Abstract
The lysogenic state of bacteriophage lambda is maintained by CI repressor, which negatively regulates two promoters to block lytic gene expression. Expression of CI is itself controlled by positive and negative feedback as CI binds to OR to regulate the PRM promoter. In addition to direct interactions with operator DNA, CI tetramers bound at OL and OR can come together to form an octamer, looping the DNA that lies between them and allowing OL to assist with negative regulation of PRM. We used a fluorescent reporter protein to measure the CI concentration for a set of constructs that differ in their ability to assume various forms of the looped structure. Based on the observed steady-state fluorescence for these constructs, the presence of OL increases PRM activation unless both operators can be fully occupied. By calculating the probabilities for the underlying operator configurations present in each construct, two different models for the mechanism of enhanced activation allow us to predict that when the DNA is looped, PRM activation can be 2- to 4-fold higher than is possible for unlooped DNA. Based on our results, transcriptional regulation for lambda's lysogenic/lytic switch includes both activation and repression due to DNA looping.
Keywords: bacteriophage lambda, gene regulation, flow cytometry
Over the last several decades, phage lambda has been an important model system for studying gene regulation, in part because it has two very different modes of growth in its Escherichia coli host. In the lytic mode, the phage uses the host cellular machinery for large-scale production of new phage, which are then released by cell lysis for another round of infection. In the lysogenic mode, the phage DNA is integrated into the host genome and passed on to each daughter cell as the infected cell grows and divides. The phage is maintained in this quiescent state by a single protein, CI, which prevents transcription from the early lytic phage promoters. The lytic genes continue to be repressed until the host cell suffers DNA damage and activates RecA, which switches the cell to the lytic growth mode by catalyzing degradation of CI (1). Spontaneous switching from the lysogenic state to lytic growth is very rare; in the absence of RecA, phage particles are found in lysogenic E. coli cultures at very low frequencies (2), and most contain mutations in the regulatory elements of the phage DNA (3). Not only is the lysogenic state very stable, but it efficiently switches to the lytic mode when induced (2, 4). The regulatory mechanisms that provide simultaneous sensitivity and stability for this genetic switch are not yet fully understood.
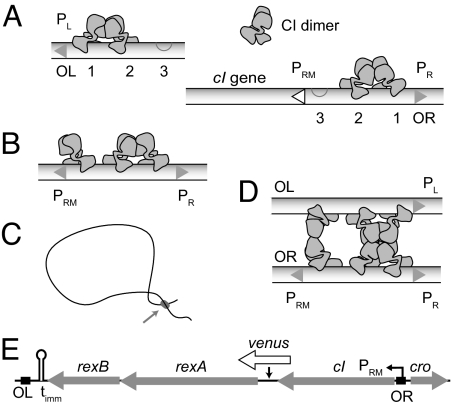
There is, however, a wealth of genetic and biochemical data to draw on as we explore this question (1). CI stabilizes the lysogenic state by binding to the operators OL and OR, repressing PL and PR, the promoters that lead to lytic growth (Fig. 1). OL and OR each contain three binding sites for CI and are spaced ≈2.3 kb apart. When they bind to the highest affinity sites of each operator, dimers of CI prevent RNA polymerase (RNAP) from initiating transcription from the lytic promoters. Cooperativity between adjacent dimers facilitates CI binding so that the second sites are readily occupied. When CI is bound to OR2, its own promoter, PRM, is activated ≈10-fold (5). The third site of OR has a much lower affinity for CI, but when OR3 is occupied, PRM transcription is blocked. This feedback repression is enhanced by a long-range DNA loop that forms between OL and OR (6), which can orient OL opposite OR in such a way that a CI dimer weakly bound to OR3 is stabilized by interacting with a CI dimer bound to OL (7). It is not yet known how the loop affects PRM activation.
Fig. 1.
Lysogenic regulation. (A) The six binding sites for CI comprise the operators OL and OR. CI dimers bind cooperatively to the two highest affinity sites and repress PL and PR. While repressing PR, CI bound to OR2 simultaneously activates PRM. (B) When there is sufficient CI present to occupy OR3, PRM is repressed. (C) A long-range DNA loop can form between CI tetramers bound at OL and OR. (D) A diagram of the loop site, showing cooperativity between OL3 and OR3 enhancing repression of PRM. Many other looped configurations are possible. Although shown as parallel, the orientation of the two DNA strands is not known. Shapes of CI dimers are based on the tetramer model of (37). (E) The immunity region contains the PRM transcript, which begins with the start codon of cI and terminates at timm. In our constructs, a fluorescent protein was inserted between cI and rexA.
There are many ways that CI dimers can bind to OL and OR at lysogenic levels of CI, so a range of operator configurations will be present within a population of cells or in a single cell over time. When a lysogenic cell is in a steady state, the number of CI molecules produced per cell division matches the mean number present per cell. Because CI production in a lysogen is autoregulated, the measured CI concentration both predicts and depends on the probabilities of activated and repressed configurations; the mean amount of CI present in the cell leads to an overall rate of production that maintains this mean concentration. We designed a set of constructs with well defined changes in the operator DNA so that, for each, the probabilities of activated and repressed configurations depend on the CI concentration in a unique way, and thus each maintains a different steady-state. By measuring the CI concentration for each population and modeling the underlying operator states, we found that a DNA loop between OL and OR can increase PRM activation >2-fold.
