Microbes employ several catalytic strategies to transform conformationally flexible peptide chains into rigidified scaffolds that possess antibiotic or toxin activity. Prominent examples include the biosynthesis of the β-lactam antibiotics of the penicillin and cephalosporin families (1) and the maturation of vancomycin (2) where distinct structural modifications to the nascent peptide chains confer physiological function. In this issue of PNAS, Lee et al. (3) provide the first insight into the chemical structure of streptolysin S (SLS), a hemolytic toxin produced by the human pathogen Streptococcus pyogenes. Its peptide backbone undergoes remarkable posttranslational tailoring, resulting in heterocycle formation and cytolytic activity. Lee et al. further show that a variety of prokaryotes harbor analogous maturation machinery, which suggests widespread use of heterocyclization for altering peptide shape/flexibility and creating functional toxins. This work builds on previous examples where enzymes morph peptide frameworks of both ribosomal and nonribosomal origin.
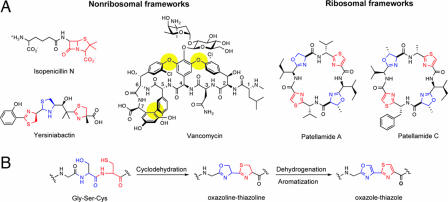
One famous strategy for constraining peptide flexibility and supplying antibiotic function is the bis-cyclization of the l-δ-(α-aminoadipoyl)–l-cysteinyl–d-valine (ACV) tripeptide to isopenicillin N by isopenicillin N synthetase (IPNS) in penicillin/cephalosporin biosynthesis (1). IPNS, a mononuclear nonheme Fe(II) oxygenase, creates the four–five fused ring system of isopenicillin N (Fig. 1) in one catalytic cycle. Formation of the five-membered thiolane and four-membered β-lactam rings rigidifies the ACV tripeptide scaffold and affords a suicide substrate for peptidogylcan cross-linking transpeptidases that inhibits bacterial cell wall biosynthesis. Further tailoring of the isopenicillin N core provides the various penicillin and cephalosporin family members.
Fig. 1.
Nonribosomal and ribosomal heterocyclic peptides. (A) Isopenicillin N, yersiniabactin, and vancomycin are products of NRPS machinery. Patellamides A and C are ribsomally derived peptides. (B) Conversion of a Gly-Ser-Cys tripeptide into a oxazole–thiazole pair via an oxazoline–thiazoline intermediate. This two-step process of cyclodehydration and aromatization occurs in microcin, patellamide, and now SLS posttranslational modification.
An equally remarkable oxygen-based rigidification strategy occurs in the maturation of vancomycin, which contains a heptapeptide scaffold (Fig. 1) (2). The sequential action of three cytochrome P450-type oxygenases produces the dome-shaped architecture of the cross-linked vancomycin aglycone. The linkages formed include two aryl-ether connections between the side chains of residues 2–4 and 4–6, and a carbon–carbon bond between the aromatic side chains of residues 5–7. Cross-linking of the vancomycin heptapeptide is essential for its antibiotic function. The rigidified framework recognizes, binds to, and sequesters the N-acyl–d-Ala–d-Ala termini of immature peptidoglycan strands, and blockade of bacterial cell wall biosynthesis results.
The penicillins, cephalosporins, and vancomycins are prominent members of the nonribosomal family of antibiotics. They are synthesized in the cytoplasm on nonribosomal peptide synthetase (NRPS) assembly lines. Their amino acid sequences are determined by multimodular protein thiotemplating rather than by mRNA (4). Another hallmark of nonribosomal peptide synthesis logic is the heterocyclization of X-Cys and X-Ser dipeptide moieties, which occurs during thiotemplated peptide chain elongation. Cyclodehydration of X-Cys and X-Ser dipeptides forms thiazolines and oxazolines, respectively, and also rigidifies the peptide backbone. The five-membered thiazoline and oxazoline rings are commonly found in siderophores, high-affinity Fe(III) chelators produced by some periods during periods of nutrient deprivation, where they provide (now) basic nitrogen donor atoms for Fe(III) coordination (5, 6). Further enzymatic tailoring of such cyclized peptide backbones alters their redox state and function. For instance, reduction of thiazoline to thiazolidine occurs in the maturation of the siderophore yersiniabactin (Fig. 1) (7), and oxidation of thiazoline rings to planar aromatic thiazoles occurs in the tailoring of the antitumor antibiotic bleomycin (8). During its maturation, a Cys-Cys residue pair is cyclodehydrated and oxidized to a bithiazole moiety, which is a DNA intercalator. Resculpting of the Cys-Cys dipeptide into the planar, intercalating bithiazole constitutes remarkable re-engineering of the bleomycin peptide backbone.
