Abstract
The T cell leukemia 3 (Tlx3) gene has been implicated in specification of glutamatergic sensory neurons in the spinal cord. In cranial sensory ganglia, Tlx3 is highly expressed in differentiating neurons during early embryogenesis. To study a role of Tlx3 during neural differentiation, mouse embryonic stem (ES) cells were transfected with a Tlx3 expression vector. ES cells stably expressing Tlx3 were grown in the presence or absence of a neural induction medium. In undifferentiated ES cells, there was no significant difference in gene expression in the presence or absence of Tlx3, even after ES cells were cultured for an extensive time period. In contrast, expression levels of Mash1, Ngn1, and NeuroD were significantly higher in Tlx3-expressing cells after neural induction for 4 days compared with those in cells expressing the control vector. At 7 days after neural induction, whereas expression of the proneural genes was down-regulated, VGLUT2, GluR2, and GluR4 were significantly increased in ES cell-derived neurons expressing Tlx3. The sequential and coordinated expression of the proneural and neuronal subtype-specific genes identifies Tlx3 as a selector gene in ES cells undergoing neural differentiation. In addition, the differential effects of Tlx3 overexpression in undifferentiated ES cells compared with ES cell-derived neurons suggest that Tlx3 exerts context-dependent transcriptional signals on its downstream target genes. The context-dependent function of Tlx3 as a selector gene may be used to establish a novel strategy to conditionally generate excitatory glutamatergic neurons from ES cells to cure various types of neurodegenerative disorders.
Keywords: glutamatergic neurons, mouse, proneural genes, sensory ganglia
The T cell leukemia (Tlx) genes belong to a family of homeobox genes and are expressed in a variety of tissues including the spinal cord, spleen and branchial arches and in a restricted region of the developing nervous system during embryogenesis. Three family members, Tlx1/HOX11, Tlx2/HOX11-L1/Enx, and Tlx3/HOX11-L2/Rnx, have been identified based on their homology to human HOX11, a putative proto-oncogene involved in human T cell leukemia (1, 2). Target inactivation of these genes in mice has revealed their distinctive phenotypes: mutant mice lacking Tlx1 exhibit asplenogenesis (3), whereas null mutation in Tlx2 results in hyperganglionic megacolon (4). Tlx3 null mice die within 1 day after birth because of central respiratory failure (2). In addition, Tlx3 mutant mice exhibit abnormalities in nervous system functions in the ventral medulla and display improper development of somatic sensory neurons in the dorsal spinal cord and abnormalities in the formation of the primary visceral sensory neurons in the brainstem (5). Tlx3 is expressed in, and is required for, specification of glutamatergic neurons in the dorsal spinal cord. Forced expression of Tlx3 was sufficient to suppress GABAergic differentiation and induce formation of a glutamatergic neuron phenotype by control of ladybird homeobox homolog 1 (Lbx1) (6). In the dorsal spinal neurons overexpressing Tlx3, expression of paired related homeobox protein-like 1 (Prrxl1), glutamate receptor 2 (GluR2), and vesicular glutamate transporter 2 (VGLUT2) was significantly up-regulated. These compelling findings identify Tlx3 as a genetic switch that selects a glutamatergic over a GABAergic transmitter phenotype in distinct cell populations in the central nervous system (6, 7). Given the essential role of Tlx3 as a selector gene in vivo, we wished to test whether Tlx3 promotes glutamatergic neuronal specification from mouse embryonic stem (ES) cells. We also wanted to test whether a glutamatergic cell fate choice can be made in ES cells without a neural cell fate commitment. In other words, we wanted to test whether neural transmitter phenotype acquisition can take place without neural differentiation. To answer these questions, we established an in vitro transgenic system, by which Tlx3 is stably expressed in undifferentiated ES cells that subsequently undergo neural differentiation. This system allowed us to analyze the effects of forced Tlx3 expression on ES cells subjected to various developmental contexts.
Results
Tlx3 Expression Is Confined to Postmitotic Neurons in Cranial Sensory Ganglia.
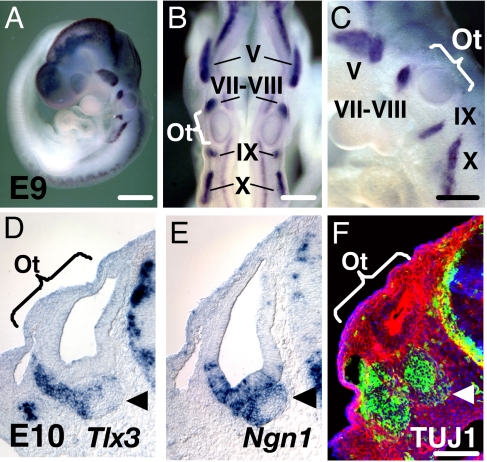
To investigate Tlx3 gross expression patterns, we performed whole-mount in situ hybridization with embryonic day (E)9 mouse embryos. Strong Tlx3 expression was confined to all cranial sensory ganglia, including the trigeminal (V), facio-acoustic (VII–VIII), glossopharyngeal (IX) nerve and associate ganglia, and the inferior ganglion of the vagus (X) nerve (Fig. 1 A–C). The hindbrain and dorsal root ganglia (DRG) were also positive for Tlx3. Transverse cryostat sections cut through the E10 head revealed strong Tlx3 signals in the vestibulocochlear ganglion (VCG) adjacent ventrally to the otocyst (Fig. 1D). Tlx3 was not expressed in the otic epithelium. Interestingly, the Tlx3 expression domain only partially overlaps with the Neurogenin1 (Ngn1) expression domain. Ngn1 was expressed in the ventral region of the otic epithelium and the VCG (Fig. 1E), whereas Tlx3 expression was confined to the VCG and migratory neuroblasts. Because all progenitor cells that give rise to sensory neurons in the inner ear are born within the otic epithelium and emigrate out of the otocyst to form VCG after acquiring a neural cell fate choice, the lack of Tlx3 expression in the otic epithelium suggests that its expression is triggered in progenitor cells after neural differentiation. To test this hypothesis, we stained some of the E10 transverse sections with an antibody against neuron-specific class III beta-tubulin (TUJ1). Consistent with the Tlx3 expression pattern, TUJ1-positive cells were detected in the VCG, but none in the otic epithelium was positive for TUJ1 (Fig. 1F). Together, our results strongly suggest that Tlx3 is expressed only in differentiating VCG neurons, most of which are believed to use glutamate as a neurotransmitter (8). These results are consistent with those of previous studies in which Tlx3 was shown to be expressed only in postmitotic neurons, but not in multipotent neural progenitors (9, 10).