Results
Immunity Region Constructs.
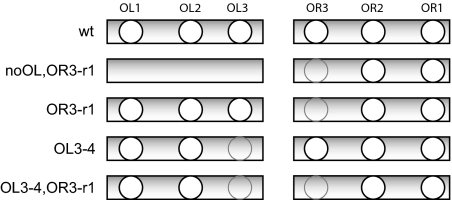
A fluorescent reporter protein cotranscribed with cI was used to estimate the CI concentration in individual cells. The constructs used in this study contain the entire immunity region of lambda, the region of DNA that spans OL and cro. The PRM transcript from which all cI is produced extends from the start codon of cI through rexA and rexB to terminate at timm (Fig. 1E). A copy of the venus fluorescent protein gene (8) fused to a strong ribosome binding site was inserted into this transcript in the noncoding region between cI and rexA. The ribosome binding site for venus favors more frequent translation initiation than is possible for the leaderless CI (9), which amplifies the signal from transcription events, improving the sensitivity of our measurements. A low copy number plasmid (1 or 2 copies per genome) (10) was used as the vector for all experiments, and E. coli strain K12 was used as the host. The five constructs contain all of the regulatory elements required to maintain lysogeny and differ by mutations or deletions in the operator regions, which affect CI binding (Fig. 2) and change the probability that PRM is activated or repressed. The WT construct has wild-type OL and OR operator sites. The noOL,OR3-r1 construct lacks all three operator sites of OL, so it cannot form the DNA loop. It also contains a mutation in OR3 that substantially reduces binding affinity so that PRM repression is impaired. This mutation also slightly increases the basal activity of PRM (11). The OR3-r1 construct is able to form loops between OL and OR, but the OR3-r1 mutation impairs PRM repression, which occurs when CI binds to OR3. The OL3-4 and OL3-4,OR3-r1 constructs carry a mutation that destroys CI affinity at the third site of OL, OL3, which prevents OL from assisting PRM repression (7).
Fig. 2.
Operator constructs. The constructs differ by site-specific changes in operator DNA that alter affinity for CI, shown here schematically. The OR3-r1 and OL3-4 mutations substantially reduce affinity for CI and are identical to those used by Dodd et al. (7, 11). Gray circles indicate mutated operator sites. For the noOL,OR3-r1 construct, the three sites of the left operator were deleted.
Fluorescence Data.
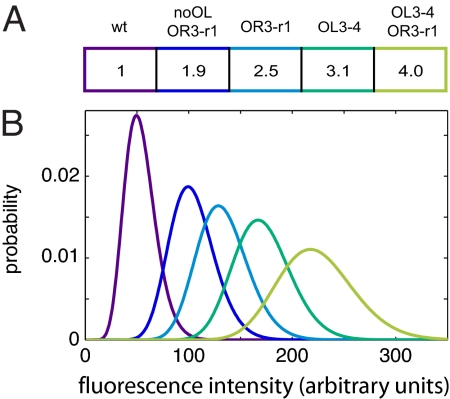
To obtain the CI concentration for each construct, we used flow cytometry to measure the Venus fluorescence per cell over a population of 100,000 cells. As anticipated, each construct has a distinct distribution of fluorescence (Fig. 3), which we take to be proportional to CI concentration (see Materials and Methods). The lowest concentration of CI is seen in the WT construct, for which the loop between OL and OR facilitates PRM repression. Simultaneously deleting the left operator and mutating OR3 to reduce binding affinity (noOL,OR3-r1) increased the CI concentration only twofold. In contrast, when the left operator was present and only OR3 was mutated (OR3-r1), the CI concentration was 2.5 times WT levels. The highest steady-state CI levels, 3.1 and 4.0, were observed when the OL3 site was mutated to remove CI affinity (OL3-4 and OL3-4,OR3-r1). These data can be understood by assuming that some operator configurations with CI bound to OL can enhance PRM transcriptional activation and that there are other operator configurations with all three OL sites bound that do not allow optimal activation. Although the data for the noOL,OR3-r1 construct is quite different from that observed in the reporter studies of Dodd et al. (see Discussion), the ratios for the CI concentration of OR3-r1 and OL3-4 mutants relative to WT are near the values obtained by Dodd et al. using gel shift assays of cell lysate from lysogens with the same point mutations: OR3-r1/WT = 2.51–2.99 and OL3-4/WT = 2.94–3.14 [95% confidence interval (7)]. The values from our mutants are: noOL,OR3-r1/WT = 1.92 ± 0.07; OR3-r1/WT = 2.45 ± 0.1; OL3-4/WT = 3.12 ± 0.08; and OL3-4,OR3-r1/WT = 4.03 ± 0.09 (95% confidence interval). Based on our data, the ability to form a DNA loop between OL and OR increases the maximum activation of PRM. To investigate how this might occur, we examined possible ways that CI can form a DNA loop and activate PRM.
Fig. 3.
