Abstract
Environmental and genetic factors can modulate aggressiveness, but the biological mechanisms underlying their influence are largely unknown. Social experience with conspecifics suppresses aggressiveness in both vertebrate and invertebrate species, including Drosophila. We searched for genes whose expression levels correlate with the influence of social experience on aggressiveness in Drosophila by performing microarray analysis of head tissue from socially isolated (aggressive) vs. socially experienced (nonaggressive) male flies. Among ≈200 differentially expressed genes, only one was also present in a gene set previously identified by profiling Drosophila strains subjected to genetic selection for differences in aggressiveness [Dierick HA, Greenspan RJ (2006) Nat Genet 38:1023–1031]. This gene, Cyp6a20, encodes a cytochrome P450. Social experience increased Cyp6a20 expression and decreased aggressiveness in a reversible manner. In Cyp6a20 mutants, aggressiveness was increased in group-housed but not socially isolated flies. These data identify a common genetic target for environmental and heritable influences on aggressiveness. Cyp6a20 is expressed in a subset of nonneuronal support cells associated with pheromone-sensing olfactory sensilla, suggesting that social experience may influence aggressiveness by regulating pheromone sensitivity.
Keywords: aggression, cytochrome P450, pheromone
Aggression is critical for the survival and reproduction of many animal species (1–3). Although aggression is an innate behavior subject to genetic influences, levels of aggressiveness are subject to environmental modifications as well. An important unanswered question is whether these influences act by independent or shared biological mechanisms. Although genes underlying heritable differences in aggressiveness are beginning to be identified (4, 5), very little is known about the molecular mechanisms underlying environmental influences on aggression.
Environmental influences on aggressiveness have been well documented in a variety of animal models. Social status established by previous agonistic experience influenced subsequent aggression-related behavior in crayfish (6) and crickets (7). Resident female Mediterranean fruit flies (Ceratitis capitata) located at a resource had a higher probability of defeating an intruder, suggesting that experience on a resource may increase aggressiveness (8). Male fruit flies (Drosophila melanogaster and Drosophila simulans) raised at high density failed to successfully defend their territories against males raised at low density, an effect potentially related to differences in body size (9).
Social experience with conspecifics is one environmental influence on aggressiveness that is common to many species, including humans (3). Socially isolated male mice are more aggressive than group-housed males (10). Similar phenomena have been reported in rats (11), cichlid fish (Haplochromis burtoni) (12), and other vertebrate species. Effects of social experience on aggressiveness have also been described in invertebrates. Hoffman (13) reported that male D. melanogaster held in isolation exhibited more aggressive behaviors and required less time to establish their territories than males held in groups, suggesting that social experience suppresses aggressiveness in the fruit flies. Analogous observations have also been reported for female D. melanogaster (14).
Taken together, these data suggest that the effect of social experience on aggressiveness is shared among many species. However, the nature of the molecular mechanisms mediating this effect and whether they are evolutionarily conserved are poorly understood. Here, we have used fruit flies, D. melanogaster, a genetically tractable organism in which aggression has been well characterized (15–17) to investigate the molecular basis of the influence of social experience on aggressiveness.
Results
Group Housing Suppresses Aggressiveness in a Reversible Manner.
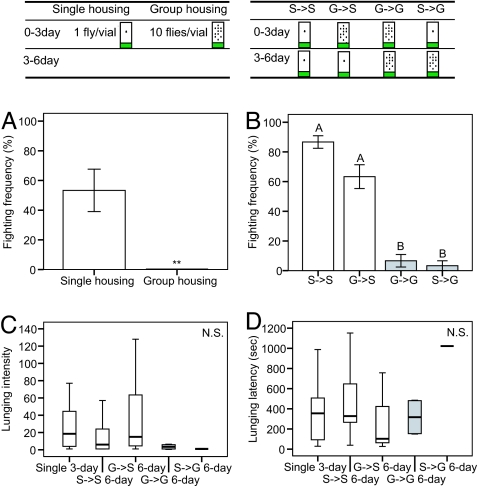
Flies raised in isolation after eclosion are more aggressive than those raised in groups (13). To quantify more easily this behavioral difference, we modified a fight chamber (18) to permit multiplex analysis of aggressiveness [supporting information (SI) Fig. S1]. A pair of male flies of similar age and social experience (raised in isolation shortly after eclosion or in groups of 10 male flies for 3 days before the test) was transferred into a fighting arena containing a small food patch. Consistent with earlier reports (17), we observed that lunging behavior, in which one fly rears up on its hind legs and charges the other fly, was the predominant form of aggression (18). Therefore, we counted lunges as a measure of aggressiveness during a 20-min observation period. Three different parameters were measured: (i) the fighting frequency, defined as the percentage of fly pairs that exhibited at least one lunge (Fig. 1 Lower A and B); (ii) the lunging intensity, defined as the average number of lunges, calculated for all pairs that exhibited at least one lunge (Fig. 1 Lower C); and (iii) the average latency to the first lunge (Fig. 1 Lower D). Flies single-housed for 3 days exhibited a mean fighting frequency of ≈50% (53.3 ± 14.3%) during the 20-min observation period, whereas group-housed flies did not exhibit any lunges (Fig. 1 Lower A, P < 0.01). A similar difference was observed between flies group- or single-housed for 6 days (but transferred to a new vial at day 3) (Fig. 1 Lower B; G → G vs. S → S, P < 0.05). These data confirm that, in flies, as in other animals including mice, social experience suppresses aggressiveness.
Fig. 1.
Social experience influences Drosophila aggressiveness. (Upper) Experimental manipulations. (Lower) (A) Mean fighting frequencies in 3-day-old, single-housed and group-housed flies (mean ± SEM; n = 6 experiments each containing five fly pairs; ∗∗, P < 0.01). (B) Mean fighting frequencies in 6-day-old, S → S, G → S, G → G, and S → G flies (mean ± SEM. n = 6 experiments each containing five pairs. Significant differences (P < 0.05) are indicated by letters above bars). (C) Median lunging intensities of 3-day-old, single-housed flies, and 6-day-old, S → S, G → S, G → G, and S → G flies [n = 16, 25, 19, 2, and 1 pairs, respectively; N.S., not significantly different (P > 0.05)]. (D) Median lunging latencies of 3-day-old, single-housed flies and 6-day-old, S → S, G → S, G → G, and S → G flies [n = 16, 25, 19, 2, and 1 pairs, respectively; N.S. (P > 0.05)]. Comparisons between groups were made by using the Mann–Whitney U test (A), ANOVA followed by post hoc test (B) and Kruskal–Wallis ANOVA (C and D).