Fig. 1.
Tlx3 expression is confined to cranial sensory ganglia. (A–C) Whole mount in situ hybridization for Tlx3 in an E9 embryo. High-magnification photographs of the embryo shown in dorsal (B) and lateral (C) views. (D) Tlx3 expression in the E10 VCG (arrowhead). Cranial transverse sections collected through the rostral otocyst at E10. (E) Ngn1 expression in the E10 otocyst and VCG (arrowhead). (F) TUJ1 expression (green) in the E10 VCG. The section was counterstained with phalloidin (red) and DAPI (blue). Ot, otocyst; V, trigeminal ganglion; VII–VIII, facio–acoustic ganglion; IX, glossopharygeal ganglion; X, inferior ganglion of the vagus nerve; VCG, vestibulocochlear ganglion. (Scale bars: 500 μm in A; 250 μm in B and C; 100 μm in F; scale bar in F also applies to D and E.)
Forced Expression of Tlx3 Does Not Alter the ES Cell Phenotype or Interfere with Neural Differentiation from ES Cells.
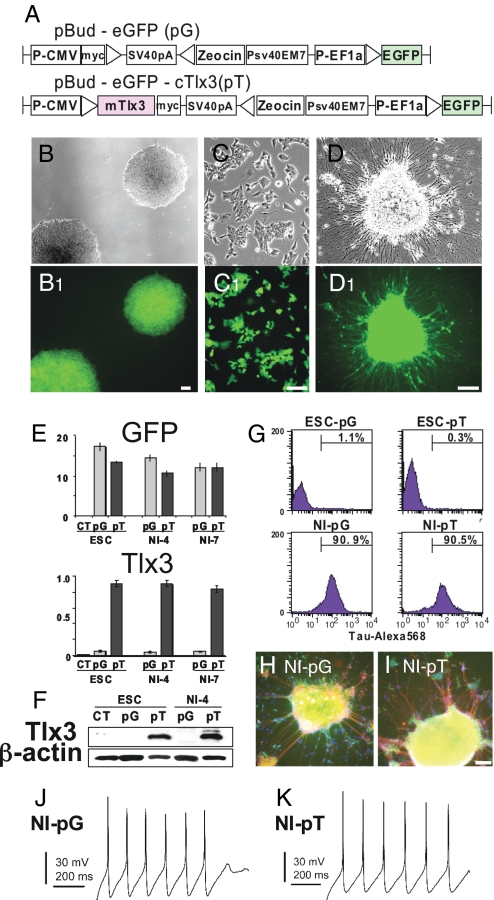
To elucidate the role for Tlx3 during neuronal differentiation, we constructed a Tlx3 expression vector and expressed it in mouse ES cells. We chose mouse ES cells as a model system to study the effects of Tlx3 expression on neural development based on the following reasons. First, Tlx3 is not constitutively expressed in mouse ES cells (Fig. 2 E and F), making them an ideal cell system to elucidate Tlx3 gain-of-function effects. Second, we have used an efficient neural induction protocol, by which more than 80% of mouse ES cells reportedly give rise to neurons (11), and by which we have demonstrated that temporal expression patterns of early and mature neural markers in mouse ES cells during the neural induction recapitulate those during normal neural development in vivo [supporting information (SI) Fig. S1]. Thus, we believe that the majority of mouse ES cells can be directed to differentiate into true neurons by means of our neural induction protocol. Third, we have established a powerful ES cell-based expression system, in which a transgene can be introduced and stably expressed even after passages and forced differentiation (Fig. 2 B–D). Using this system, we are able to evaluate the effects of transgene expression on ES cells at various points during neural differentiation. Undifferentiated ES cells were transfected with pBud-eGFP-cTlx3 (pT) or pBud-eGFP (pG) control vector by using an Amaxa nucleofector kit (Fig. 2A). Transfected ES cells were expanded and selected by Zeocin (Fig. 2B). Virtually 100% of Zeosin-resistant cells exhibited intense GFP fluorescence (Fig. 2 B1, C1, and D1), demonstrating that our transfection/selection method yields highly purified ES cells stably expressing the expression constructs. To check the efficacy of transgene expression, we analyzed Tlx3 expression in Zeosin-resistant cells by RT-PCR (Fig. 2E). High-level Tlx3 expression was detected in ES cells expressing pBud-eGFP-cTlx3 but not in untreated ES cells or ES cells expressing pBud-eGFP. A set of ES cell markers, including Oct4, Nanog, Rex1, and Sox2, was detected in these cells regardless of the presence or absence of Tlx3, strongly suggesting that forced Tlx3 expression did not promote differentiation of ES cells (Fig. S2). After dissociation of GFP-positive colonies (Fig. 2C), cells were incubated in a differentiation medium for 5 days to induce formation of embryoid bodies (EBs). EBs were then plated on poly-d-lysine/laminin-coated dishes and grown in the presence of a neural induction medium supplemented with brain derived neurotrophic factor (BDNF). Within 2 days after the start of neural induction, cells underwent extensive process outgrowth (Fig. 2D). Our Western blot analyses revealed that early (Musashi1, HuC, and NeuN) and mature neuronal markers (TUJ1, Calretinin, Synaptophysin, NSE, and Tau) are induced sequentially during the course of neural induction, whereas the ES cell markers, Oct4 and Sox2, became undetectable (Fig. S1). In addition, the percentage of ES-derived cells expressing multiple neuronal surface markers, including tau, CD24, and CD56, increased dramatically after culture in