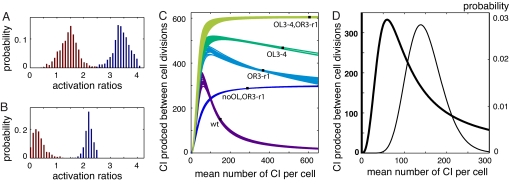
Fluorescence data. (A) Mean levels of each operator mutant relative to the WT construct, rounded. Exact values and error estimates are given in Results. The WT construct, which has good repression because of intact OL, has the lowest level of CI. All other mutants are impaired in repression, and, of these, the lowest level of CI is observed in the noOL,OR3-r1 construct, which cannot loop because OL was deleted. (B) A representative set of deconvolved fluorescence probability histograms for the five constructs, colored and ordered as in A.
Operator Configurations.
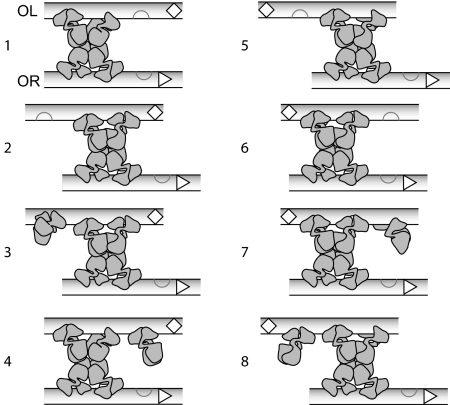
Previous studies allow us to relate our experimental data to microscopic configurations of CI bound to the six operator sites. Taking into account pairwise cooperativity between CI dimers, there are nine possible binding configurations for each operator (12, 13): one configuration with no bound CI, three configurations with a single dimer bound, three with two dimers bound, and two with all three sites occupied. This gives a total of 81 unlooped operator configurations [supporting information (SI) Dataset S1]. Tetramers of CI bound at OL and OR can form a DNA loop (6), so, like Dodd et al. (7), we allowed any of the four operator configurations with a tetramer of cooperatively bound dimers to form a loop. For the large distance (>2 kb of DNA) between the sites, the operators should be equally able loop in either possible orientation of OL relative to OR; when looped, OL1 may be opposite either OR1 or OR3. This gives 32 looped configurations (Fig. S1 and Fig. S2), for a total of 113 configurations.
Activation States.
These configurations can be assigned to activation states based on data from previous studies, data from our constructs, and a model for the underlying mechanism of looped activation. When CI is bound to OR2, PRM is activated (14); but, when OR3 is occupied, PRM is repressed (15). Unlooped configurations were categorized as unactivated if OR2 and OR3 are not occupied (18 configurations), activated if OR2 is occupied with OR3 unoccupied (18 configurations), and repressed if OR3 is occupied (45 configurations). Of the looped configurations, 24 are repressed because OR3 is occupied. The eight remaining looped configurations (Fig. 4) have OR2 occupied and are expected to be activated to some degree. Each of these looped species may have a unique activation rate, but our data are not complete enough to analyze each configuration separately. For simplicity, we assume that the activated configurations can be grouped into three distinct activation states. All unlooped activated DNA are thought to have same activation level, so we call these configurations “act1.” The highest levels of CI were observed when OL was present but only two sites could be occupied, so we expect an additional activation state that includes such a configuration, designated “act2.” When all three sites of OL are intact, substantially less CI is produced, so “act3” could include configurations with all three sites bound. There are still many ways to assign configurations to these three states, reflecting different mechanisms by which the loop between OL and OR enhances activation. The actual mechanism is unclear at this point, but further insight can still be gained by exploring possible scenarios. We therefore analyzed our data, using two different models to assign the eight configurations of Fig. 4 to act1, act2, and act3.
Fig. 4.
Activated looped configurations. In a lysogen, OL and OR can likely interact equally well in two orientations. There are eight looped configurations with OR3 unoccupied, and each is thus expected to be activated to some degree, although each may have a different activation level. The UP element adjacent to OL3 is indicated by an open diamond, and PRM is shown as an open triangle. Again, the DNA strands are represented as parallel, but the actual orientations in a cell are not known.
Upstream Promoter (UP) Element Activation.
One possible mechanism (suggested by an anonymous reviewer) is that an UP element near OL3 (16) is accessible to the α subunit of RNAP in some looped configurations, increasing the promoter strength. Based on this mechanism, we propose that the four looped states with the UP element oriented away from PRM (loops 5–8 of Fig. 4) have transcriptional activity similar to unlooped activated configurations, so they were also assigned to act1. Because the highest CI levels were seen in the constructs for which OL3 was mutated so that it is not occupied, the activated configuration with OL3 unoccupied and the UP element oriented toward PRM (loop 1) was assigned to act2. The remaining three configurations (loops 2–4) were designated act3. Additional ways of grouping these configurations according to the UP element model are compared in Fig. S3.
Looped Octamer Activation.
Another possible mechanism is that the additional octamer contacts that CI makes to form a loop might involve conformational changes that lead to enhanced activation of RNAP by CI. In this case, the orientation of the two strands would not necessarily specify the activation state. The decreased levels of CI produced when OL3 is occupied could be interpreted to mean that CI bound to the third site of OL is large enough to interfere with RNAP binding to the promoter on the opposite side of the loop. Based on this mechanism, the six configurations with only two occupied OL sites or with the third site of OL oriented away from PRM (loops 1–3, 5, 6, and 8) were designated act2. The remaining two configurations with the third occupied site of OL in proximity to PRM (loops 4 and 7) were assigned to act3. Additional ways of using this mechanism to group configurations are given in Fig. S3B.