The effect of social experience on aggressiveness was reversible. When flies were single-housed for 3 days, followed by 3 days of group housing before the test, the fighting frequency was as low as that of flies group-housed for 6 days (Fig. 1 Lower B, G → G vs. S → G, P > 0.05). Conversely, when flies were group-housed for 3 days, followed by 3 days of single housing, their aggressiveness was approximately as high as that of flies single-housed for 6 days (Fig. 1 Lower B, S → S vs. G → S, P > 0.05). The median lunging intensity and latency among pairs exhibiting at least one lunge were not significantly different across all social conditions (Fig. 1 Lower C and D, P > 0.21, P > 0.12).
Cyp6a20 Shows Differential Expression Levels in Single- vs. Group-Housed Flies.
To investigate the molecular basis of social influences on aggressiveness, we performed comparative gene expression profiling on heads from 6-day-old group- vs. single-housed male flies. Using criteria of fold change >1.25 and P < 0.002, we identified 141 probe sets that were up-regulated and 48 probe sets that were down-regulated in single- vs. group-housed males (Dataset S1). The differentially expressed genes fell into diverse ontological and biological categories, including neurotransmitter metabolism, immunity, and olfaction.
While this study was underway, a report (4) appeared describing a microarray comparison, using head mRNA, between Drosophila strains selected for increased aggressiveness (AggrI and AggrII) and strains selected for decreased aggressiveness (NeutrI and NeutrII). Multiple differentially expressed genes were identified, which reflect allelic variations underlying heritable differences in aggressiveness. To determine whether there is any commonality in the molecular mechanisms through which aggressiveness is modified by heritable and environmental factors, we compared the differentially expressed genes identified in our social experience experiments, with those identified by selective breeding. Applying the same criteria (fold change >1.25; P < 0.002), we identified Cyp6a20, a cytochrome P450 gene, as the only gene similarly regulated in both datasets. Cyp6a20 was expressed at relatively lower levels both in AggrI&II vs. NeutrI&II (4) and socially isolated vs. group-housed flies. An independent microarray experiment identified many genes differentially expressed in flies selected for increased aggressiveness, but Cyp6a20 was not among them (5). However, that study differed in several important details from the methods used in our own experiments and those described in ref. 4, including the conditions under which flies were tested and the criteria used to define aggressive behavior.
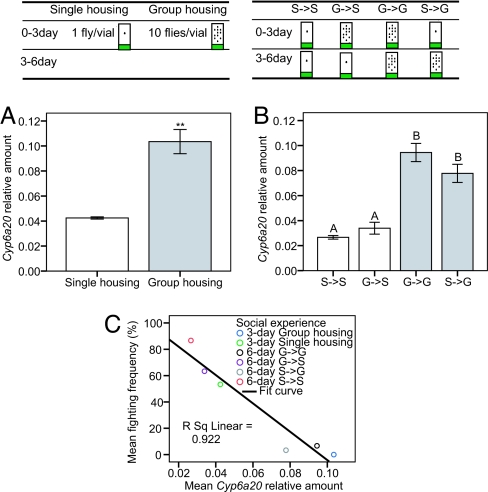
These data led us to further investigate the role of Cyp6a20 in the effect of social experience on aggressiveness. We first confirmed the correlation between Cyp6a20 expression levels and social experience, using quantitative RT-PCR. Cyp6a20 expression was almost 3-fold lower in flies single-housed for 3 days than in group-housed flies of same age (Fig. 2 Lower A, P < 0.01). Furthermore, in flies switched from single housing to group housing after 3 days or vice versa, the levels of Cyp6a20 expression changed in parallel with, but in the opposite direction to, the changes in aggressiveness caused by these social manipulations (Fig. 2 Lower B). There is, on one hand, a positive correlation between levels of Cyp6a20 expression and social experience (Fig. 2 Lower A and B) and, on the other hand, a negative correlation between social experience and aggressiveness (Fig. 1), suggesting the Cyp6a20 expression levels might be negatively correlated with aggressiveness. Indeed, a plot of Cyp6a20 mRNA levels vs. aggressiveness was well fit by a linear regression function, with a correlation coefficient equal to −0.96 (Fig. 2 Lower C).
Fig. 2.
Cyp6a20 expression is correlated with social experience. (Upper) Experimental manipulations. (Lower) (A) Relative levels of Cyp6a20 mRNA (normalized to Ddc mRNA levels) in 3-day-old, single-housed and group-housed flies (mean ± SEM; n = 4; ∗∗, P < 0.01). (B) Relative levels of Cyp6a20 mRNA in 6-day-old, S → S, G → S, G → G, and G → S flies (mean ± SEM; n = 4; significant differences are indicated by letters above each bar). (C) Negative correlation between relative levels of Cyp6a20 mRNA and fighting frequency. The linear regression plot (R2 = 0.922) is compiled by using the data in A and B and Fig. 1 Lower A and B. Comparisons between groups were made by using student's t test (A), or ANOVA followed by a post hoc test (B).
Cyp6a20 Mediates the Suppressive Effect of Group Housing on Aggressiveness.