the neural induction medium for 7 days (Figs. 2 G–I and Fig. S3). Although the percentage of CD56-positive cells was lower than expected, this might be primarily due to the fact that ES-cell derived neurons were not fully differentiated at neural induction day 7 (12). Our single-cell current-clamp experiments (Fig. 2 J and K) recorded action potentials in 67% of pT cells (n = 21) and 68% of pG cells (n = 19), and both the pT and pG cells expressed voltage-dependent inward sodium currents, indicating that ES cell-derived neurons had acquired functional properties of true neurons. No difference was observed between pT and pG cells in terms of sodium current amplitude, action potential amplitude, resting membrane potential, or membrane input resistance (data not shown). These results are consistent with those of a previous study (11), in which > 80% of mouse ES cells were shown to differentiate into neurons with the two-step neural induction protocol.
Fig. 2.
Mouse ES cells can be efficiently directed to differentiate into neurons and stably express a transgene after neural differentiation. (A) Mouse Tlx3 expression construct pBud-eGFP-cTlx3 (pT) and control construct pBud-eGFP (pG) used in this study. (B–D) ES cells stably expressing the EGFP-tagged expression construct after Zeosin selection (B), dissociation (C), and neural induction (D). (E) Quantitative RT-PCR for EGFP and Tlx3 in ES cells expressing the Tlx3 expression vector (pT) or control vector (pG) before neural induction, 4 days (NI-4) or 7 days (NI-7) after neural induction. CT, untranfected ES cells. (F) Western blot analysis for Tlx3 in undifferentiated ES cells or ES-derived cells at neural induction day 4 (NI-4) expressing pT or pG. (G) Flow cytometry profiles of Tau-Alexa568 fluorescence in pG- or pT-expressing ES-derived cells before and after neural induction (NI). (H and I) Pan-neural marker Tau (red) is expressed in both pG-expressing (H) or pT-expressing (I) cells. Cellular nuclei were stained with DAPI (blue). (J and K) ES-derived neurons expressing pG-generated (J) and pT-generated (K) action potentials in response to current injection. Cells were biased to a resting membrane potential of −65 to −70 mV, and action potentials were elicited with 1-sec-long current injections of 2–50 pA. (Scale bars: 100 μm.)
To check the stability of the expression constructs and transgene expression in ES cell-derived neurons, we evaluated Tlx3 transcript levels. An equivalent level of Tlx3 mRNA was detected in ES cell-derived neurons expressing pT when compared with undifferentiated ES cells expressing the same construct. There was no significant difference in the Tlx3 level in cells between neural induction days 4 and 7 (Fig. 2E). It should be noted that Tlx3 was barely detectable in ES cell-derived neurons expressing pG, indicating that neural differentiation by itself does not up-regulate Tlx3 expression (Fig. 2E). Western blot analysis was performed to ascertain that Tlx3 protein was synthesized in ES cells overexpressing the Tlx3 expression construct. A strong immunoreactive band at ≈35 kDa was detected in cells expressing pT in both undifferentiated and neurally induced conditions (Fig. 2F). However, the immunoreactive band was absent in cells expressing pG and in untransfected ES cells. To ascertain that forced Tlx3 expression does not interfere with neural differentiation or maturation, we checked expression of several pan-neural marker genes. All HuC, Calretinin, Synaptophysin, NSE, and Tau were detected in cells expressing either Tlx3 or control vector at days 4 and 7 after the start of neural induction (Fig. S2). These findings, along with the observation of no significant difference in the percentage of CD24-positive or CD56-positive cells between cells expressing the Tlx3 construct and those expressing the control construct at neural induction day 7 (Fig. S3), strongly suggest that Tlx3 does not alter the neuronal phenotype in ES-derived cells undergoing neural differentiation.
Forced Expression of Tlx3 Exerts Context-Dependent Effects on ES-Derived Neurons.