For either mechanism, we infer that looped DNA with only two sites of OL occupied (act2) provides the highest activation, but to estimate the magnitude, we must first calculate the probability of finding each state in our five constructs.
Activation State Probabilities.
To examine these mechanisms in more detail, we first calculated the probability of each operator configuration at the measured level of CI. The probability of each configuration can be calculated from the measured [CI], using the method of Shea and Ackers (17):
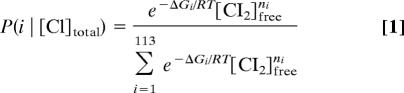 |
where ΔGi is the free-energy for forming the ith configuration, R is the gas constant, T is temperature in Kelvin, [CI]total is the total concentration of CI monomers per cell obtained from flow cytometry data (see Materials and Methods). The [CI2]free for a given [CI]total was calculated taking into account the equilibrium of free dimer with monomers and with specifically and nonspecifically bound states. The parameters used to calculate ΔGi are given in Table 1. Representative probabilities for unactivated, unlooped activated, repressed, and eight looped activated states are given in Dataset S2.
Table 1.
Parameter free energies
| Parameter | ΔG(kcal/mol) | ± |
|---|---|---|
| OR1* | −12.5 | 0.5 |
| OR2* | −10.5 | 0.5 |
| OR3* | −9.5 | 0.5 |
| OR3-r1†‡ | −6.6 | 0.5 |
| OR12coop* | −2.7 | 0.5 |
| OR23coop* | −2.9 | 0.5 |
| OL1§ | −13.0 | 0.5 |
| OL2§ | −11.2 | 0.5 |
| OL3§ | −12.0 | 0.5 |
| OL3–4‡ | see ΔGns | |
| OL12coop§ | −2.7 | 0.5 |
| OL23coop§ | −2.0 | 0.5 |
| Δ Goct†‡ | −0.5 | 0.5 |
| Δ Gtet†‡ | −3.0 | 0.5 |
| Δ Gns‡¶ | −4.1 | 0.9 |
We varied all parameters in increments of 0.1 kcal/mol over the range given in the table, even though the reported uncertainty estimates for several of the experimental values were smaller than 0.5 kcal/mol.
*From ref. 13.
†From estimates calculated in ref. 7.
‡Values that have not been directly measured.
§From data presented in figure 1B of ref. 12, recalculated with a dimerization free energy ΔGdim = −11.0 kcal/mol (36) and setting OL12coop = OR12coop = −2.7 kcal/mol. The values for OL cooperativity were not unambiguously determined by Senear et al. (12), but the chosen values reflect our assumptions.
¶From estimates calculated in ref. 35.
CI-Dependent Transcription Activation.
We used the configuration probabilities to calculate activation levels by assuming that the steady-state amount of CI was produced entirely by act1, act2, and act3. Unactivated transcription is considered to be negligible, because these configurations are rare at the observed CI concentrations and basal activity is only 10% of act1 (5). The concentration of CI for each construct is proportional to the probability of the operator DNA being in each activated state, weighted by the activation levels of act2 and act3 relative to act1, denoted as A2 and A3, respectively:
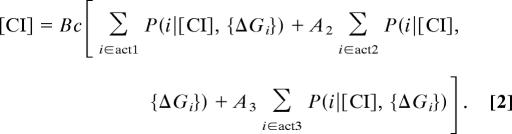 |
where c is a multiplicative factor that corrects for the changes in basal PRM activity of constructs bearing the OR3-r1 mutation: c = 1.09 for constructs with this mutation, and c = 1 for all others (11). B is a proportionality constant that represents the basal rate of mRNA production from PRM, the degradation and dilution rate constants for mRNA and protein, the rate constant for protein production from mRNA template, and cellular volume (see SI Text for a mathematical description of B). B is assumed to be identical for all constructs.
Relative Activation Levels.
To determine the magnitude of A2 and A3 for the two models, we sampled activation values calculated from Eq. 2, using combinations of free-energy parameters drawn randomly from the ranges listed in Table 1, and chose parameter sets that were able to reproduce the data within experimental error. For the UP element model, A2 ≈ 3.3, and A3 ≈ 1.5 (Fig. 5A). For the looped octamer activation model, A2 ≈ 2.2, and A3 ≈ 0.2 (Fig. 5B). A2 is well defined for a given activation model, but the values obtained for A3 depend on the exact assignment of the looped activation states (Fig. S3). To determine which of the many parameters used in the calculation have the largest effect on these activation values, we did sensitivity analysis by calculating the correlation between each free-energy parameter and the values obtained for A2 and A3. We found that the calculated activation levels were most sensitive to the binding free energies of ΔGoct, ΔGns, OR12coop and OR2 (Fig. S4 and Fig. S5). Based on this method of calculation, the predicted probability for each activation state varies widely, because many of the parameters are interdependent (data not shown). To get a better estimate of the probability of each activation state over a physiological range of CI concentrations, we repeated the analysis varying only the parameters for which direct experimental estimates are not available, OR3-r1, ΔGoct, ΔGtet, and ΔGns. Representative datasets are given in Fig. S6 and Dataset S2. The two models predict equivalent total activation curves for the set of constructs used in this study (Fig. 5C). A comparison of these curves with activation data from ref. 7 is shown in Fig. S7.