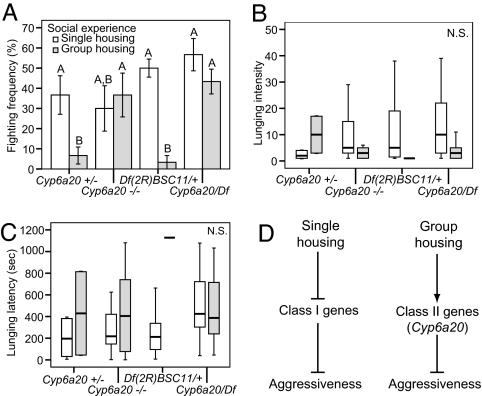
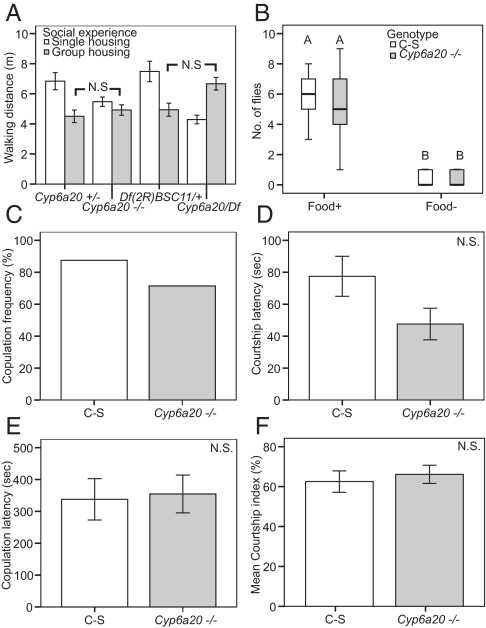
If Cyp6a20 plays a role in mediating the effect of social experience, then flies deficient in Cyp6a20 should exhibit higher fighting frequencies under group-housing but not single-housing conditions. Indeed, group-housing flies bearing a homozygous P-element insertion in the Cyp6a20 locus showed a significantly higher fighting frequency than group-housed Cyp6a20+/− flies (Fig. 3A, gray bars; Cyp6a20−/− vs. Cyp6a20+/−, P < 0.05), whereas there was no significant difference between these genotypes under single-housing conditions (Fig. 3A, white bars, Cyp6a20−/− vs. Cyp6a20+/−, P > 0.05). Furthermore, the fighting frequency of group-housed Cyp6a20−/− mutant flies was as high as that of single-housed Cyp6a20−/− mutant flies, whereas heterozygous Cyp6a20+/− flies (like wild-type Canton-S flies) showed a significantly reduced fighting frequency under group-housing conditions (Fig. 3A). There was no statistically significant difference in locomotor activity between Cyp6a20+/− and Cyp6a20−/− flies under group-housing conditions as measured by the total distance traveled during a 20-min filming period (Fig. 4A). Furthermore, group-housed Cyp6a20−/− flies exhibited normal odor-guided behavior (Fig. 4B) and normal courtship behavior toward wild-type virgin females (Fig. 4 C–F), arguing that the mutation in Cyp6a20 does not cause general deficits in olfaction or social behavior.
Fig. 3.
Cyp6a20 mutants exhibit increased aggressiveness only under group-housing conditions. (A) Mean fighting frequencies of 6-day-old, single-housed and group-housed flies, of the indicated genotypes [mean ± SEM; n = 6 experiments each containing five pairs; significant differences (P < 0.05) are indicated by letters above the bars]. Comparison between groups was made by using ANOVA followed by a post hoc test. (B) Median lunging intensities of 6-day-old, single- or group-housed flies of the indicated genotypes (n = 10, 2, 9, 11, 15, 1, 17, 13 pairs, respectively; N.S., P > 0.05). (C) Median lunging latencies of 6-day-old, single- or group-housed flies of different genotypes (n = 10, 2, 9, 11, 15, 1, 17, 13 pairs, respectively; N.S., P > 0.05). Comparisons between groups were made by using Kruskal-Wallis ANOVA. (D) Two classes of negative genetic regulators of aggressiveness and their interaction with social experience. Class I genes, when mutated, increase aggressiveness under single-housing conditions, where their expression levels are normally relatively lower, but this phenotype is not observed in group-housing. Class II genes, when mutated, increase aggressiveness under group- but not single-housing conditions, overriding the effect of social experience to suppress aggressiveness. Cyp6a20 is a class II gene.
Fig. 4.
Group-housed Cyp6a20 mutants exhibit normal locomotor, olfactory and courtship behavior. (A) Mean walking distances of 6-day-old, single-housed and group-housed flies of the indicated genotypes (n = 20; N.S., P > 0.05). Comparisons between groups were made by using ANOVA followed by post hoc test. (B) Median number of flies of the indicated genotypes (n = 15) trapped in food-containing (food+) vs. empty (food−) traps. Significant differences are indicated by letters above the boxes). Comparisons between groups were made by using Kruskal–Wallis ANOVA followed by a post hoc test. (C–F) Cyp6a20 mutants have normal courtship behavior. (C) Percentage of fly pairs of the indicated genotypes that copulated in 30 min. (D) Mean courtship latency of flies of the indicated genotypes (n = 21 and 20, respectively; N.S., P > 0.05). (E) Mean copulation latency of flies of the indicated genotypes (n = 21 and 20, respectively; N.S., P > 0.05). (F) Mean courtship index of flies of the indicated genotypes (n = 21 and 20, respectively; N.S., P > 0.05). For D–F, comparisons between groups were made by using Student's t test.
To confirm that the selective increase in aggressiveness under group-housing conditions was indeed caused by the P-element insertion in the Cyp6a20 locus, we tested the Cyp6a20 insertion over a deficiency spanning the Cyp6a20 gene, Df(2R)BSC11 (4). Like Cyp6a20−/− flies, Cyp6a20Df/− mutant flies showed a significantly higher fighting frequency than Cyp6a20Df/+ hemizygous flies under group-housing conditions (Fig. 3A, gray bars; Df(2R)BSC11/+ vs. Cyp6a20/Df, P < 0.05), whereas there was no significant difference between these genotypes under single-housing conditions (Fig. 3A, white bars; Df(2R)BSC11/+ vs. Cyp6a20/Df, P > 0.05). In addition, the fighting frequency of Cyp6a20Df/− flies under group-housing conditions was as high as that under single-housing conditions (Fig. 3A). There was no statistically significant difference in locomotor activity between Cyp6a20Df/+ and Cyp6a20Df/− flies under group-housing conditions (Fig. 4A). The lunging intensity and latency remained unchanged in all of the four genotypes (Fig. 3B and C, P > 0.11; P > 0.08). Previous experiments have shown that the levels of Cyp6a20 mRNA in Cyp6a20−/− and Cyp6a20Df/− flies are only 8–15% of those in Cyp6a20+/− and Cyp6a20Df/+ flies (4). Taken together, these data suggest that the phenotype of the P-element insertion indeed reflects a reduction in Cyp6a20 expression or function, although rescue experiments will be required to formally confirm this. Thus, in flies with reduced levels of Cyp6a20 expression, group housing is much less effective in suppressing aggressiveness (for more detailed analysis, see Fig. S2 and SI Text). These genetic data, when taken together with the observation that Cyp6a20 mRNA levels are up-regulated by group housing (Fig. 2 Lower A and B), suggest that Cyp6a20 mediates the effect of social experience to suppress aggressiveness (Fig. 3D).
Cyp6a20 Is Specifically Expressed in a Subset of Olfactory Sensory Organs.