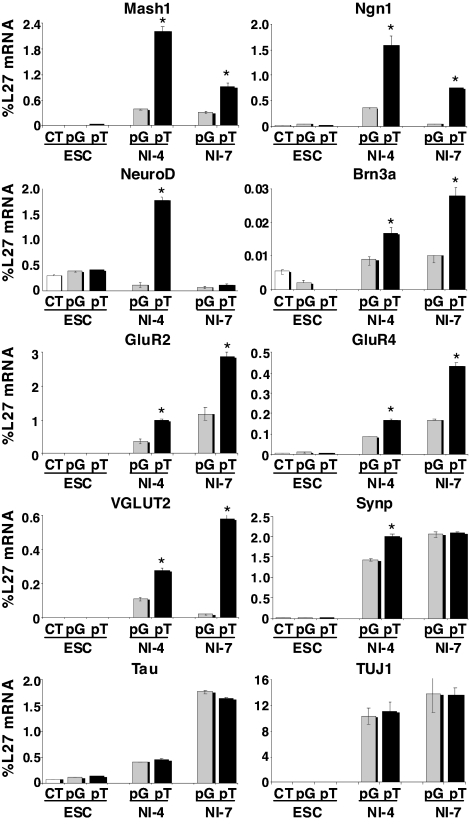
Although ectopic expression of Tlx3 did not alter the ES cell phenotype or neural differentiation from ES cells, we examined potential alterations in gene expression in response to Tlx3 by performing quantitative RT-PCR analysis for Mash1, Ngn1, NeuroD, Brn3a, GluR2, GluR4, VGLUT2, Synaptophysin, Tau, and TUJ1 (Fig. 3). In undifferentiated ES cells, there were no significant differences in gene expression in the presence or absence of Tlx3. In striking contrast, the expression levels of Mash1, Ngn1, and NeuroD in Tlx3-expressing ES-derived cells grown in neural induction medium for 4 days were ≈5- to ≈10-fold higher than those in ES-derived cells expressing the control vector. Moreover, the expression level of the transcription factor Brn3a, the AMPA receptors, GluR2, and GluR4, and VGLUT2 in Tlx3-expressing ES-derived cells became significantly higher than that in control cells after 4 days of neural induction, and this trend continued at neural induction day 7. In striking contrast, expression levels of an array of GABAergic neuronal subtype markers, including Pax2, Gad1, Gad2, Grik2, and Viaat (7), in Tlx3-expressing cells were significantly lower than those in control cells at neural induction day 7 (Fig. S4). Pan-neural markers Tau and TUJ1 were up-regulated in ES cells after neural induction, but there were no significant differences in their expression levels between ES-derived cells expressing Tlx3 and those expressing the control vector.
Fig. 3.
Proneural and glutamatergic marker genes are up-regulated in Tlx3-expressing ES-derived cells after neural induction. Quantitative RT-PCR analysis for transcription factors (Mash1, Ngn1, NeuroD, and Brn3a), pan-neural markers (Tau, TUJ1, Synaptophysin), glutamate receptors (GluR2, GluR4), and the transporter (VGLUT2) in ES cells expressing the Tlx3 expression vector (pT) in comparison with those expressing the control vector (pG). Undifferentiated ES cells or ES-derived cells grown in a neural induction medium for 4 days (NI-4) or 7 days (NI-7). Values are mean ± SD. *, P ≤ 0.05.
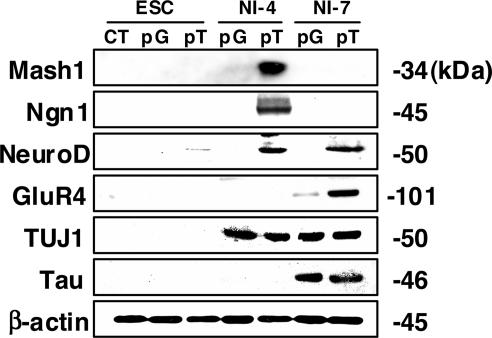
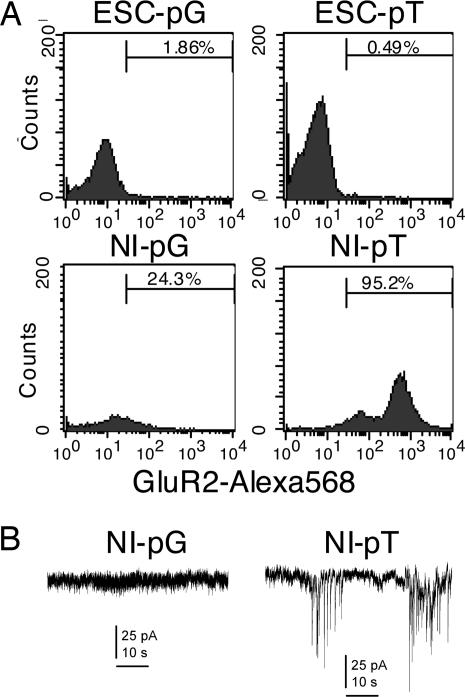
To evaluate protein expression levels in the presence or absence of Tlx3, we performed Western blot analysis (Fig. 4). None of the proneural (Mash1, Ngn1, NeuroD), glutamatergic (GluR4) or neural marker (TUJ1, Tau) proteins examined was detected in undifferentiated ES cells regardless of the presence or absence of the Tlx3 expression or control construct. However, Mash1, Ngn1, and NeuroD proteins were detected in Tlx3-expressing ES-derived cells grown in neural induction medium for 4 days but not in those expressing the control vector. Interestingly, the expression of Mash1 and Ngn1 appears to be temporal because they were not detectable in ES-derived cells grown in neural induction medium for 7 days. In contrast, NeuroD expression was detected in ES-derived cells 7 days after the start of neural induction. GluR4 was not detected in ES-derived cells expressing Tlx3 4 days after the start of neural induction, whereas it became detectable after 7 days of neural induction. GluR4 was present in control cells at 7 days of neural induction, but its level was significantly lower than in cells expressing Tlx3. In contrast to the obvious differences in the expression level of the proneural and glutamatergic marker proteins between Tlx3-expressing ES-derived cells and control cells, no significant difference was detected in the expression of the pan-neuronal markers, TUJ1 and Tau. TUJ1 became detectable in both Tlx3-expressing cells and nonexpressing cells at 4 days after the start of neural induction and retained its expression at neural induction day 7 as well. Tau was also detectable in both Tlx3-expressing cells and nonexpressing cells at approximately the same level at neural induction day 7. Flow cytometric analyses were performed to compare the number of cells expressing glutamate receptors between Tlx3-expressing cells and control cells (Fig. 5A). GluR2 was detected only in a very small fraction of undifferentiated ES cells in the presence (0.49%) or absence (1.86%) of Tlx3. However, the percentage of GluR2-expressing cells in ES cell-derived neurons with the Tlx3 construct (95.25%) was much greater than those with the control construct (24.33%) after 7 days of neural induction. To evaluate functional properties of ES cell-derived neurons in the presence or absence of Tlx3, we recorded spontaneous excitatory postsynaptic currents (EPSCs). Robust EPSCs were recorded from 33% of ES cell-derived neurons expressing Tlx3 (n = 9) but were not detectable from those expressing the control construct (n = 10) (Fig. 5B).