Fig. 5.
Modeled activation of PRM. (A) Histograms of A2 (blue) and A3 (red), the activation levels of act2 and act3 relative to act1, shown for parameter sets that predict ratios of CI within error of experimental data, calculated by using the UP element model to assign activation states. (B) Histograms of A2 and A3 calculated assuming activation through a looping interaction. (C) Theoretical activation curves for the five constructs (both models yield a set of curves that are essentially identical). Measured CI levels from this study (filled squares) occur where the mean production rates match the mean cellular levels. (D) The activation curve predicted for a wild-type lysogen (thick line) and a deconvolved fluorescence histogram from the WT construct, converted to number of CI per cell (thin line).
Discussion
Our data and the details of activation outlined above are not consistent with the full set of activation curves reported in the pioneering work by Dodd et al. or the model they developed, which predicts that DNA looping lowers the transcription rate by approximately a fourth (11) (see also ref. 18). We attribute this to differences in mRNA stability for some of the constructs used in their study. In the previous work, the constructs for measuring promoter activity in the presence and absence of OL did not contain the full lambda DNA sequence between OR and OL (11, 18). Instead, OR was cloned in front of the lac transcript. When present, the left operator was placed before the normal rho-independent transcription termination site. High-affinity sites for DNA-binding proteins can stall elongating transcription complexes such that RNAP must be removed from the stall site by premature transcript termination (19, 20). With as few as 50 CI monomers per cell, the probability that at least one of the left operator sites is occupied is >96% (this study). It is possible that, in constructs with OL placed before the terminator, transcripts are terminated prematurely and thus lack the stabilizing stem-loop structure characteristic of rho-independent termination (21), leading to lower protein concentrations. We tested this hypothesis by removing the stem-loop structure of the timm terminator for the PRM transcript in one of our constructs. As anticipated, we saw a substantial decrease in Venus fluorescence (Fig. S8). Our model is based on data from constructs that contain intact timm terminators and therefore all produce transcripts with identical stability.
Mechanisms.
Although our data clearly indicate that DNA looping can increase transcription activation, further studies are needed to clarify the underlying mechanism. From previous studies, activation of PRM is understood to occur by a direct interaction between the amino-terminal domain (NTD) of a CI bound at OR2 and specific sites in the σ and α subunits of RNAP (22, 23). These contacts activate transcription by stimulating isomerization of bound RNAP to an open complex capable of transcription initiation (24). This mechanism is based on in vitro experiments, using only right operator DNA, which corresponds to act1 in our models. At least two models for enhanced transcription activation due to looping are consistent with our data. For the first model, the UP element located near OL3 could further activate PRM by providing an additional site where the second α subunit of RNAP could interact. The α subunits of RNAP contain a linker domain that gives considerable flexibility in binding locations, suggesting that it might be able to interact with a site on the opposite side of the loop. Interestingly, when RNAP is bound to PRM, the locations of the two α subunits change when CI is bound nearby, with one α subunit unaccounted for; this opens the possibility that it could interact with OL when the DNA is looped (23). If the UP element is the only site where such an interaction could occur, deleting it in an otherwise wild-type construct would be expected to have the same result as deleting the entire left operator. A second possibility is that the additional octamer contacts that CI makes to form a loop (25) might involve conformational changes that affect the interactions with RNAP at OR2. When the NTD of CI binds to operator DNA, changes in the carboxyl-terminal domain (CTD) of CI that are sensitive to the number and identity of adjacent sites have been detected (26, 27), which suggests conformational changes for octamerization could potentially include the region of the NTD that interacts with RNAP. Our data fits both models well, so additional experiments are needed to determine whether either (or possibly neither) of these microscopic interpretations of looped activation is correct.
Implications for Lambda Regulation.
Although our data do not resolve the detailed mechanism for how the loop enhances PRM activation, they still provide insight into the behavior of a lysogen. Based on the distribution of fluorescence intensity measured in our WT construct, which reflects the CI concentrations found in a lysogenic population or a single lysogen over time, the lysogenic state is expected to be very stable. The probability distribution overlaps with the activation curve such that cells with the lowest levels of CI have high transcription rates (Fig. 5D), which will drive the CI concentration back toward the mean value. This is consistent with our observation that the CI level in the WT construct does not spontaneously fall to <10% of the mean lysogenic value, the threshold for switching to the lytic state (4); from our data and assuming a mean of 150 monomers per cell, we estimate that ≈50 CI monomers is the lowest value accessible to a lysogenic cell. Our analysis predicts that at this level, the probability that PR would be de-repressed (i.e., OR1 is unoccupied) is ≈0.2, and it falls rapidly as the CI level increases. At the same time, the mean PRM activation rate is high, so PR transcription is expected be transient and rare. This is consistent with quantitative PCR measurements of cro transcript levels in a population of cells containing our WT construct (see SI Text). Under these conditions, the only way for a lysogen to switch to the lytic state is by catalyzed degradation of CI, which occurs when RecA is activated as part of the SOS response to DNA damage. Rates for producing new CI molecules and for RecA-catalyzed degradation vary in complex ways as a function of total CI levels, but to successfully clear CI from the cell, at any given CI concentration, the rate of cleavage must presumably be faster than its rate of production from PRM. Even so, catalyzed degradation of CI only needs to persist long enough to free PR and allow cro to be produced; when cro is present in sufficient quantity to repress PRM, the cell can fully commit to lytic growth (28). In the absence of SOS signals of cellular distress, the steep activation curve effectively buffers against spontaneous lytic induction, and a low average rate of CI production is sufficient to maintain lysogeny, allowing the phage to place a minimal load on the host cell as it is quietly maintained in the genome.