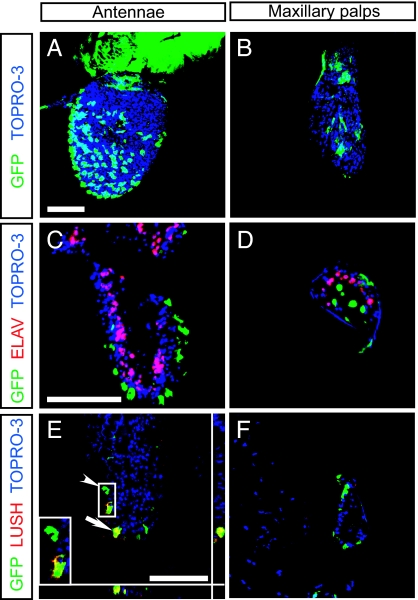
As an initial step toward investigating the mechanism of action of Cyp6a20, we investigated where the gene is expressed, using an enhancer trap line, P[GawB]NP2593, in which Gal4 is integrated into the Cyp6a20 locus. In P[GawB]NP2593/UAS-mCD8GFP adult flies, the reporter was expressed in the antennae and maxillary palps, the two main Drosophila olfactory sensory organs (Fig. 5 A and B). Scattered GFP signal was also seen in the brain (data not shown). Thus, like other P450 genes described in Drosophila (19), Cyp6a20 appears antennal-enriched. We wished to verify that the Gal4-targeted GFP expression faithfully recapitulated the expression pattern of endogenous Cyp6a20 mRNA. However, the small number and inaccessibility of GFP+ cells in the antennae precluded a comparison of Cyp6a20 mRNA levels between GFP+ and GFP− cells, and Cyp6a20 expression was undetectable by in situ hybridization (data not shown). We therefore examined the expression of Cyp6a20 mRNA in larvae, where strong GFP expression was observed specifically in the salivary gland of P[GawB]NP2593/UAS-mCD8GFP specimens (Fig. S3A). RT-PCR experiments performed on larval tissues confirmed that Cyp6a20 transcripts were enriched in salivary gland (Fig. S3B). Thus, the P[GawB]NP2593 insertion in Cyp6a20 correctly reports expression in larval tissues, making it reasonably likely that the same holds true for the adult, although further experiments will be required to confirm this.
Fig. 5.
Cyp6a20 expression in olfactory sensory organs. (A and B) GFP expression in the antennae (A) and palps (B). Whole-mount antennae and palps were stained with rabbit anti-GFP (green). (C and D) GFP+ cells are nonneuronal. Frozen sections were stained with rabbit anti-GFP (green) and rat anti-ELAV (red). (E and F) a subset of GFP+ cells in the antennae (E), but not in the palps (F), coexpressed LUSH. Frozen sections were stained with chick anti-GFP (green) and rabbit anti-LUSH (red). Confirmation of colabeling of one cell (white arrow) by z-series analysis is shown below and to the right of E. Inset in E is a higher magnification view of the boxed region (arrowhead), illustrating GFP+ LUSH− and GFP+ LUSH+ cells. In all images, TOPRO-3 (blue) was used for nuclear staining. (Scale bars, 50 μm.)
Intriguingly, the GFP+ cells in the adult antennae were preferentially if not exclusively associated with trichoid sensilla, which are thought to be involved in pheromone detection (20). Such trichoid sensillar-specific expression has been described for two P450 genes in the moth Mamestra brassicae (21) but has not been reported in Drosophila. More surprisingly, although it is often assumed that antennal-specific P450 enzymes are expressed by olfactory receptor neurons, GFP+ cells in both the antennae and palps did not coexpress ELAV, a neuronal marker (22) (Fig. 5 C and D). However, a subset of GFP+ cells coexpressed LUSH (Fig. 5 E and F), an odorant binding protein that marks a subpopulation of nonneuronal support cells (23). Thus, Cyp6a20 is a P450 gene specifically expressed in a subset of olfactory sensory support cells.
Discussion
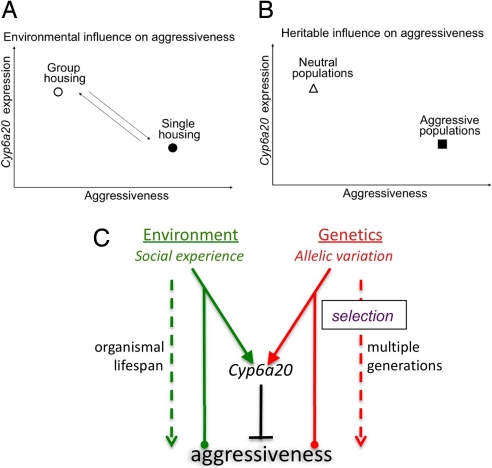
Both genes and environment can influence aggressiveness; however, whether there is a commonality to the underlying biological mechanisms has not been clear. Using Drosophila as a model system, we show that an evolutionarily conserved environmental influence on aggressiveness, social experience, is associated with changes in gene expression. Detailed analysis of one of the regulated transcripts, Cyp6a20, indicates that it is up-regulated by social experience in a manner that correlates with the effects of social experience to suppress aggressiveness. Genetic experiments confirm that Cyp6a20 is a negative regulator of aggressiveness (4) but reveal that its influence is only observed under conditions of group housing, where its expression is relatively higher. These data suggest that Cyp6a20 is required to mediate the effect of group housing to suppress aggressiveness. Cyp6a20 was the only gene in our dataset in common with a set identified in an independent expression profiling analysis of Drosophila populations selected for differential levels of aggressiveness (4). Taken together, these data suggest that Cyp6a20 represents a common genetic target of heritable and environmental influences on aggressive behavior in fruit flies (Fig. 6C). Whether Cyp6a20 is the only such target remains to be determined.
Fig. 6.
Cyp6a20 is a common genetic target of environmental and heritable influences on aggressive behavior. (A) Social experience influences aggressiveness in a reversible manner (bidirectional arrows), mediated by differential expression of Cyp6a20. (B) Genetic selection over multiple generations establishes neutral and aggressive populations with differential levels of aggressiveness, which correlate with differential Cyp6a20 expression (4). (C) Cyp6a20 regulation constitutes a common molecular target of environmental and genetic influences on aggressiveness. Circular arrowheads indicate that both positive and negative influences are possible. Environmental influences act on a time scale of the lifespan of the organism (Left), whereas genetic influences act over multiple generations as a consequence of selection (natural or artificial) (Right).
Social Experience Influences Aggressiveness by Regulating Gene Expression.
Prior social experience with conspecifics influences numerous aspects of animal behavior. Social isolation causes behavioral abnormalities in rodents, including anxiety and hyperaggressiveness (10, 24, 25). D. melanogaster reared as groups exhibit circadian rhythm coherence (26), and longer periods of daytime sleep (27). Social experience has also been shown to regulate courtship behavior in fruit flies (28), as well as aggression (13, 14). Further investigations are required to determine the extent to which the effects of social experience on these different behaviors are mediated by divergent or common pathways.