Fig. 4.
Proneural and glutamatergic marker proteins are induced in Tlx3-expressing ES-derived cells after neural induction. Western blot analysis for sensory-neuron marker proteins (Mash1, Ngn1, NeuroD, GluR4) as well as pan-neural marker proteins (TUJ1 and Tau) in ES-derived cells expressing the Tlx3 expression vector (pT) or those expressing the control vector (pG).
Fig. 5.
ES cell-derived neurons expressing Tlx3 exhibit an excitatory neuronal phenotype. (A) Flow cytometry profiles of GluR2-Alexa568 fluorescence in pT- or pG-expressing ES-derived cells before and after neural induction (NI). The majority of NI-pT cells express GluR2, whereas only 24.3% of NI-pG cells are positive for GluR2. (B) Large spontaneous excitatory postsynaptic currents (EPSCs) were observed in some of ES cell-derived neurons expressing the Tlx3 expression vector (Right). By contrast, EPSCs were not obvious in ES cell-derived neurons expressing the control vector (Left).
Discussion
The data presented here demonstrate that Tlx3 expression is induced in postmitotic progenitor cells in cranial sensory ganglia only after they acquire neural competence. When Tlx3 is introduced into mouse ES cells that do not constitutively express Tlx3, a set of proneural and glutamatergic marker genes and proteins was selectively up-regulated. Interestingly, however, the Tlx3-induced up-regulation of these genes and proteins was context-dependent. Tlx3 had no effect on expression of the genes in undifferentiated mouse ES cells, whereas it induced significant up-regulation of several proneural and glutamatergic genes in ES cells that were forced to differentiate into neurons. Moreover, the Tlx3-induced up-regulation of target genes was sequential: Up-regulation of proneural genes preceded that of glutamatergic genes by ≈3 days. Previous in vivo studies presented evidence that Tlx3, in collaboration with Tlx1, promotes glutamatergic specification at the expense of GABAergic specification in postmitotic neuronal progenitors in the dorsal spinal cord. The present results, demonstrating that Tlx3 instructively promotes expression of several AMPA receptors and a glutamate transporter while suppressing expression of GABA synthesizing enzymes, a kainate receptor, and an inhibitory amino acid transporter, are consistent with those of previous studies and further support a function of Tlx3 as a selector gene during neural differentiation. However, our results demonstrate that Tlx3 can also control expression of proneural genes, such as Mash1, Ngn1, and NeuroD. These results seem to conflict with those of a previous study in which Tlx3 was considered to be regulated by Mash1 (13). Although Mash1 expression in the hindbrain precedes Tlx3 expression (14), there does not appear to be a direct relationship between cells expressing Mash1 and Tlx3. Ectopic Tlx3 expression was observed in some, but not all, hindbrain progenitor cells misexpressing Mash1 (15). In addition, Tlx3 expression was lost in some, but not all, caudal hindbrain neurons in Mash1-deficient mice (16). Interestingly, in the forebrain, where Tlx3 is not expressed, the loss of Ngn1/2 functions results in ectopic generation of GABAergic neurons (17). These results implicate that Ngns, too, can regulate a glutamatergic neurotransmitter phenotype. Also, neither Ngn1 nor Mash1 appears to directly regulate Tlx3 expression, because both Mash1-positive ventral and Ngn1-positive dorsal regions in the forebrain are devoid of Tlx3. It is possible that Tlx3 functions to promote neurogenic activity by maintaining expression of proneural genes until neural progenitor cells are terminally differentiated into neuronal subtypes.