By measuring the in vivo CI concentration in our constructs and drawing on the elegant experimental and theoretical work that has been done over the last several decades, we were able to show that the interaction of OL and OR via a DNA loop can either activate or repress PRM. These findings bring us closer to explaining the notable stability and efficiency of bacteriophage lambda's genetic switch. As more experiments and simulations are done to explore the basis and the outcome of this activation behavior, lambda will continue to be an important system for understanding the role of DNA loops in gene regulation.
Materials and Methods
Strains and Plasmids.
E. coli DH5α was routinely used as a cloning host. Fluorescence experiments were done on E. coli K12λ−F− (MG1655) transformed with the indicated plasmids. All immunity region clones were first constructed in the high copy number plasmid pLOI2403 [obtained from L. Ingram (University of Florida, Gainesville, FL)] (29). Primer sequences and details of molecular cloning are presented in SI Text. Each high-copy clone was verified by sequencing, then the cro-OR-cI-venus-rexA-rexB-OL fragment was subcloned into the low-copy-number plasmid pKLJ12 (10). For testing the effect of deleting the timm terminator, a PCR-based mutagenesis method was used (30).
Culture Conditions.
Cells were grown in EZ Rich Defined medium (EZRDM) (Teknova) (31) with 10 mM glucose for flow cytometry and in Luria broth for cloning. Growth was at 37°C with good aeration. High-copy pLOI2403 plasmids and low-copy pKLJ12 plasmids were maintained with 100 and 50 μg/ml ampicillin, respectively.
Flow Cytometry.
Samples were started from overnight EZRDM cultures at 104-fold dilution and incubated at 37°C. Cells were harvested every 10 min between 3 and 5 h of growth after dilution. The OD600 ranged from 0.15 to 3.5, with a cell doubling time of 26 min. Immediately upon harvest, spectinomycin was added at a concentration of at least 200 μg/ml to inhibit further protein synthesis. Samples were protected from light and stored at 4°C until analysis. Forward scatter (FSC), side scatter (SSC), and cellular fluorescence (FL1) were collected by using a FACSCalibur flow cytometer (BD Biosciences). Datasets of 100,000 events were collected at the lowest flow rate, ≈12 μl/s. All FSC, SSC, and FL1 data were collected in linear mode. Detector settings are in SI Text. Flow cytometry FCS data files were imported into MatLab, using code from the script “FCS data reader” written by L. Balkay (University of Debrecen, Debrecen, Hungary), obtained from the MatLab Central file exchange web site (www.mathworks.com). We selected a data point that had the least variation in FSC and SSC between constructs and filtered the data to represent cells that differ in volume by only 5–7.5% then deconvolved the data to remove instrument noise, but we found that the ratios of the fluorescence intensities for the mutant constructs calculated in this way and ratios calculated from the mean of the raw histograms were indistinguishable (see SI Text).
Estimating CI Levels.
The Venus fluorescent intensities were converted to CI concentration by assuming that the concentration of CI in our WT construct was the same as a lysogenic cell, which is supported by quantitative PCR of cI mRNA (see SI Text). The mean number of CI monomers in lysogenic E. coli cell was taken to be ≈150 molecules per cell (32, 33). The estimated mean cell volume, v̄ = 2.0 μm3, was calculated from the 26 min doubling time of the cells (34). Using this value, a single molecule per cell corresponds to a concentration of 0.83 nM.
Operator Configurations and Activation States.
The probability of each operator configuration for each mutant construct was calculated from Eq. 1, where the relationship between [CI]total, the total number of CI monomers per cell, and [CI2], the number of dimers available to bind operator DNA (35), was obtained by solving
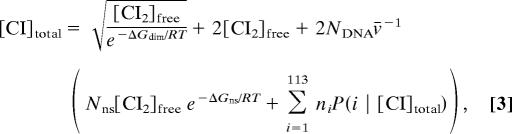 |
where the free energy of dimerization, ΔGdim = −11.0 kcal/mol (36), and NDNA = 3.3 is the copy number of both the genomic DNA and the plasmid, calculated from the doubling time (34). Nns = 4.64 × 106 is the number of nonspecific binding sites, i.e., the E. coli genome size, and ΔGns is the average nonspecific binding energy, which we varied from −5.0 to −3.2 kcal/mol (35) in increments of 0.1 kcal/mol. We used the linsolve function in MatLab to calculate A2 and A3 for each randomly chosen parameter set by using Eq. 2. Because activation factors are proportional to the rate of mRNA production, we discarded any negative activation values. We calculated the error of each parameter set as the root mean squared difference between the observed and predicted ratios of CI levels in the mutants relative to the WT construct. The sensitivity of A2 and A3 to the free energy parameters was determined by calculating the coefficient of linear correlation, r = σxy/(σxσ y), for each parameter.
Supplementary Material
Acknowledgments.