In cichlid fish, social interactions can regulate the brain expression of genes encoding neuropeptides (29) and steroid hormone receptors (30), but it has been difficult to extend such observations from correlation to causality. Here, we have demonstrated that extended male-male social interactions regulate gene expression in Drosophila, a system affording facile genetic manipulations. Recent studies have shown that rapid-onset changes in gene expression accompany male–female courtship in Drosophila (31). (We have not yet examined the influence of male–female interactions in group housing on aggressiveness.) Further investigation of these genes may lead to a more comprehensive understanding of the effect of social experience on animal behaviors.
Under our stringency conditions, we identified 141 probe sets exhibiting higher expression in socially isolated than in group-housed flies and 48 probe sets exhibiting higher expression under group-housing conditions. Because aggressiveness is higher in socially isolated flies (13), genes in the first category are candidate positive regulators of aggressiveness, whereas those in the second category are candidate negative regulators. Loss-of-function mutations in candidate negative regulators should increase aggressiveness, as shown for Cyp6a20 mutants (4). The present analysis confirms this but reveals that flies homozygous for a hypomorphic allele of Cyp6a20 only show increased aggressiveness under group-housing conditions.
Interestingly, flies bearing mutations in two other candidate negative regulators of aggressiveness that we identified in a similar analysis, using a different fly strain, exhibited increased aggressiveness under single-housing conditions, but their aggressiveness could still be suppressed by group housing (L.W. and D.J.A., unpublished observations). These data suggest that there are at least two classes of genes that negatively regulate aggressiveness and whose expression levels are relatively higher in group-housed compared with single-housed flies: (i) genes, such as Cyp6a20, a hypomorphic allele of which overrides the effect of group-housing to suppress aggressiveness but which does not increase aggressiveness under single-housed conditions (Fig. 3 A and D, class II); and (ii) genes such as those identified in our second screen, hypomorphic mutations in which cause increased aggressiveness under single-housing conditions but not under group-housing conditions (Fig. 3D, class I). In principle, a third category of negative regulators may promote constitutively increased aggressiveness when mutated, under both single- and group-housing conditions, but we have not yet identified exemplars of this class. Recent studies have implicated serotonin, octopamine, and neuropeptide F in the control of aggressiveness in Drosophila. Genes related to these neuromodulatory pathways were not among those identified in our screen (18, 32–34).
Environmental and Heritable Influences on Aggressiveness.
Genes can influence behavior both through polymorphic variation, on which natural selection can act, and by environmentally regulated changes in expression that occur within the lifetime of an individual. For example, naturally occurring polymorphisms in the foraging gene, which encodes a guanosine 3′, 5′-monophosphate (cGMP)-dependent protein kinase (PKG), cause modifications of feeding behavior in D. melanogaster (35), whereas developmental changes in the expression of its honey bee ortholog, Amfor, modulate feeding behavior during the life history of single individuals (36). It is not yet known whether naturally occurring polymorphisms in Cyp6a20 itself or rather in genes that encode upstream regulators of its expression underlie the differences in Cyp6a20 transcript levels between the Neutr and Aggr strains selected in ref. 4. Nevertheless our results, taken together with the genetic selection experiments in ref. 4, identify a common genetic target of environmental and heritable influences on aggressive behavior within a single species (Fig. 6).
Studies have shown that increased aggressiveness promotes enhanced mating success (37). This raises the question of why, if aggressive behavior provides a general selective advantage, the ability of social experience to suppress aggressiveness is not eventually lost over many generations and replaced by constitutively aggressive populations. One explanation is that there may be positive selection for the ability of social experience to suppress aggressiveness. For example, under conditions where food resources are scarce and flies tend to feed in groups, individuals engaged in ongoing aggressive activity, despite this enriched social experience, might divert their energy and attention from feeding and reproductive behavior, thereby reducing their likelihood of reproductive success.
Control of Aggressiveness by Social Regulation of Cytochrome P450s.
Social experience suppresses aggressiveness in many species, leaving open the question of whether the underlying molecular mechanisms are also conserved. Although Cyp6a20 does not have a clear vertebrate ortholog and appears to function in an insect-specific olfactory pathway (see the next paragraph), it encodes a cytochrome P450, which encompasses a large family of proteins with diverse enzymatic activities (38). In vertebrates, one member of this family is aromatase, which converts testosterone to estrogen and is required for inter-male aggressiveness (39, 40). Interestingly, the expression of brain aromatase has been shown to be regulated by social experience and other environmental influences (41, 42). Although flies lack testosterone, it is interesting that, in addition to Cyp6a20, five additional cytochrome P450 genes exhibited significant differential expression, by our criteria, between single-housing vs. group-housing conditions (Dataset S1). Thus, the general role of aromatases in mediating environmental influences on aggressiveness may be conserved, even if the pathways in which they act are not.
The antennal-specific expression of Cyp6a20, taken together with its up-regulation by group housing and its functional role as revealed by genetics, suggests that social experience may cause changes in olfactory sensitivity via regulation of Cyp6a20. Consistent with this interpretation, studies have shown that the effect of social experience on male-male interactions requires pheromonal perception (28). Whether changes in Cyp6a20 expression caused by social experience indeed influence pheromonal sensitivity and, if so, in what direction, is not yet clear. Pharmacologic inhibition of antennal-enriched cytochrome P450s eliminates pheromone sensitivity in some insects (43), suggesting a requirement for these proteins for maintaining olfactory sensitivity. If Cyp6a20 were a positive regulator of pheromone sensitivity, then its increased expression under group-housing conditions might enhance sensitivity to an aggression-suppressing pheromone, resulting in a lower level of aggressiveness. Such a model would be consistent with the suggestion that inhibitory pheromones are used to suppress male-male interactions (28). Alternatively, as suggested in ref. 4, Cyp6a20 may function to decrease sensitivity to an aggression-promoting pheromone. Distinguishing these hypotheses will require identification of the relevant pheromones and their functional role in aggression.
Materials and Methods
Fly Stocks and Rearing Conditions.
