylogenetic analysis of 16S rRNA sequences indicates that B. coahuilensis is closely related to other marine Bacillus spp. (4), in agreement with the theory of an ancient marine origin of these ponds. We sequenced the genome of B. coahuilensis to gain insight on the origin and genomic adaptation of this bacterium to the extremely P-limited oligotrophic environment of the Churince pond in CCB.
Fig. 1.
Marine origin of B. coahuilensis, isolated from a pond in the Chihuahuan desert. (A) Sierra de San Marcos is a prominent mountain system in the middle of CCB where >400 ponds sustain most of the biodiversity. The geomorphological origin of CCB has been recently reviewed (2). (B) Churince system (shown with a red triangle in A) consists of a springhead that feeds a 2-km-long stream with an intermediate lagoon terminating at a large shallow desiccation lagoon. (C) Phylogenomic reconstruction. Maximum likelihood phylogenomic reconstruction by using Tree-Puzzle (29) was carried out with 20 universally conserved COGs from the sequenced Bacillus spp. and closely related species (Table S3). Maximum likelihood bootstrap percentage support values are only indicated for major nodes; numbers in red represent tentative timescale, in million years, calculated with the method proposed by Battistuzzi et al. (30). Genome size (represented as bars) and GC content of each Bacillus spp. genome is shown to the right.
Results
General Genome Features.
Sequencing of the B. coahuilensis genome was accomplished by using a hybrid strategy of high-coverage pyrosequencing (29×) and a low-coverage Sanger sequencing (6×) (supporting information (SI) Table S1). The assembled genome size is 3.35 Megabases (Mb), making it the smallest genome reported to date for Bacillus spp. (Fig. 1C). Its genome is composed of a predicted 3,640 coding genes (5) and has a GC content of 37.5% (Table S2), with an average 1,100 orthologues shared with all 14 Bacillus genomes currently available (including Bacillus sp. NRRL B-14911, a recently sequenced marine Bacillus from the Gulf of Mexico) and 905 unique B. coahuilensis genes.
By using a phylogenomic approach (6), we reconstructed a phylogeny through a concatenated alignment of 20 universally distributed genes from the Clusters of Orthologous Groups (COGs) that are considered largely unsusceptible to horizontal gene transfer (6), and that are present in the 14 Bacillus spp. genomes (Table S3). This analysis revealed that B. coahuilensis is basal to the other 14 Bacillus spp. genomes and more related to the marine strain NRRL B-14911 than any other Bacillus spp. Additionally, the short phylogenetic branch length suggests that the B. coahuilensis genome is associated with a low substitution rate compared with the other genomes, despite its small size (Fig. 1C). An extensive 16S analysis shows that B. coahuilensis clearly forms a unique group, but also supports a phylogenetic relation to marine isolates (Fig. S1). However, at the genome level we found no signals that would suggest the presence of bacteria closely related to B. coahuilensis in the Global Ocean Sampling database (7) (data not shown). Taken together, these results suggest that B. coahuilensis is ancestral to the other sequenced Bacillus spp. and that the presence of this species in the CCB did not originate from a recent migration through spore disbursal from present marine habitats.
Lipid Profile Adaptation to a Low-Phosphorous Environment.