It is currently unknown how Tlx3 controls transcription of its target genes in a context-dependent manner. However, Tlx1, like other HOX proteins, was believed to activate its target genes through binding to specific DNA sequences. Thus, it is possible that Tlx3 recognizes and selectively binds neuron-specific promoter sequences in the regulatory region of its target genes. An alternative to the specific homeodomain-DNA binding is possible involvement of cofactors in transactivation of target genes. A group of non-HOX homeodomain proteins is known to function as cofactors for HOX proteins and modulate their DNA binding affinity and specificity. Of these are Pbx and Meis proteins, both of which are in the TALE (three amino acid loop extension) family of transcription factors. Pbx3, in particular, is highly expressed in the developing nervous system and mutant mice deficient for Pbx3 display neural phenotypes that resemble defects seen in Tlx3-deficient mice (18). Furthermore, in vitro assays demonstrated the ability of Pbx3 to form a DNA-binding complex with Tlx3 as well as Meis. Finally, the degree of transcriptional activation by Tlx3 was significantly increased in the presence of Pbx3 and Meis compared with the absence of these TALE proteins (18). Because Pbx proteins have been implicated in retinoid-dependent neuronal differentiation (19), it is possible that interplay between Tlx3 and Pbx3 underlie the selective actions of Tlx3 on ES cell-derived neurons over undifferentiated ES cells. A third possible mechanism responsible for the context-dependent effects of Tlx3 involves the transport factor importin-α. A recent study presented compelling evidence that the switching of importin-α subtypes from importin-α1 to -α5 occurs in ES cells during neural differentiation (20). Because the nuclear transport of transcription factors from the cytoplasm is regulated by transport factors, and because each of the importin-α factors can import only a specific set of transcription factors, it is possible that Tlx3 has a significantly higher affinity to importin-α5 than to -α1.
The ability of Tlx3 to trigger sequential activation of proneural and glutamatergic genes, along with its specific effects on ES cell-derived neurons, presents significant therapeutic values. Much like what is seen during nervous system development, Tlx3 was able to first induce a set of proneural genes, followed by activation of neural subtype-specific genes. One of the current major efforts in stem cell biology is to establish a means to trigger a cascade of genetic programs that recapitulate a coordinated order of gene expression seen during embryonic development in vivo. In this regard, it appears feasible to generate a desirable number of excitatory glutamatergic neurons in a selective region in the nervous system by transplanting Tlx3-expressing ES cells and selectively exposing them to neural induction signals. If genetic manipulation to introduce Tlx3 in ES cells accompanies any risks to the human health, then we would need to identify a signaling protein that promotes Tlx3 expression. We previously demonstrated that a conditioned medium prepared from the E10 mouse hindbrain/somite/otocyst can up-regulate Ngn1, NeuroD, Brn3a, GluR4, and VGLUT2—the same set of genes up-regulated in ES cell-derived neurons by Tlx3—in adult pluripotent progenitor cells after neural induction (21). Interestingly, Tlx3 expression was also induced in these neural competent progenitor cells grown in the hindbrain/somite/otocyst conditioned medium (T.K., unpublished observations). Thus, yet-to-be identified soluble protein(s) in the embryonic microenvironment appears to have the ability to positively control Tlx3 expression in a certain cellular context. Determining the identity of the Tlx3-inducing factor, thus, will likely lead to the establishment of an efficient method to generate excitatory glutamatergic sensory neurons from ES cells as well as somatic stem cells.
In summary, the present study demonstrates that Tlx3 triggers a sequential activation of proneural and neural subtype-specific genes in ES cells only after they become committed to a neural lineage. The context-dependent functions of Tlx3 as a glutamatergic selector gene could be used to generate excitatory glutamatergic neurons from ES cells in a selective area of the brain at a desirable time. Future investigations should aim to elucidate the molecular mechanisms underlying the context-dependent functions of Tlx3 and identification of a signaling protein (or proteins) that promotes Tlx3 expression.
Materials and Methods
Construction of Plasmid Vectors.
pBud-eGFP was constructed by inserting a PCR-amplified copy of EGFP from pIRES2-EGFP (Clontech) into the KpnI-XhoI site of the pBudCE4.1 vector (Invitrogen) such that expression would be driven by the elongation factor 1α promoter (EF1 α). Primers were designed that corresponded to sequences overlapping the EGFP translation start and stop codon and that included restriction sites for KpnI and XhoI sequences. DNA bands of the expected respective size were isolated with the Gel Extraction kit (Qiagen). An EcoRI-BamHI fragment containing the full-length Tlx3 cDNA from FAMTOM clone (AK141870; RIKEN Brain Science Institute) was inserted into the pcDNA3.1(−) vector (Invitrogen). The Tlx3 full-length sequence clone (pFLCl-Tlx3) was purchased from the Institute of Physical and Chemical Research (RIKEN). The pBud-eGFP-cTlx3 (pT) was reconstructed by inserting the XbaI-BamHI fragment including Tlx3 from pcDNA-Tlx3 into the area following the cytomegalovirus promoter of pBud-eGFP (pG).
ES Cell Culture and Transfection.
R1 murine ES cells were maintained and passaged as described previously (22). Briefly, ES cells were plated on gelatin-coated tissue culture plates and grown in high-glucose DMEM supplemented with 15% FBS (Invitrogen), 1.0 mM sodium pyruvate (Stemcell Technologies), 10 mM nonessential amino acids (Stemcell Technologies), 0.01% penicillin streptomycin (Stemcell Technologies), 2.0 mM l-glutamine (Stemcell Technologies), 1,000 units/ml leukemia inhibiting factor (Chemicon), and 0.055 mM 2-mercaptoethanol. After a few passages, ES cells were transfected with 4 μg of the construct-containing plasmid (pBud-eGFP-cTlx3) or control plasmid (pBud-eGFP) by using the Mouse ES Cell Nucleofector kit (Amaxa) according to the manufacture's instructions. After transfection, cells were incubated in ES cell maintenance medium with 50 μg/ml Zeosin (Invitrogen).