All fly stocks were reared in plastic vials containing yeast, corn syrup, and agar medium at 25°C, 60% humidity, and a 12-h light:12-h dark cycle. Newly eclosed males were reared either individually (single housing) or at 10 flies (group housing) per vial [2.4 cm (diameter) × 9.4 cm (height)] for 3 or 6 days before performing the behavioral assay. Wild-type Canton-S (CS) flies were used for all experiments unless otherwise indicated. Cyp6a20−/− was introgressed into the CS background from y1w67c23; P{y[+mDint2] w[BR.E.BR] = SUPor-P}KG04665 as described in ref. 4. Cyp6a20+/− flies were generated by crossing Cyp6a20−/− males with Canton-S females. Cyp6a20Df/+ (Df(2R)BSC11/+) flies were generated by crossing Df(2R)BSC11/SM6a males with Canton-S females. Cyp6a20Df/− (Cyp6a20/Df) flies were generated by crossing Df(2R)BSC11/SM6a males and Cyp6a20−/− females. P[GawB]NP2593 flies were from the Drosophila Genetics Resource Center, Kyoto Institute of Technology, Kyoto, Japan.
Aggression Assay.
A multiplex aggression apparatus containing five arenas was constructed as illustrated in Fig. S1. Two males of the same age and social experience but from different vials were introduced into each well by gentle aspiration without anesthesia. The behavior of five pairs was video-captured for 20 min and analyzed manually, by counting the number of lunges in each arena. Lunging behavior was defined as described in ref. 17. The temperature and humidity of the apparatus was set to ≈25°C and ≈40%, respectively. Aggression assays were performed between 5 p.m. and 12 a.m.
Microarray Analysis.
Three biological replicates were performed for both single-housing and group-housing conditions. For each replicate, 20 heads were isolated from 6-day-old, single-housed or group-housed male flies that were collected and frozen at 5 pm-6 pm on different days. Total RNA was prepared by using TRIzol as described in ref. 44. The following steps, including RNA quality test, reverse transcription, cRNA labeling, fragmentation and hybridization (Affymetrix Drosophila Genome Array 2.0) were performed by the Millard and Muriel Jacobs Genetics and Genomics Laboratory at California Institute of Technology. The raw data from all arrays are available online at http://www.ncbi.nlm.nih.gov/project/geo under series GSE6994.
Quantitative RT-PCR.
Four biological replicates were performed for each group indicated in Fig. 2. For each replicate, 20 heads were isolated from flies that were collected and frozen at 5 pm-6 pm on different days. Total RNA was prepared by using Qiagen RNeasy Micro Kit. For each biological replicate, three RT-PCR (technical) replicates were performed.
Other Behavioral Assays.
Locomotor activity of flies was measured by using a customized program written in Matlab (Dankert et al., in preparation). For each measurement, a pair of 6-day-old male flies was introduced into a fighting arena with a food patch and agar, videotaped for 20 min and analyzed. To facilitate the automatic tracking of fly trajectories, the fighting arena used here was square-shaped instead of round-shaped as Fig. S1. For food trap assay, male flies were group-housed (10 flies/vial) for 5 days and wet-starved for an additional day on 1% agar medium before the test. On the day of the test, 10 flies were carefully aspirated into a plastic cylinder with the same dimensions as the fighting arena (Fig. S1) containing two odor traps (one with food and one without food). The trap was made by inserting a P1000 pipette tip into a 5-ml glass serum bottle [1.6 cm (diameter) × 3.4 cm (height)], containing 1 ml of standard fly food, or, as a control, 1% agar. After 2 hours, the number of flies trapped in each trap was counted and analyzed. For courtship assay, male flies were raised at 10 flies/vial for 5–6 days before testing. On the day of the test, one male fly of a given genotype, and a wild-type (C-S) virgin female of a similar age were carefully aspirated into a square-shaped fighting arena and videotaped for 30 min. Fly pairs that did not perform copulation were not included in the analysis. Three parameters were analyzed: courtship latency (the latency to the first courtship behavior exhibited toward the female), copulation latency (the latency of copulation), and courtship index (the percentage of time spent on courtship, including copulation, during the first 10 min of videotaping).
Statistical Analysis of Behavioral Data.
For two-group comparisons, Student's t test (Fig. 4 D–F) or the Mann–Whitney U test (Fig. 1A) were performed for parametrically or nonparametrically distributed data. For comparisons of more than two groups, ANOVA (Figs. 1B, 3A, and 4A) or Kruskal–Wallis ANOVA (Figs. 1 C and D, 3 B and C, and 4B) was performed for parametrically or nonparametrically distributed data, respectively. We used Student–Newman–Keuls test (Fig. 3A) or Dunnett's C test (Figs. 1B and 4 A and B) as post hoc tests after ANOVA (and Kruskal–Wallis ANOVA for Fig. 4B) to identify significantly different groups with or without the assumption of homogeneity of variance, respectively. Significance levels for ANOVA and Kruskal–Wallis ANOVA were set to 0.05. Bar graphs are used to illustrate comparisons of means, with error bars representing SEM. Boxplots are used to illustrate comparisons of medians, with the lower and upper edges of the boxes representing the 25% and 75% quantiles, respectively, and the whiskers representing the 5% and 95% quantiles. The only exception was Fig. 1A, in which a bar graph (mean ± SEM) is used for illustrative purposes, whereas the statistical comparison was made between medians. This was done for the consistency with the other “fighting frequency” graphs (Figs. 1B and 3A).
Statistical Analysis of Microarray and Quantitative RT-PCR Data.
Array data were analyzed by using the Rosetta Resolver platform through the default processing pipeline, including normalization, grouping, and intergroup comparison. A fold-change of 1.25 and a P value of 0.002 were used to identify differently expressed genes (Dataset S1). For quantitative RT-PCR, Student's t test was performed for the comparison between two normally distributed datasets (Fig. 2A). ANOVA followed by Student–Newman–Keuls post hoc test was used for the comparison between four normally distributed datasets with homogeneity of variance assumed (Fig. 2B).
Immunohistochemistry.
Whole-mount antibody staining was adapted from ref. 45. Three- to 6-day-old male P[GawB]NP2593/UAS-mCD8GFP flies were anesthetized and dissected in PBS. Antennae (both second and third segments) and palps were fixed in 4% paraformaldehyde in PBS for 30 min at room temperature (RT), washed two times for 10 min in PBS, incubated in PBS containing 5% Triton X-100 (5% PBT) for 5 min, washed three times for 10 min in 0.3% PBT, blocked for 1 h in 0.3% PBT containing 5% heat inactivated normal goat serum (0.3% PBT/S), and incubated with primary antibody in 0.3% PBT/S overnight at 4°C. On the following day, samples were incubated in 0.3% PBT/S at RT for 1 h after washing three times for 10 min in 0.3% PBT. Samples were then incubated with secondary antibody and TOPRO-3 in 0.3% PBT/S for 2 h at RT in dark, washed three times for 5 min in 0.3% PBT, mounted in Vectashield (Vector Laboratories), and imaged on confocal microscope (Leica).