Cyclodehydration of X-Cys and X-Ser peptide linkages is not restricted to peptides synthesized on NRPS assembly lines. Prokaryotic ribosomal protein products can undergo the same types of modification. The best-studied example of a ribosomal protein-to-heterocycle morphing system is the posttranslational modification of McbA, a 69-residue pro-toxin produced by some types of enterobacteria, which affords the peptide toxin microcin B17 (MccB17) (9–12). Three tailoring enzymes, McbBCD, modify the nascent peptide and create four thiazole and four oxazole moieties from six glycines, four serines, and four cysteines. In vitro characterization of the McbBCD proteins verified cyclodehydration and desaturation activity, and validated the two-step process of Gly-Cys/Gly-Ser dipeptide cyclodehydration followed by flavoprotein-mediated desaturation/aromatization (Fig. 1). After heterocyclization, removal of the first 26 residues by a signal peptidase occurs. Of the 43 residues present in the mature toxin, 14 of them are used to generate the eight heterocycles, which include a Gly-Ser-Cys moiety that yields an oxazole–thiazole pair. Mature MccB17 targets DNA gyrase and inhibits DNA replication.
The mcbABCD genes, encoding the substrate protein and the three posttranslational tailoring enzymes, are clustered. This gene organization is preserved in other systems that morph ribosomal peptides into heterocycles (3, 13). The microcin B17 operon therefore serves as paradigm for several recently discovered examples of ribosomal protein tailoring that occur during maturation.
Of special note is the work of Schmidt et al. (13) to trace the origins of the patellamides (Fig. 1), heterocycle-containing cyclic peptides, isolated from didemnid extracts. Patellamides A and C are octapeptides that each contain two thiazole and two oxazoline rings. They arise from a ribosomally synthesized 71-residue precursor protein, which undergoes proteolytic cleavage, macrocyclization, epimerization, heterocyclization, and dehydrogenation to yield the active cytotoxins. The pat gene cluster, identified during the sequencing of the Prochloron didemni genome, contains seven genes patA–G. Several of these genes encode for the precursor protein (patE), a protease (patA) for cleavage of PatE, and tailoring enzymes (patDG) responsible for formation of the thiazole and oxazoline rings from X-Cys and X-Ser dipeptides. The proposed maturation steps, which involve cyclodehydration and aromatization, are reminiscent of those enacted by the mcbBCD gene products in MccB17 maturation. The Schmidt group suggests that the patellamide morphing strategy may be general for the conversion of other cyanobacterial pro-proteins into peptide heterocycles. These analyses augur that other cyclic thiazole-containing peptide antibiotics, including thiostrepton (14) and GE2270 (15), may arise from similar posttranslational modifications.
The contribution of Lee et al. (3) reported in this issue of PNAS can be placed in the context delineated above. Their study addresses streptolysin S (SLS), a hemolytic toxin and virulence factor from the human pathogenic bacterium Streptococcus pyogenes, which is responsible for human infections that range from pharyngitis to life-threatening necrotizing fasciitis (16). Despite a long-standing interest in its mechanism of action, the structure of SLS has remained obscure for decades. The current work describes a significant advance, the cloning of a SLS-associated gene locus (17) with the organization sagABCD. Three of the protein products, SagBCD, have homology with McbBCD from the MccB17 gene cluster. SagB shares homology with McbC (flavin-dependent dehydrogenase), SagC with McbB (zinc-dependent cyclodehydratase), and SagD with McbD (ATPase). The sagA gene encodes the 53-residue pro-toxin.
Heterocycles are a recurring motif in Nature's medicinal chemistry toolbox.
As it happens, the structure of mature SLS remains unknown. Because Lee et al. (3) were unable to detect SagA by mass spectrometry, they used McbA, the 69-residue MccB17 precursor protein, to assay SagBCD activity. The SagBCD complex processes McbA and installs up to four heterocycles into its framework. SagBCD could also convert a maltose binding protein fusion of SagA into a cytolytic product. These studies indicate that SagBCD will convert SagA into a thiazole- and/or oxazole-containing membrane-disrupting toxin. This work sets the stage for the in vitro scale-up and isolation of unfused SLS to determine the number and placement of heterocycles in its peptide backbone.
Bioinformatic analysis of other prokaryotic genomes conducted by Lee et al. (3) indicates the presence of homologous operons in Clostridium botulinum, Listeria monocytogenes, and Staphylococcus aureus RF122, among others. Yet to be elucidated are the circumstances in which these pathogens express and morph the pro-proteins into heterocyclic peptides and whether the mature toxins have physiological targets beyond cell membrane disruption (e.g., SLS) and DNA gyrase (e.g., MccB17).
Given the cyanobacterial studies of Schmidt et al. (13) and the current work from Jack Dixon's group on SLS (3), the posttranslational resculpting of peptide backbones into planar heterocyclic frameworks appears to be much more widespread than initially appreciated. Because some of the predicted gene clusters for heterocyclic metabolites contain additional kinds of tailoring enzymes (e.g., acetyltransferases and methyltransferases) (3), we anticipate that Nature will further morph the peptide frameworks and peripheries to achieve potent and target-specific toxins.