Some of the transfected ES cells were subjected to neural differentiation according to a previously established procedure, with minor modifications (11). Cells were dissociated into single cells by using 0.25% trypsin-EDTA and resuspended in differentiation medium containing G-MEM (Invitrogen), 5% Knockout serum replacement (Invitrogen), 2.0 mM l-glutamine, 1.0 mM sodium pyruvate, 0.1 mM nonessential amino acids, 0.01% penicillin streptomycin, and 0.1 mM 2-mercaptoethanol. Cells were cultured in bacteria plates for 5 days at a concentration of 5 × 104 cells per ml to allow embryoid body (EB) formation. Differentiation medium was changed at day 3 of the serum-free suspension culture. EBs were plated en bloc on tissue culture plates or chamber slides double-coated with poly-d-lysine (200 μg/ml) and mouse laminin (10 μg/ml) at a concentration of 1–2 × 102 EBs per cm2 and cultured for 2 days in differentiation medium. After 2 days, the medium was changed to neural induction medium containing G-MEM, 1% N2, 2 mM glutamine, 1 mM pyruvate, 0.1 mM nonessential amino acids, 0.1 mM 2-mercaptoethanol, 0.01% penicillin streptomycin and 10 ng/ml BDNF (PeproTech). Neural induction cultures were maintained for 4 or 7 days before extraction of RNA or proteins, electrophysiological recordings, flow cytometric analyses, or fixation for immunohistochemistry.
Electrophysiology.
Whole cell patch-clamp recordings were conducted at ≈21°C by using an EPC-10 amplifier, and data were acquired by using the Pulse program (HEKA Electronics). Fire-polished electrodes (4.0–5.0 MΩ) were fabricated from 1.5 mm capillary glass by using a P-97 puller (Sutter Instruments). The pipette solution contained: 140 mM KCl, 5 mM MgCl2, 5 mM EGTA, 2.5 mM CaCl2, 4 mM ATP, 0.3 mM GTP, and 10 mM Hepes, pH 7.3 (adjusted with KOH). The bathing solution contained: 140 mM NaCl, 1 mM MgCl2, 5 mM KCl, 2 mM CaCl2, 10 mM Hepes, and 10 mM glucose, pH 7.3 (adjusted with NaOH). Voltage-clamp and current-clamp data were analyzed by using the Pulsefit (HEKA Electronics), Origin (OriginLab) and Microsoft Excel software programs.
In Situ Hybridization.
ICR mouse embryos at E9 and E10 were fixed with 4% paraformaldehyde overnight. Some of the embryos were embedded in OCT compound, and transverse cryostat sections at 10 μm throughout the head were collected and stored at −80°C. Sense and anti-sense digoxigenin-UTP-labeled riboprobes for Tlx3 or Ngn1 were prepared by linearization of a mouse Tlx3 or Ngn1 cDNA with restriction enzymes, followed by in vitro transcription with the DIG-RNA Labeling kit (Roche). All hybridization steps were conducted at room temperature with rocking, unless otherwise indicated. Whole-mount samples were treated with proteinase K (10 μg/ml) and subsequently prehybridized in hybridization buffer. Proteinase K digestion was stopped by incubation of samples in glycine (2 mg/ml) in PBST for 10 min. Samples were then postfixed in 4% paraformaldehyde/0.2% gluteraldehyde for 20 min. Samples were prehybridized in hybridization buffer (50% formamide, 1% SDS, 5× SSC, 50 μg/ml Escherichia coli tRNA, 50 μg/ml heparin) for 1 h at 70°C. Samples were subsequently incubated overnight in fresh hybridization buffer with riboprobe (1 μg/ml) at 70°C. Thereafter, the samples were washed and treated with RNase A (100 μg/ml) for 1 h at 37°C to remove unbound riboprobe, blocked 1 h in 1% Blocking Reagent (Roche), and incubated with alkaline phosphate-conjugated anti-digoxygenin antibody (Roche) overnight at 4°C. After extensive washing to remove unbound antibody, hybridization signals were visualized by BM Purple (Roche). Control experiments using sense probes were performed for each probe to check the specificity of hybridization signals. Some of the E10 cryostat sections were processed for immunohistochemistry, as described in SI Methods.
RT-PCR, Western blotting, and flow cytometry are described in SI Methods.
Supplementary Material
Acknowledgments.
We thank Medhane Cumbay for his assistance with plasmid construction, Qiufu Ma (Dana–Farber Cancer Institute, Boston) for the Neurogenin1 probe, and Jeffrey Clarke and Angela Thompson for their assistance with ES cell culture. This work was supported by National Institutes of Health grants R01DC007390 (to E.H.), R01HL082981 (to R.J.C), R01NS053422 (to T.R.C.), and T32DC000012 (to D.A.Z.) and by the Deafness Research Foundation (to T.K. and E.H.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0708704105/DCSupplemental.