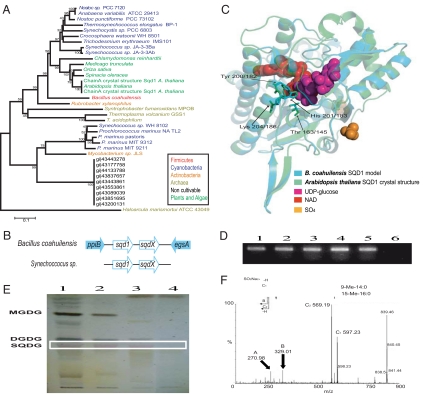
Membrane phospholipids constitute ≈30% of the total phosphate in most organisms. In plants and cyanobacteria subjected to phosphorous deprivation, phospholipids can be replaced by non-P lipids (such as sulfo- and galactolipids) to maintain membrane functionality and integrity, and release P to sustain other P-requiring cellular processes (8). Interestingly, genes encoding sulfoquinovose synthase (sqd1) and glycosyltransferase (sqdX), the two key enzymes in the synthesis of sulfolipids, are present in B. coahuilensis. Because the sulfoquinovose synthesis operon is absent in all other known Bacillus spp. genomes, this finding suggests that the adaptation of B. coahuilensis to the extremely low P concentration of CCB includes the acquisition of these genes through horizontal gene transfer (HGT). The B. coahuilensis genes are closely related to cyanobacterial sqd1 and sqdX (Fig. 2A and Fig. S2), and the operon arrangement is identical to that in Synechococcus sp. PC7942, where these genes participate in the synthesis of sulfolipids (9) (Fig. 2B). The structural prediction of the B. coahuilensis Sqd1 protein is remarkably similar to the crystallized structure of Arabidopsis thaliana SQD (1.2 Å; Fig. 2C); in particular, residues important for the interactions with NAD+ and UDP-ribose are conserved in B. coahuilensis. Reverse transcription PCR (RT-PCR) experiments revealed that, in contrast to plants and cyanobacteria, sqd1 is constitutively expressed and not induced by P limitation (Fig. 2D). Thin layer chromatography revealed a spot in B. coahuilensis lipid extracts with migration similar to that of pure sulfolipids, comparable to that present in A. thaliana and in a cyanobacteria spp. (isolated from CCB) but absent in lipid preparations from Bacillus sp. NRRL B-91411 (Fig. 2E). Mass spectrometry analysis confirmed the presence of sulfolipids in B. coahuilensis (Fig. 2F and Fig. S3). The remarkable acquisition of constitutively expressed genes allowing B. coahuilensis to replace membrane phospholipids with sulfolipids is in agreement with genomic adaptations to extreme phosphate limitation.
Fig. 2.
Acquisition of sulfoquinovose synthesis capabilities through HGT, an adaptation of B. coahuilensis to a phosphorus-limiting environment. (A) Neighbor-joining phylogenetic reconstruction of SQD1 (31). (B) The operon structure of the sqd1 and sqdX resembles that of the Synecchococcus genes (9). Flanking genes are ppi (peptidyl-prolyl cis-trans isomerase B) and egsA (glycerol-1-phosphate dehydrogenase) (not to scale). (C) Modeling (32) to the A. thaliana SQD (PDB ID code 1I24) protein shows conservation of the NAD and UDP-glucose-binding residues. Diagrammic representation was done by using PyMol (http://www.pymol.org). (D) Expression of the sqd1 gene. RT-PCR was carried out from RNA of B. coahuilensis grown under different phosphate concentrations (lanes 1 to 5, RT-PCR products obtained from cells cultured with 0.001, 0.005, 0.05, 0.5, and 5 mM phosphate, respectively; lane 6, control without reverse transcriptase). (E) TLC analysis reveals the presence of a probable SQDG band in B. coahuilensis, A. thaliana, Cyanobacteria sp. (lanes 1 to 3) but absent in Bacillus sp. NRRL B-14911 (lane 4). (F) Mass spectroscopic analysis confirmed the identity of the sulfoquinovoside (Fig. S3).
Light Sensing in a High-Radiation Environment.