We thank C. Bertozzi for use of the flow cytometer; A. Arkin for helpful comments on the manuscript; W. Chang for help with cloning and the initial cytometry experiments; S. Banani for help in the early stages of cloning; R. Calendar (University of California, Berkeley, CA), L. Ingram, J. Keasling (University of California, Berkeley, CA), and the Yeast Resource Center, University of Washington, for strains and plasmids; I. B. Dodd (University of Adelaide, Adelaide, Australia) for graciously providing datasets for PRM activation curves; and three anonymous reviewers who offered insightful and constructive comments that led to the present form of this article. This work was supported by a National Science Foundation Graduate Research Fellowship (to L.M.A.), Department of Energy Contact DE-AC03-76SF00098, and University of California, Berkeley.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0705570105/DCSupplemental.
References
- 1.Ptashne M. 3rd Ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2004. A Genetic Switch: Phage Lambda Revisited. [Google Scholar]
- 2.Little JW, Shepley DP, Wert DW. Robustness of a gene regulatory circuit. EMBO J. 1999;18:4299–4307. doi: 10.1093/emboj/18.15.4299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aurell E, Brown S, Johanson J, Sneppen K. Stability puzzles in phage lambda. Phys Rev. 2002;65:e051914. doi: 10.1103/PhysRevE.65.051914. [DOI] [PubMed] [Google Scholar]
- 4.Bailone A, Levine A, Devoret R. Inactivation of prophage-lambda repressor in vivo. J Mol Biol. 1979;131:553–572. doi: 10.1016/0022-2836(79)90007-x. [DOI] [PubMed] [Google Scholar]
- 5.Meyer BJ, Ptashne M. Gene-regulation at the right operator (OR) of bacteriophage-lambda. 3. Lambda-repressor directly activates gene-transcription. J Mol Biol. 1980;139:195–205. doi: 10.1016/0022-2836(80)90304-6. [DOI] [PubMed] [Google Scholar]
- 6.Revet B, von Wilcken-Bergmann B, Bessert H, Barker A, Muller-Hill B. Four dimers of lambda repressor bound to two suitably spaced pairs of lambda operators form octamers and DNA loops over large distances. Curr Biol. 1999;9:151–154. doi: 10.1016/s0960-9822(99)80069-4. [DOI] [PubMed] [Google Scholar]
- 7.Dodd IB, Shearwin KE, Perkins AJ, Burr T, Hochschild A, Egan JB. Cooperativity in long-range gene regulation by the lambda CI repressor. Genes Dev. 2004;18:344–354. doi: 10.1101/gad.1167904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nagai T, Ibata K, Park ES, Kubota M, Mikoshiba K, Miyawaki A. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat Biotechnol. 2002;20:87–90. doi: 10.1038/nbt0102-87. [DOI] [PubMed] [Google Scholar]
- 9.Shean CS, Gottesman ME. Translation of the prophage-lambda CI transcript. Cell. 1992;70:513–522. doi: 10.1016/0092-8674(92)90175-c. [DOI] [PubMed] [Google Scholar]
- 10.Jones KL, Keasling JD. Construction and characterization of F plasmid-based expression vectors. Biotechnol Bioeng. 1998;59:659–665. [PubMed] [Google Scholar]
- 11.Dodd IB, Perkins AJ, Tsemitsidis D, Egan JB. Octamerization of lambda CI repressor is needed for effective repression of P-RM and efficient switching from lysogeny. Genes Dev. 2001;15:3013–3022. doi: 10.1101/gad.937301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Senear DF, Brenowitz M, Shea MA, Ackers GK. Energetics of cooperative protein DNA interactions—comparison between quantitative deoxyribonuclease footprint titration and filter binding. Biochemistry. 1986;25:7344–7354. doi: 10.1021/bi00371a016. [DOI] [PubMed] [Google Scholar]
- 13.Koblan KS, Ackers GK. Site-specific enthalpic regulation of DNA-transcription at bacteriophage-lambda OR. Biochemistry. 1992;31:57–65. doi: 10.1021/bi00116a010. [DOI] [PubMed] [Google Scholar]
- 14.Meyer BJ, Maurer R, Ptashne M. Gene-regulation at the right operator (OR) of bacteriophage-lambda. 2. OR1, OR2, and OR3—their roles in mediating the effects of repressor and cro. J Mol Biol. 1980;139:163–194. doi: 10.1016/0022-2836(80)90303-4. [DOI] [PubMed] [Google Scholar]
- 15.Maurer R, Meyer BJ, Ptashne M. Gene-regulation at the right operator (OR) of bacteriophage-lambda. J Mol Biol. 1980;139:147–161. doi: 10.1016/0022-2836(80)90302-2. [DOI] [PubMed] [Google Scholar]
- 16.Giladi H, Murakami K, Ishihama A, Oppenheim AB. Identification of an UP element within the IHF binding site at the P(L)1-P(L)2 tandem promoter of bacteriophage lambda. J Mol Biol. 1996;260:484–491. doi: 10.1006/jmbi.1996.0416. [DOI] [PubMed] [Google Scholar]
- 17.Shea MA, Ackers GK. The or control-system of bacteriophage-lambda—a physical-chemical model for gene-regulation. J Mol Biol. 1985;181:211–230. doi: 10.1016/0022-2836(85)90086-5. [DOI] [PubMed] [Google Scholar]