Antibody staining of frozen sections was adapted from ref. 46. Three- to 6-day-old male P[GawB]NP2593/UAS-mCD8GFP flies were anesthetized, aligned by using a Martin Heisenberg-style fly collar, mounted in frozen Tissue Tek OCT, sectioned at 14 μm, fixed in 4% paraformaldehyde in PBS at RT for 7 min, washed two times for 10 min in PBS, penetrated in 0.1% PBT for 30 min, blocked in 0.1% PBT/S for 30 min, and incubated with primary antibodies in 0.1% PBT/S overnight at 4°C. On the following day, slides were washed three times for 10 min in 0.1% PBT, blocked in 0.1% PBT/S for 30 min, incubated with secondary antibodies and TOPRO-3 in 0.1% PBT/S for 2 h in dark, washed three times for 5 min in 0.1% PBT, mounted in Vectashield, and imaged on confocal microscope (Leica).
Antibodies were used diluted as follows: rabbit anti-GFP (Invitrogen; 1:800), chicken anti-GFP (Chemicon; 1:300), rabbit anti-LUSH [D. P. Smith (University of Texas Southwestern Medical Center, Dallas, TX); 1:20], rat anti-ELAV (DSHC 7E8A10; 1:10), Alexa488/Cy3-conjugated secondary antibodies (Molecular Probes; 1:500), and TOPRO-3 (1:2,000).
Supplementary Material
Acknowledgments.
Microarray data were generated and analyzed with the help of the Millard and Muriel Jacobs Genetics and Genomics Laboratory at Caltech. We thank H. Dierick and R. Greenspan for providing the Cantonized Cyp6a20 mutant, S. Hoyer and M. Heisenberg for providing their aggression arena and sharing unpublished data, D. P. Smith for providing LUSH antibody, G. Carvalho for providing the Df(2R)BSC11/SM6a strain, C. Bargmann for helpful comments on the manuscript, and D.J.A. laboratory members for technical assistance and helpful discussions. H.D. is supported by a fellowship from the Alexander von Humboldt Society. D.J.A. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE6994).
This article contains supporting information online at www.pnas.org/cgi/content/full/0801327105/DCSupplemental.
References
- 1.Kravitz EA, Huber R. Aggression in invertebrates. Curr Opin Neurobiol. 2003;13:736–743. doi: 10.1016/j.conb.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 2.Tecott LH, Barondes SH. Behavioral genetics: Genes and aggressiveness. Curr Biol. 1996;6:238–240. doi: 10.1016/s0960-9822(02)00466-9. [DOI] [PubMed] [Google Scholar]
- 3.Loeber R, Hay D. Key issues in the development of aggression and violence from childhood to early adulthood. Annu Rev Psychol. 1997;48:371–410. doi: 10.1146/annurev.psych.48.1.371. [DOI] [PubMed] [Google Scholar]
- 4.Dierick HA, Greenspan RJ. Molecular analysis of flies selected for aggressive behavior. Nat Genet. 2006;38:1023–1031. doi: 10.1038/ng1864. [DOI] [PubMed] [Google Scholar]
- 5.Edwards AC, Rollmann SM, Morgan TJ, Mackay TFC. Quantitative genomics of aggressive behavior in Drosophila melanogaster. PLOS Genet. 2006;2:1386–1395. doi: 10.1371/journal.pgen.0020154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yeh SR, Fricke RA, Edwards DH. The effect of social experience on serotonergic modulation of the escape circuit of crayfish. Science. 1996;271:366–369. doi: 10.1126/science.271.5247.366. [DOI] [PubMed] [Google Scholar]
- 7.Stevenson PA, Dyakonova V, Rillich J, Schildberger K. Octopamine and experience-dependent modulation of aggression in crickets. J Neurosci. 2005;25:1431–1441. doi: 10.1523/JNEUROSCI.4258-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Papaj DR, Messing RH. Asymmetries in physiological state as a possible cause of resident advantage in contests. Behaviour. 1998;135:1013–1030. [Google Scholar]
- 9.Hoffmann AA. Territorial encounters between Drosophila males of different sizes. Anim Behav. 1987;35:1899–1901. [Google Scholar]
- 10.Matsumoto K, Pinna G, Puia G, Guidotti A, Costa E. Social isolation stress-induced aggression in mice: A model to study the pharmacology of neurosteroidogenesis. Stress. 2005;8:85–93. doi: 10.1080/10253890500159022. [DOI] [PubMed] [Google Scholar]
- 11.Luciano D, Lore R. Aggression and social experience in domesticated rats. J Comp Physiol Psychol. 1975;88:917–923. doi: 10.1037/h0076439. [DOI] [PubMed] [Google Scholar]
- 12.Ferno A. The effect of social isolation on the aggressive and sexual behaviour in a cichlid fish, Haplochromis burtoni. Behaviour. 1978;65:1–2. doi: 10.1163/156853978x00521. [DOI] [PubMed] [Google Scholar]
- 13.Hoffmann AA. The influence of age and experience with conspecifics on territorial behavior in Drosophila melanogaster. J Insect Behav. 1990;3:1–12. [Google Scholar]
- 14.Ueda A, Kidokoro Y. Aggressive behaviors of female Drosophila melanogaster are influenced by their social experience and food resources. Physiol Entomol. 2002;27:21–28. [Google Scholar]
- 15.Skrzipek VKH, Kroner B, Hager H. Aggression bei Drosophila melanogaster—laboruntersuchungen. Z Tierpsychol. 1979;39:87–103. [Google Scholar]
- 16.Hoffmann AA. A laboratory study of male territoriality in the sibling species Drosophila melanogaster and D. simulans. Anim Behav. 1987;35:807–818. [Google Scholar]
- 17.Chen S, Lee AY, Bowens NM, Huber R, Kravitz EA. Fighting fruit flies: A model system for the study of aggression. Proc Natl Acad Sci USA. 2002;99:5664–5668. doi: 10.1073/pnas.082102599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoyer SC, et al. Octopamine in male aggression of Drosophila. Curr Biol. 2008;18:156–167. doi: 10.1016/j.cub.2007.12.052. [DOI] [PubMed] [Google Scholar]
- 19.Wang Q, Hasan G, Pikielny CW. Preferential expression of biotransformation enzymes in the olfactory organs of Drosophila melanogaster, the antennae. J Biol Chem. 1999;274:10309–10315. doi: 10.1074/jbc.274.15.10309. [DOI] [PubMed] [Google Scholar]