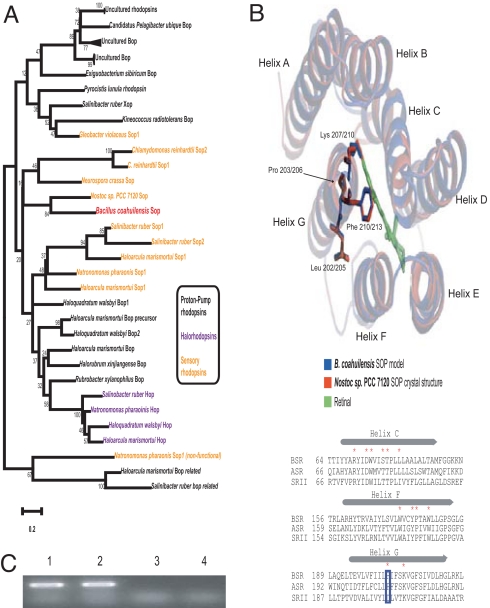
The presence of genes encoding Bacteriorhodopsin (BR) in B. coahuilensis is reminiscent of the abundance of BR genes in marine environmental samples (7), suggesting an additional adaptation of marine bacteria. The phylogeny of B. coahuilensis sensory BR (BSR) showed that its closest orthologue is the Anabaena sp. PCC7120 rhodopsin (ASR) (Fig. 3A). ASR was the first eubacterial SR identified (10) and suggested to function as a photosensory receptor. The structural prediction of BSR and its comparison with crystallized ASR and SR of Natronomonas pharaonis (11, 12) indicates that all three proteins contain seven conserved transmembrane helices and a Lys residue that binds retinal (Fig. 3B). Alignment of ASR and BSR shows that both have a Pro residue instead of the Asp (BSR203 and ASR206) residue present in all other microbial rhodopsins (10). Evidence for HGT of rhodopsins has recently been obtained from whole-genome sequencing and metagenomic projects, and is now thought to be a frequent event in marine bacteria from the photic zone and extreme saline environments (13–15). The retinal chromophore of rhodopsin is synthesized as a cleavage product of carotenoids; thus, the combination of carotenoid synthesis and rhodopsin genes has been suggested to be sufficient for rhodopsin function (14). The genome of B. coahuilensis also contains genes encoding crtB (phytoene synthase) and crtICA2 (phytoene dehydrogenases) that could be involved in retinal biosynthesis (Fig. S4). RT-PCR experiments showed that the expression of bsr is constitutive and not light-dependent (Fig. 3C). The high-radiation exposure prevalent in shallow waters of CCB could explain the selection pressure responsible for the maintenance and constitutive expression of the bsr gene.
Fig. 3.
B. coahuilensis contains a sensory bacteriorhodopsin possibly acquired from cyanobacteria through an ancient HGT event. (A) Neighbor-joining tree showing the phylogenetic diversity of rhodopsins (31). (B) Bsr possesses all of the residues involved in retinal binding and was modeled (32) to the predicted structures of Anabaena and Natronomonas pharaonis. Alignment of segments of SR from B. coahuilensis (BSR, BM4401574), Anabaena sp. PCC7120 (ASR, PDB ID code 1XIO), and Natronomonas pharaonis DSM2160 (SRII, PDB ID code 1JGJ) show conservation of residues in the retinal-binding pocket (marked with asterisks) except for a Pro residue (positions BSR203 and ASR206), which is an Asp residue in all other microbial rhodopsins (blue rectangle; alignment adapted from ref. 10). Diagrammatic representation was done by using PyMol (http://www.pymol.org). (C) bsr is expressed in B. coahuilensis grown either under white light or in the dark. RT-PCR was carried out by using RNA obtained from bacteria grown under dark or white-light conditions (lanes 1 and 2, respectively). Lanes 3 and 4 are controls without reverse transcriptase.
Nucleotide composition analysis (16) identified numerous genomic islands containing genes likely to be acquired by HGT; several have been annotated as HGT genes in B. subtilis and B. halodurans (Fig. S5). Interestingly, our nucleotide composition analysis did not identify bsr and sqd1 as HGT genes. We also determined the Codon adaptation index (CAI) (Fig. S6) for bsr and sqd1 which is of 0.719 and 0.735, respectively, indicating that these genes have been present in B. coahuilensis long enough to undergo amelioration to the average CAI (0.714).
Nitrogen Cycle Strategies and Feeding Capabilities.
To gain insight into the nutritional requirements and metabolic capabilities of B. coahuilensis within the CCB microbial community, we analyzed genes involved in ABC transport systems, one of the largest paralogous families present in the genome of this bacterium. Hidden Markov model (HMM) profiles were built for each ABC importer family (17) and searched against 11 Bacillus spp. genomes. The analysis retrieved a total of 1,038 import systems. The two most abundant families among all Bacillus spp. were metal (MET) and osmoprotectant (OTCN) importers (Fig. S6), which seems to be a characteristic for the group. B. coahuilensis returned 63 import systems, thus being one of the genomes with a less absolute number of import systems (Table S4), below the expected values for the group. Nonetheless, the best represented families in B. coahuilensis are also MET and OTCN. B. coahuilensis possesses a very high proportion of iron-siderophore (ISVH) importers relative to genome size, a feature shared with the water-column marine bacilli Oceanobacillus iheyensis, and Bacillus sp. NRRL B-14911. In addition, an operon coding for the ferric-enterobactin synthesis and transport system fepBCD is shared with O. iheyensis. It also encodes an Iron(III)-dicitrate and ferric enterobactin transporters, suggesting that marine bacilli and B. coahuilensis are actively scavenging for iron.