References
- 1.Wen XY, Tang S, Breitman ML. Genetic mapping of two mouse homeobox genes Tlx-1 and Tlx-2 to murine chromosomes 19 and 6. Genomics. 1994;24:388–390. doi: 10.1006/geno.1994.1634. [DOI] [PubMed] [Google Scholar]
- 2.Shirasawa S, et al. Rnx deficiency results in congenital central hypoventilation. Nat Genet. 2000;24:287–290. doi: 10.1038/73516. [DOI] [PubMed] [Google Scholar]
- 3.Greene WK, Bahn S, Masson N, Rabbitts TH. The T-cell oncogenic protein HOX11 activates Aldh1 expression in NIH 3T3 cells but represses its expression in mouse spleen development. Mol Cell Biol. 1998;18:7030–7037. doi: 10.1128/mcb.18.12.7030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shirasawa S, Yunker AM, Roth KA, Brown GA, Horning S. Enx (Hox11L1)-deficient mice develop myenteric neuronal hyperplasia and megacolon. Nat Med. 1997;3:646–650. doi: 10.1038/nm0697-646. [DOI] [PubMed] [Google Scholar]
- 5.Qian Y, et al. Formation of brainstem (nor)adrenergic centers and first-order relay visceral sensory neurons is dependent on homeodomain protein Rnx/Tlx3. Genes Dev. 2001;15:2533–2545. doi: 10.1101/gad.921501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng L, et al. Lbx1 and Tlx3 are opposing switches in determining GABAergic versus glutamatergic transmitter phenotypes. Nat Neurosci. 2005;8:1510–1515. doi: 10.1038/nn1569. [DOI] [PubMed] [Google Scholar]
- 7.Cheng L, et al. Tlx3 and Tlx1 are post-mitotic selector genes determining glutamatergic over GABAergic cell fates. Nat Neurosci. 2004;7:510–517. doi: 10.1038/nn1221. [DOI] [PubMed] [Google Scholar]
- 8.Safieddine S, Wenthold RJ. The glutamate receptor subunit delta1 is highly expressed in hair cells of the auditory and vestibular systems. J Neurosci. 1997;17:7523–7531. doi: 10.1523/JNEUROSCI.17-19-07523.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Logan C, Wingate RJ, McKay IJ, Lumsden A. Tlx-1 and Tlx-3 homeobox gene expression in cranial sensory ganglia and hindbrain of the chick embryo: Markers of patterned connectivity. J Neurosci. 1998;18:5389–5402. doi: 10.1523/JNEUROSCI.18-14-05389.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qian Y, Shirasawa S, Chen CL, Cheng L, Ma Q. Proper development of relay somatic sensory neurons and D2/D4 interneurons requires homeobox genes Rnx/Tlx-3 and Tlx-1. Genes Dev. 2002;16:1220–1233. doi: 10.1101/gad.982802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Watanabe K, et al. Nat directed differentiation of telencephalic precursors from embryonic stem cells. Neuroscience. 2005;8:288–296. doi: 10.1038/nn1402. [DOI] [PubMed] [Google Scholar]
- 12.Pruszak J, Sonntag KC, Aung MH, Sanchez Pernaute R, Isacson O. Markers and methods for cell sorting of human embryonic stem cell-derived neural cell populations. Stem Cells. 2007;25:2257–2268. doi: 10.1634/stemcells.2006-0744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mizuguchi R, et al. Ascl1 and Gsh1/2 control inhibitory and excitatory cell fate in spinal sensory interneurons. Nat Neurosci. 2006;9:770–778. doi: 10.1038/nn1706. [DOI] [PubMed] [Google Scholar]
- 14.Jasoni CL, Walker MB, Morris MD, Reh TA. A chicken achaete-scute homolog (CASH-1) is expressed in a temporally and spatially discrete manner in the developing nervous system. Development. 1994;120:769–783. doi: 10.1242/dev.120.4.769. [DOI] [PubMed] [Google Scholar]
- 15.Hornbruch A, et al. A BMP-mediated transcriptional cascade involving Cash1 and Tlx-3 specifies first-order relay sensory neurons in the developing hindbrain. Mech Dev. 2005;122:900–913. doi: 10.1016/j.mod.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 16.Pattyn A, Guillemot F, Brunet JF. Delays in neuronal differentiation in Mash1/Ascl1 mutants. Dev Biol. 2006;295:67–75. doi: 10.1016/j.ydbio.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 17.Fode C, et al. A role for neural determination genes in specifying the dorsoventral identity of telencephalic neurons. Genes Dev. 2000;14:67–80. [PMC free article] [PubMed] [Google Scholar]
- 18.Rhee JW, et al. Pbx3 deficiency results in central hypoventilation. Am J Pathol. 2004;165:1343–1350. doi: 10.1016/S0002-9440(10)63392-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qin P, Haberbusch JM, Zhang Z, Soprano KJ, Soprano DR. Pre-B cell leukemia transcription factor (PBX) proteins are important mediators for retinoic acid-dependent endodermal and neuronal differentiation of mouse embryonal carcinoma P19 cells. J Biol Chem. 2004;279:16263–16271. doi: 10.1074/jbc.M313938200. [DOI] [PubMed] [Google Scholar]
- 20.Yasuhara N, et al. Triggering neural differentiation of ES cells by subtype switching of importin-alpha. Nat Cell Biol. 2007;9:72–79. doi: 10.1038/ncb1521. [DOI] [PubMed] [Google Scholar]
- 21.Kondo T, Johnson SA, Yoder MC, Romand R, Hashino E. Sonic hedgehog and retinoic acid synergistically promote sensory fate specification from bone marrow-derived pluripotent stem cells. Proc Natl Acad Sci USA. 2005;102:4789–4794. doi: 10.1073/pnas.0408239102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Y, et al. Murine embryonic stem cell differentiation is promoted by SOCS-3 and inhibited by the zinc finger transcription factor Klf4. Blood. 2005;105:635–637. doi: 10.1182/blood-2004-07-2681. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.