- 20.van der Goes van Naters W, Carlson JR. Receptors and neurons for fly odors in Drosophila. Curr Biol. 2007;17:606–612. doi: 10.1016/j.cub.2007.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maibeche-Coisne M, Merlin C, Francois M-C, Porcheron P, Jacquin-Joly E. P450 and P450 reductase cDNAs from the moth Mamestra brassicae: Cloning and expression patterns in male antennae. Gene. 2004;346:195–203. doi: 10.1016/j.gene.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 22.Robinow S, White K. The locus elav of Drosophila melanogaster is expressed in neurons at all developmental stages. Dev Biol. 1988;126:294–303. doi: 10.1016/0012-1606(88)90139-x. [DOI] [PubMed] [Google Scholar]
- 23.Kim M-S, Repp A, Smith DP. LUSH odorant-binding protein mediates chemosensory responses to alcohols in Drosophila melanogaster. Genetics. 1998;150:711–721. doi: 10.1093/genetics/150.2.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barrot M, et al. Regulation of anxiety and initiation of sexual behavior by CREB in the nucleus accumbens. Proc Natl Acad Sci USA. 2005;102:8357–8362. doi: 10.1073/pnas.0500587102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Champagne FA, Curley JP. How social experiences influence the brain. Curr Opin Neurobiol. 2005;15:704–709. doi: 10.1016/j.conb.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 26.Levine JD, Funes P, Dowse HB, Hall JC. Resetting the circadian clock by social experience in Drosophila melanogaster. Science. 2002;298:2010–2012. doi: 10.1126/science.1076008. [DOI] [PubMed] [Google Scholar]
- 27.Ganguly-Fitzgerald I, Donlea J, Shaw PJ. Waking experience affects sleep need in Drosophila. Science. 2006;313:1775–1781. doi: 10.1126/science.1130408. [DOI] [PubMed] [Google Scholar]
- 28.Svetec N, Ferveur J-F. Social experience and pheromonal perception can change male-male interactions in Drosophila melanogaster. J Exp Biol. 2005;208:891–898. doi: 10.1242/jeb.01454. [DOI] [PubMed] [Google Scholar]
- 29.White SA, Nguyen T, Fernald RD. Social regulation of gonadotropin-releasing hormone. J Exp Biol. 2002;205:2567–2581. doi: 10.1242/jeb.205.17.2567. [DOI] [PubMed] [Google Scholar]
- 30.Burmeister SS, Kailasanath V, Fernald RD. Social dominance regulates androgen and estrogen receptor gene expression. Horm Behav. 2007;51:164–170. doi: 10.1016/j.yhbeh.2006.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carney G. A rapid genome-wide response to Drosophila melanogaster social interactions. BMC Genomics. 2007;8:288–297. doi: 10.1186/1471-2164-8-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baier A, Wittek B, Brembs B. Drosophila as a new model organism for the neurobiology of aggression? J Exp Biol. 2002;205:1233–1240. doi: 10.1242/jeb.205.9.1233. [DOI] [PubMed] [Google Scholar]
- 33.Dierick HA, Greenspan RJ. Serotonin and neuropeptide F have opposite modulatory effects on fly aggression. Nat Genet. 2007;39:678–682. doi: 10.1038/ng2029. [DOI] [PubMed] [Google Scholar]
- 34.Certel SJ, Savella MG, Schlegel DCF, Kravitz EA. Modulation of Drosophila male behavioral choice. Proc Natl Acad Sci USA. 2007;104:4706–4711. doi: 10.1073/pnas.0700328104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Osborne KA, et al. Natural behavior polymorphism due to a cGMP-dependent protein kinase of Drosophila. Science. 1997;277:834–836. doi: 10.1126/science.277.5327.834. [DOI] [PubMed] [Google Scholar]
- 36.Ben-Shahar Y, Robichon A, Sokolowski MB, Robinson GE. Inssuence of gene action across different time scales on behavior. Science. 2002;296:741–744. doi: 10.1126/science.1069911. [DOI] [PubMed] [Google Scholar]
- 37.Dow MA, Schilcher FV. Aggression and mating success in Drosophila melanogaster. Nature. 1975;254:511–512. doi: 10.1038/254511a0. [DOI] [PubMed] [Google Scholar]
- 38.Robin C, Daborn PJ, Hoffmann AA. Fighting fly genes. Trends Genet. 2006;23:51–54. doi: 10.1016/j.tig.2006.12.005. [DOI] [PubMed] [Google Scholar]
- 39.Matsumoto T, Honda S-i, Harada N. Alteration in sex-specific behaviors in male mice lacking the aromatase gene. Neuroendocrinology. 2003;77:416–424. doi: 10.1159/000071313. [DOI] [PubMed] [Google Scholar]
- 40.Toda K, et al. Targeted disruption of the aromatase P450 gene (Cyp19) in mice and their ovarian and uterine responses to 17beta-oestradiol. J Endocrinol. 2001;170:99–111. doi: 10.1677/joe.0.1700099. [DOI] [PubMed] [Google Scholar]
- 41.Black MP, Balthazart J, Baillien M, Grober MS. Socially induced and rapid increases in aggression are inversely related to brain aromatase activity in a sex-changing fish, Lythrypnus dalli. Proc R Soc B. 2005;272:2435–2440. doi: 10.1098/rspb.2005.3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Soma KK, Schlinger BA, Wingfield JC, Saldanha CJ. Brain aromatase, 5α-reductase, and 5β-reductase change seasonally in wild male song sparrows: Relationship to aggressive and sexual behavior. J Neurobiol. 2003;56:209–221. doi: 10.1002/neu.10225. [DOI] [PubMed] [Google Scholar]
- 43.Maibeche-Coisne M, Nikonov AA, Ishida Y, Jacquin-Joly E, Leal WS. Pheromone anosmia in a scarab beetle induced by in vivo inhibition of a pheromone-degrading enzyme. Proc Natl Acad Sci USA. 2004;101:11459–11464. doi: 10.1073/pnas.0403537101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guan Z, Saraswati S, Adolfsen B, Littleton JT. Genome-wide transcriptional changes associated with enhanced activity in the Drosophila nervous system. Neuron. 2005;48:91–107. doi: 10.1016/j.neuron.2005.08.036. [DOI] [PubMed] [Google Scholar]
- 45.Manoli DS, et al. Male-specific fruitless specifies the neural substrates of Drosophila courtship behaviour. Nature. 2005;436:395–400. doi: 10.1038/nature03859. [DOI] [PubMed] [Google Scholar]
- 46.Vosshall LB, Wong AM, Axel R. An olfactory sensory map in the fly brain. Cell. 2000;102:147–159. doi: 10.1016/s0092-8674(00)00021-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.