The proportion of polar amino acid and opine (PAO), as well as d-l-Methionine (DLM) importers relative to the total number of transporters and genome size is greater in B. coahuilensis than in any other Bacillus. It also has one of the lowest ratios of oligopeptide (OPN) importers, suggesting a specialization for the preferential acquisition of single amino acids over oligopeptides. Experimental results show that B. coahuilensis has an absolute requirement for 8 aa (four polar and three hydrophobic) and a partial requirement for another 5 aa (three hydrophobic and two polar), confirming that this bacterium depends on amino acid import (Table S4 and Fig. S6). This overrepresentation of single-amino acid importers is shared with the recently sequenced water-living beta-proteobacteria Minibacterium massiliensis (18), suggesting that this feature might be common in the reduced genomes of aquatic free-living bacteria.
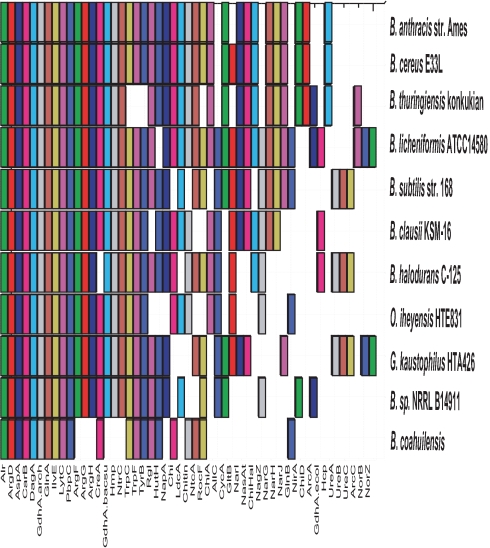
HMM profiles were built for 86 genes involved in the N2 cycle (data not shown) and were searched against all sequenced Bacillus spp. genomes. This revealed that B. coahuilensis has the lowest number of N2 cycle enzymes of all sequenced Bacillus spp., lacking an inorganic N pathway and most of the urea cycle and urea degradation pathways (Fig. 4). It only possesses the canonical transaminases, an ammonia transporter and a d-Ala transporter, thus not being capable of performing the entire N cycle on its own. B. coahuilensis codes for rocF, an arginase mainly found in Firmicutes, but not for any other urea-cycle enzyme. All sequenced Bacillus spp. code for a complete urea cycle (argFGH and rocF), with the notable exception of O. iheyensis. Moreover, growth of B. coahuilensis is impaired, but not completely arrested, when grown without arginine, and this amino acid is one of those transported by the overrepresented import family PAO. B. coahuilensis also lacks the other gene that could provide a source of arginine from citrulline, arcA. This suggests that arginine is an important amino acid in this bacterium, and that it is most likely synthesizing arginine by employing an unknown pathway. It was reported for example, that Pseudomonas spp. synthesizes arginine by an arginine racemase and a d-arginine dehydrogenase (19).
Fig. 4.
B. coahuilensis lacks many of the genes coding for enzymes involved in nitrogen metabolism. Hidden Markov models for the bacterial enzymes involved in N2 metabolism were constructed to detect these genes in all of the sequenced Bacillus spp. Bars in different colors denote the presence of a gene predicted to code for a given enzyme.
In summary, B. coahuilensis exhibits numerous auxotrophies and high dependence on the N2 cycle, which is mainly carried out in the CCB ponds by cyanobacteria, and is therefore also likely to be highly dependent on the microbial community within this environment. This is in contrast with the genome from the marine strain NRRL B-14911, which exhibits partial requirement for only 2 aa and appears well suited to perform most of its N2 cycle on its own (Fig. 4 and Table S5).
Genome Size Evolution and Essential Genes.