- 18.Michalowski CB, Short MD, Little JW. Sequence tolerance of the phage lambda P-RM promoter: Implications for evolution of gene regulatory circuitry. J Bacteriol. 2004;186:7988–7999. doi: 10.1128/JB.186.23.7988-7999.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pavco PA, Steege DA. Elongation by it Escherichia coli RNA-polymerase is blocked in vitro by a site-specific DNA-binding protein. J Biol Chem. 1990;265:9960–9969. [PubMed] [Google Scholar]
- 20.Roberts J, Park JS. Mfd, the bacterial transcription repair coupling factor: Translocation, repair and termination. Curr Opin Microbiol. 2004;7:120–125. doi: 10.1016/j.mib.2004.02.014. [DOI] [PubMed] [Google Scholar]
- 21.Grunberg-Manago M. Messenger RNA stability and its role in control of gene expression in bacteria and phages. Annu Rev Genet. 1999;33:193–227. doi: 10.1146/annurev.genet.33.1.193. [DOI] [PubMed] [Google Scholar]
- 22.Jain D, Nickels BE, Sun L, Hochschild A, Darst SA. Structure of a ternary transcription activation complex. Mol Cell. 2004;13:45–53. doi: 10.1016/s1097-2765(03)00483-0. (2004) [DOI] [PubMed] [Google Scholar]
- 23.Kedzierska B, et al. The C-terminal domain of the Escherichia coli RNA polymerase subunit plays a role in the CI-dependent activation of the bacteriophage lambda PM promoter. Nucleic Acids Res. 2007;35:2311–2320. doi: 10.1093/nar/gkm123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fong R SC, Woody S, Gussin GN. Modulation of P(RM) activity by the lambda-P(R) promoter in both the presence and absence of repressor. J Mol Biol. 1993;232:792–804. doi: 10.1006/jmbi.1993.1432. [DOI] [PubMed] [Google Scholar]
- 25.Bell CE, Lewis M. Crystal structure of the lambda repressor c-terminal domain octamer. J Mol Biol. 2001;314:1127–1136. doi: 10.1006/jmbi.2000.5196. [DOI] [PubMed] [Google Scholar]
- 26.Deb S, Bandyopadhyay S, Roy S. DNA sequence dependent and independent conformational changes in multipartite operator recognition by lambda-repressor. Biochemistry. 2000;39:3377–3383. doi: 10.1021/bi9919955. [DOI] [PubMed] [Google Scholar]
- 27.Ghosh K, Chattopadhyaya R. Papain does not cleave operator-bound lambda repressor: Structural characterization of the carboxy terminal domain and the hinge. J Biomol Struct Dyn. 2001;18:557–567. doi: 10.1080/07391102.2001.10506688. [DOI] [PubMed] [Google Scholar]
- 28.Schubert RA, Dodd IB, Egan JB, Shearwin KE. Cro's role in the CI-cro bistable switch is critical for lambda's transition from lysogeny to lytic development. Genes Dev. 2007;21:2461–2472. doi: 10.1101/gad.1584907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martinez-Morales F, Borges AC, Martinez K, Shanmugam KT, Ingram LO. Chromosomal integration of heterologous DNA in Escherichia coli with precise removal of markers and replicons used during construction. J Bacteriol. 1999;181:7143–7148. doi: 10.1128/jb.181.22.7143-7148.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zheng L, Baumann U, Reymond JL. An efficient one-step site-directed and site-saturation mutagenesis protocol. Nucleic Acids Res. 2004;32:e115. doi: 10.1093/nar/gnh110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Neidhardt F, Bloch P, Smith D. Culture medium for enterobacteria. J Bacteriol. 1974;119:736–747. doi: 10.1128/jb.119.3.736-747.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Levine A, Bailone A Devoret R. Cellular-levels of the prophage-lambda and 434-repressors. J Mol Biol. 1979;131:655–661. doi: 10.1016/0022-2836(79)90014-7. [DOI] [PubMed] [Google Scholar]
- 33.Reichardt L, Kaiser AD. Control of lambda repressor synthesis. Proc Natl Acad Sci USA. 1971;68:2185–2189. doi: 10.1073/pnas.68.9.2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Donachie WD, Robinson AC. Neidhardt F. Vol 2. Washington, DC: American Society for Microbiology; 1987. Escherichia coli and Salmonella typhimurium; pp. 1578–1593. [Google Scholar]
- 35.Bakk A, Metzler R. Nonspecific binding of the O-R repressors CI and cro of bacteriophage lambda. J Theor Biol. 2004;231:525–533. doi: 10.1016/j.jtbi.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 36.Koblan KS, Ackers GK. Energetics of subunit dimerization in bacteriophage-lambda CI repressor—linkage to protons, temperature, and KCl. Biochemistry. 1991;30:7817–7821. doi: 10.1021/bi00245a022. [DOI] [PubMed] [Google Scholar]
- 37.Chattopadhyaya R, Ghosh K. A comparative three-dimensional model of the carboxyl-terminal domain of the lambda repressor and its use to build intact repressor tetramer models bound to adjacent operator sites. J Struct Biol. 2003;141:103–114. doi: 10.1016/s1047-8477(02)00627-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.