Heterocycles are a recurring motif in Nature's medicinal chemistry toolbox of bioactive secondary metabolites. Further investigations of the recently uncovered gene clusters for heterocyclic peptide biosynthesis, in addition to the discovery of new tailoring systems, will help elucidate the mechanisms of action of these toxins/antibiotics. The lessons gained from such endeavors will also provide a guide for the combinatorial biosynthesis of novel variants to optimize the future generations of antibiotics.
Footnotes
The authors declare no conflict of interest.
See companion article on page 5879.
References
- 1.Schofield CJ, et al. Proteins of the penicillin biosynthesis pathway. Curr Opin Chem Biol. 1997;7:857–864. doi: 10.1016/s0959-440x(97)80158-3. [DOI] [PubMed] [Google Scholar]
- 2.Hubbard BK, Walsh CT. Vancomycin assembly: Nature's way. Angew Chem Int Ed. 2003;42:730–765. doi: 10.1002/anie.200390202. [DOI] [PubMed] [Google Scholar]
- 3.Lee SW, et al. Discovery of a widely distributed toxin biosynthetic gene cluster. Proc Natl Acad Sci USA. 2008;105:5879–5884. doi: 10.1073/pnas.0801338105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fischbach MA, Walsh CT. Assembly-line enzymology for polyketide and nonribosomal peptide antibiotics: Logic, machinery, and mechanisms. Chem Rev. 2006;106:3468–3496. doi: 10.1021/cr0503097. [DOI] [PubMed] [Google Scholar]
- 5.Crosa JH, Walsh CT. Genetics and assembly line enzymology of siderophore biosynthesis in bacteria. Microb Mol Biol Rev. 2002;66:223–249. doi: 10.1128/MMBR.66.2.223-249.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miethke M, Marahiel MA. Siderophore-based iron acquisition and pathogen control. Microb Mol Biol Rev. 2007;71:413–451. doi: 10.1128/MMBR.00012-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drechsel H, et al. Structure elucidation of yersiniabactin, a siderophore from highly virulent yersinia strains. Liebigs Ann Org Bioorg Chem. 1995;10:1727–1733. [Google Scholar]
- 8.Schneider TL, Shen B, Walsh CT. Oxidase domains in epothilone and bleomycin biosynthesis: Thiazoline to thiazole oxidation during chain elongation. Biochemistry. 2003;42:9722–9730. doi: 10.1021/bi034792w. [DOI] [PubMed] [Google Scholar]
- 9.Li YM, Milne JC, Madison LL, Kolter R, Walsh CT. From peptide precursors to oxazole and thiazole-containing peptide antibiotics: Microcin B17 synthase. Science. 1996;274:1188–1193. doi: 10.1126/science.274.5290.1188. [DOI] [PubMed] [Google Scholar]
- 10.Milne JC, Eliot AC, Kelleher NL, Walsh CT. ATP/GTP hydrolysis is required for oxazole and thiazole biosynthesis in the peptide antibiotic microcin B17. Biochemistry. 1998;37:13250–13261. doi: 10.1021/bi980996e. [DOI] [PubMed] [Google Scholar]
- 11.Milne JC, et al. Cofactor requirements and reconstitution of microcin B17 synthetase: A multienzyme complex that catalyzes the formation of oxazoles and thiazoles in the antibiotic microcin B17. Biochemistry. 1999;38:4768–4781. doi: 10.1021/bi982975q. [DOI] [PubMed] [Google Scholar]
- 12.Kelleher NL, Hendrickson CL, Walsh CT. Posttranslational heterocyclization of cysteine and serine residues in the antibiotic microcin B17: Distributivity and directionality. Biochemistry. 1999;38:15623–15630. doi: 10.1021/bi9913698. [DOI] [PubMed] [Google Scholar]
- 13.Schmidt EW, et al. Patellamide A and C biosynthesis by a microcin-like pathway in Prochloron didemni, the cyanobacterial symbiont of Lissoclinum patella. Proc Natl Acad Sci USA. 2005;102:7315–7320. doi: 10.1073/pnas.0501424102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anderson B, Hodgkin DC, Viswamitra MA. The structure of thiostrepton. Nature. 1970;225:233–235. doi: 10.1038/225233a0. [DOI] [PubMed] [Google Scholar]
- 15.Kurz M, Sottani C, Bonfichi R, Lociuro S, Selva E. Revised structure of the antibiotic GE 2270A. J Antibiot. 1994;47:1564–1567. doi: 10.7164/antibiotics.47.1564. [DOI] [PubMed] [Google Scholar]
- 16.Datta V, et al. Mutational analysis of the group A streptococcal operon encoding streptolysin S and its virulence role in invasive infection. Mol Microbiol. 2005;56:681–695. doi: 10.1111/j.1365-2958.2005.04583.x. [DOI] [PubMed] [Google Scholar]
- 17.Nizet V, et al. Genetic locus for streptolysin S production by group A streptococcus. Infect Immun. 2000;68:4245–4254. doi: 10.1128/iai.68.7.4245-4254.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]