The basal position of B. coahuilensis (Fig. 1C) suggests that it is ancestral to the other sequenced Bacillus spp. and related species. Although most of the essential genes (20) of B. subtilis are conserved in B. coahuilensis, many of those with redundant functions are absent (Table S6, Table S7, and Table S8). Genes for endospore formation are conserved, but the number of genes encoding components of the spore coat are significantly underrepresented (Table S6), although this does not compromise formation of heat-resistant spores. We also carried out an analyses of the largest paralogous gene families within B. coahuilensis compared with those in B. subtilis (21), as a means of identifying important functions that are maintained in a small genome. Although B. subtilis has 13 paralogous gene families of >5 members involved in secondary metabolites biosynthesis, transport, and catabolism, these are almost completely absent in B. coahuilensis (COG Q category, 1 gene family of >5 members). Signal transduction, in contrast, seems to be overrepresented in B. coahuilensis (COG T category, 57 gene families of >5 members) compared with the same category in B. subtilis (1 gene family of >5 members) suggesting that environmental monitoring is key to survival in the CCB environment (Fig. S7). Analysis of all known B. subtilis genes involved in the synthesis of cell wall and membrane components reveals that B. coahuilensis also lacks genes necessary for the synthesis of polyphosphate-rich teichoic and polyanion teichuronic acids, useful cell wall phosphorus storage compounds. Because teichoic acid synthesis genes are not only present in several Bacillus species, but also in Listeria spp. which are used as an outgroup for comparing the Bacillus genus (Table S7), it is likely that B. coahuilensis lost the capacity to produce teichoic acid because of the P-limited environment of CCB. Of the 45 genes that constitute the PhoP/PhoR regulon in B. subtilis, only 24 are present in B. coahuilensis (Table S8). B. coahuilensis lacks the pit gene encoding a low-affinity P transporter that is conserved in Listeria spp. and many Bacillus spp. (except for B. halodurans, B. clausi, and O. iheyensis) suggesting that pit was also lost in B. coahuilensis because it provided no advantage in the CCB low-P environment. Interestingly, the pit gene is also absent in the marine cyanobacterial genomes (22). Overall, several genomic characteristics seem to be the result of evolutionary mechanisms that caused a reduction in genome size and selected against redundant genes and genes that provided little or no advantage for survival in the oligotrophic environment of the CCB pools. However, given the basal position of B. coahuilensis in the phylogenomic analysis, it is also possible that some of the genes it is lacking, but which are present in other Bacillus spp., may represent gains of bacilli species that evolved later.
Discussion
P limitation seems to be an important driving force for the adaptations observed in the B. coahuilensis genome. This bacterium has the capability of synthesizing membrane sulfolipids, apparently acquired by HGT from a cyanobacteria. Sulfolipid SQDG is present in all higher plants, mosses, ferns, and algae, but it has also been reported in nonphotosynthetic organisms, predominantly in cyanobacteria (Fig. 3A). Discovery of this capability in the bacilli is especially interesting given the context of an environment with extremely limiting P. HGT of sqd1 and sqdX seems to be frequent in cyanobacteria and, because it involves transfer of only a couple of genes, it seems relatively simple for a bacteria to appropriate this mechanism to cope with P scarceness. Finally, genome-size reduction in an ecosystem with very low P availability, such as the CCB pools, could increase the fitness of a bacterium with a reduced P demand for nucleotide synthesis.
The predicted rhodopsin sequence of B. coahuilensis is phylogenically closer to sequences from cyanobacteria than to those reported in other bacteria and archeobacteria. Because retinal, which is derived from carotenoids, is probably readily available in many of the pigmented CCB bacteria, the acquisition of rhodopsin genes from cyanobacteria could easily lead to a new functional adaptation. B. coahuilensis genome encodes a couple of photolyases that repair UV-induced DNA lesions and could explain, along with the bsr gene, the adaptation of B. coahuilensis to the high-light fluency in the shallow and clear waters of the CCB ponds.
The incidence of both rhodopsin and sulfolipid biosynthesis genes in environmental samples is spatially restricted. We searched the CAMERA database (23) for homologue genes to the B. coahuilensis' bsr and sqd1 and found only four sample points within the Global Ocean Sampling (GOS) having both genes represented: GS027, GS031, GS031, and GS034, which are located in shallow waters of the Galapagos Islands. Two important common features between CCB ponds and the Galapagos Islands are a high-radiation environment in shallow waters and the lack of P. The fact that sqd1 and bsr coexist in two distinct geographically distant locations with common environmental characteristics is suggestive of common adaptation strategies. P starvation seems to be a general constraint in marine environments, as shown in ref. 7.
Analysis of paralogous gene families revealed a dramatic limitation in genes for secondary metabolism in B. coahuilensis possibly as a result of both genome-size reduction and adaptation to a unique niche (24, 25). The large number of auxotrophies, the limitation in N2 cycle genes (Fig. 4), and overrepresented signal transduction genes suggest that this bacterium is a specialist within the CCB water systems, dependent and tightly bound to the primary producers and prepared for sensing and responding to the extreme seasonal environmental conditions (2). This is in contrast to pandemic and generalist bacteria like B. subtilis with its arsenal of secondary metabolism genes.
Our findings show that HGT played a key role in the adaptations of B. coahuilensis. We are currently exploring whether transposons (of which there are >20 in B. coahuilensis), phages [known to be abundant in ponds of the CCB (26)], and natural competence could provide the mechanisms driving changes in the B. coahuilensis genome as well as in the other bacteria in the community. We have isolated numerous cyanobacteria in the Churince pond (27).
The strategy of sequencing a single bacterial isolate to obtain information on the adaptations of bacteria living in a highly oligotrophic environment was highly fruitful because it led to a clear identification of the phylogenetic affiliation of some ecological functions. A metagenome approach might have obscured the phylogenetic association of genes of cyanobacterial origin to firmicutes and would not have allowed us to observe the important nitrogen cycle and amino acid synthesis limitations that make this bacterium dependent on the microbial community. The small genome size, constraints in secondary metabolism, and overrepresentation of signal transduction genes are also features that could not have been deduced from a metagenomic approach. Efforts to study the metagenomics of the CCB ponds should be made to help us understand how these ancient water ecosystems in the middle of the desert are self-sufficient in their biogeochemical cycles.
Our results provide evidence that, because of the specific genome dynamics, its ancestry, and the local adaptive response, B. coahuilensis is most likely the result of adaptation of an ancient marine bacterium to a novel environment. Results that are in agreement with the geology/ontology model that predicts a marine environment for this region in the early Jurassic followed by the rising of the continent, the formation of the CCB valley, and its isolation by the surrounding Sierras in the Cretaceous period ≈70 million years ago (2). B. coahuilensis is likely a primitive bacterial component of a complex community that included Archaea and Cyanobacteria that provided genomic fodder for gene transfer and the implementation of innovative and necessary strategies for survival in an evolving ecology.
Materials and Methods
Strains.
B. coahuilensis was provided by V.S (4) and NRRL B-14911 by J.S. The media used for growth are described in SI Text.
Genome Sequencing and Annotation.
B. coahuilensis was sequenced by a hybrid Sanger/454 approach. The entire genome sequence was obtained from a combination of 16,698 end sequences (providing 6-fold coverage) from a pUC18 genomic shotgun library (2–5 kb), by using dye terminator chemistry on automated DNA sequencers (ABI3700; Applied Biosystems) and 454 technology with seven runs at a 29-fold coverage. Predicted protein-encoding genes were manually refined (see SI Text) and automatically annotated by using the BASys system (5).
Prediction of Horizontal Transfer.
The Similarity Plot (S-plot) application was used (16) to identify windows that contained regions of unusual compositional properties (RUCPs) within B. coahuilensis genome (see SI Text).
RT-PCR.
Semiquantitative RT-PCRs were carried out by using SuperScript One Step RT-PCR with Platinum Taq (Invitrogen Life Technologies) (see SI Text) from RNA isolated from strains grown in modified marine medium supplemented with phosphate. For light/dark experiments, strain was grown on Petri dishes with marine medium grown at 37°C either under white or blue light or in the dark.
Lipid Extraction and Analysis.
Lipids from Arabidopsis, Cyanobacteria spp., and B. coahuilensis were extracted (details are available on request) and analyzed by using the TLC technique as described in ref. 28. For lipid footprint analysis, individual lipids were isolated from TLC plates, and duplicates of each lipid spot were analyzed by MALDI-TOF MS technology (see SI Text).
Supplementary Material
Acknowledgments.
We thank Laura Espinosa Azuar and Antonio Cruz in technical assistance at Instituto de Ecología, Universidad Nacional Autónoma de México; Dr. Michael Travisano (University of Minnesota) and June Simpson (Cinvestav Campus Guanajuato) for their comments on the manuscript. This work was supported in part by Secretaría de Agricultura, Ganadería, Desarrollo Rural, Pesca y Alimentación (SAGARPA) Zea-2006 and Consejo Nacional de Ciencia y Tecnología (CONACyT)-Secretaria de Educatión Pública 43979 grants and Howard Hughes Medical Institute Grant 55005946 (to L.H.-E.); CONACyT IN Grants 223105 and 44673Q (to V.S. and L.E.); CONACyT-Secretaria de Medio Ambiente y Recursos Naturales Grant 2002-C01-0237 (to V.S.); Exobiology Grant NG04GJ12G (to J.S.); and graduate scholarships from CONACyT (to L.D.A., R.C., and J.C.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequence from this Whole Genome Shotgun project has been deposited in the DDBJ/EMBL/GenBank database (accession no. ABFU00000000). The version described in this article is the first version (accession no. ABFU01000000).
This article contains supporting information online at www.pnas.org/cgi/content/full/0800981105/DCSupplemental.
References
- 1.Minckley WL. Environments of the Bolson of Cuatro Ciénegas, Coahuila, México, with Special Reference to the Aquatic Biota. El Paso, TX: Texas Western Press; 1969. University of Texas: El Paso Science Series. [Google Scholar]
- 2.Souza V, et al. An endangered oasis of aquatic microbial biodiversity in the Chihuahuan desert. Proc Natl Acad Sci USA. 2006;103:6565–6570. doi: 10.1073/pnas.0601434103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Elser JJ, et al. Effects of phosphorus enrichment and grazing snails on modern stromatolitic microbial communities. Freshwater Biol. 2005;50:1808–1825. [Google Scholar]
- 4.Cerritos R, et al. Bacillus coahuilensis sp. nov. a new moderately halophilic species from different pozas in the Cuatro Ciénegas Valley in Coahuila, México. Int J Syst Evol Microbiol. 2008;58:919–923. doi: 10.1099/ijs.0.64959-0. [DOI] [PubMed] [Google Scholar]
- 5.Van Domselaar GH, et al. BASys: A web server for automated bacterial genome annotation. Nucleic Acids Res. 2005;33:W455–W459. doi: 10.1093/nar/gki593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ciccarelli FD, et al. Toward automatic reconstruction of a highly resolved tree of life. Science. 2006;311:1283–1287. doi: 10.1126/science.1123061. [DOI] [PubMed] [Google Scholar]
- 7.Rusch DB, et al. The Sorcerer II Global Ocean Sampling Expedition: Northwest Atlantic through Eastern Tropical Pacific. PLoS Biol. 2007;5:e77. doi: 10.1371/journal.pbio.0050077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dormann P, Benning C. Galactolipids rule in seed plants. Trends Plants Sci. 2002;7:112–118. doi: 10.1016/s1360-1385(01)02216-6. [DOI] [PubMed] [Google Scholar]
- 9.Benning C. Biosynthesis and function of the sulfolipid sulfoquinovosyl diacylglycerol. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:53–75. doi: 10.1146/annurev.arplant.49.1.53. [DOI] [PubMed] [Google Scholar]
- 10.Jung KH, Trivedi VD, Spudich JL. Demonstration of a sensory rhodopsin in eubacteria. Mol Microbiol. 2003;47:1513–1522. doi: 10.1046/j.1365-2958.2003.03395.x. [DOI] [PubMed] [Google Scholar]
- 11.Vogeley L, et al. Anabaena sensory rhodopsin: A photochromic color sensor at 20 A. Science. 2004;306:1390–1393. doi: 10.1126/science.1103943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luecke H, Schobert B, Lanyi JK, Spudich EN, Spudich JL. Crystal structure of sensory rhodopsin II at 2.4 angstroms: Insights into color tuning and transducer interaction. Science. 2001;293:1499–1503. doi: 10.1126/science.1062977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mongodin EF, et al. The genome of Salinibacter ruber: Convergence and gene exchange among hyperhalophilic bacteria and archaea. Proc Natl Acad Sci USA. 2005;102:18147–18152. doi: 10.1073/pnas.0509073102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frigaard NU, Martinez A, Mincer TJ, DeLong EF. Proteorhodopsin lateral gene transfer between marine planktonic Bacteria and Archaea. Nature. 2006;439:847–850. doi: 10.1038/nature04435. [DOI] [PubMed] [Google Scholar]
- 15.Venter JC, et al. Environmental genome shotgun sequencing of the Sargasso Sea. Science. 2004;304:66–74. doi: 10.1126/science.1093857. [DOI] [PubMed] [Google Scholar]
- 16.Putonti C, et al. A computational tool for the genomic identification of regions of unusual compositional properties and its utilization in the detection of horizontally transferred sequences. Mol Biol Evol. 2006;23:1863–1868. doi: 10.1093/molbev/msl053. [DOI] [PubMed] [Google Scholar]
- 17.Bouige P, Laurent D, Piloyan L, Dassa E. Phylogenetic and functional classification of ATP-binding cassette (ABC) systems. Curr Protein Pept Sci. 2002;3:541–559. doi: 10.2174/1389203023380486. [DOI] [PubMed] [Google Scholar]
- 18.Audic S, et al. Genome analysis of Minibacterium massiliensis highlights the convergent evolution of water-living bacteria. PLoS Genet. 2007;3:e138. doi: 10.1371/journal.pgen.0030138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jann A, Matsumoto H, Haas D. The fourth arginine catabolic pathway of Pseudomonas aeruginosa. J Gen Microbiol. 1988;134:1043–1053. doi: 10.1099/00221287-134-4-1043. [DOI] [PubMed] [Google Scholar]
- 20.Kobayashi K, et al. Essential Bacillus subtilis genes. Proc Natl Acad Sci USA. 2003;100:4678–4683. doi: 10.1073/pnas.0730515100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pushker R, Mira A, Rodriguez-Valera F. Comparative genomics of gene-family size in closely related bacteria. Genome Biol. 2004;5:R27. doi: 10.1186/gb-2004-5-4-r27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Su Z, et al. Computational inference of regulatory pathways in microbes: An application to phosphorus assimilation pathways in Synechococcus sp. WH8102. Genome Inform. 2003;14:3–13. [PubMed] [Google Scholar]
- 23.Seshadri R, Kravitz SA, Smarr L, Gilna P, Frazier M. CAMERA: A community resource for metagenomics. PLoS Biol. 2007;5:e75. doi: 10.1371/journal.pbio.0050075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mira A, Pushker R. The silencing of pseudogenes. Mol Biol Evol. 2005;22:2135–2138. doi: 10.1093/molbev/msi209. [DOI] [PubMed] [Google Scholar]
- 25.Mira A, Pushker R, Rodriguez-Valera F. The Neolithic revolution of bacterial genomes. Trends Microbiol. 2006;14:200–206. doi: 10.1016/j.tim.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 26.Desnues C, et al. Biodiversity and biogeography of phages in modern stromatolites and thrombolites. Nature. 2008;452:340–343. doi: 10.1038/nature06735. [DOI] [PubMed] [Google Scholar]
- 27.Falcon LI, Cerritos R, Eguiarte LE, Souza V. Nitrogen fixation in microbial mat and stromatolite communities from Cuatro Cienegas. Mexico Microb Ecol. 2007;54(2):363–373. doi: 10.1007/s00248-007-9240-3. [DOI] [PubMed] [Google Scholar]
- 28.Hartel H, Dormann P, Benning C. DGD1-independent biosynthesis of extraplastidic galactolipids after phosphate deprivation in Arabidopsis. Proc Natl Acad Sci USA. 2000;97:10649–10654. doi: 10.1073/pnas.180320497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schmidt HA, Strimmer K, Vingron M, von Haeseler A. TREE-PUZZLE: Maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics. 2002;18:502–504. doi: 10.1093/bioinformatics/18.3.502. [DOI] [PubMed] [Google Scholar]
- 30.Battistuzzi FU, Feijao A, Hedges SB. A genomic timescale of prokaryote evolution: Insights into the origin of methanogenesis, phototrophy, and the colonization of land BMC. Evol Biol. 2004;4:44. doi: 10.1186/1471-2148-4-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kumar S, Tamura K. Nei M MEGA3: Integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- 32.Schwede T, Kopp J, Guex N, Peitsch MC. SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Res. 2003;31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.