Abstract
Describing at a genomic scale how mutations in different genes influence one another is essential to the understanding of how genotype correlates with phenotype and remains a major challenge in biology. Previous studies pointed out the need for accurate measurements of not only synthetic but also buffering interactions in the characterization of genetic networks and functional modules. We developed a sensitive and efficient method that allows such measurements at a genomic scale in yeast. In a pilot experiment (41 genome-wide screens), we quantified the fitness of 140,000 double deletion strains relative to the corresponding single mutants and identified many genetic interactions. In addition to synthetic growth defects (validated experimentally with factors newly identified as genetically interfering with mRNA degradation), most of the identified genetic interactions measured weak epistatic effects. These weak effects, rarely meaningful when considered individually, were crucial to defining specific signatures for many gene deletions and had a major contribution in defining clusters of functionally related genes.
Keywords: epistasis, functional genomics, genetic screen, mRNA decapping, Saccharomyces cerevisiae
Following the completion of the genomic sequence for Saccharomyces cerevisiae, a systematic gene deletion library was built as a tool to link genes to functions and phenotypes. Yet, the phenotypic consequences of single deletions are rarely sufficient to define the function of genes. Moreover, very little is known of the phenotypic influences that different mutations have on each other. A large panel of responses can be observed when combining mutations, from aggravating to neutral, buffering, and even alleviating effects. Several high-throughput genetic screen methods, such as SGA (synthetic genetic array), dSLAM, and SLAM (synthetic lethality analyzed by microarray) (1–3), analyze the growth defect of combining a given query mutation with every gene deletion from the library of tagged nonessential yeast knockouts. These approaches are useful in identifying the strong synthetic defects that are seen for only a minor fraction of all of the possible gene deletion pairs (≈0.5%) (4). However, they are not suited to evaluating more general buffering relationships between genes. Yet, recent studies have demonstrated the importance of accurate measurements of the complete spectrum of genetic interactions to define functional gene modules (5, 6). Broader quantitative measurements of genetic interactions are obtained in epistatic miniarrays (E-MAPs) (5, 7) but at the expense of coverage, because the E-MAP results depend on high-density genetic interaction matrixes made possible by focusing on logically connected gene subsets. Here, we present a method that we call GIM for “genetic interaction mapping,” which is not limited to a subset of genes, and allows sensitive and quantitative measurements of the complete spectrum of genetic interactions.
Double mutant populations are efficiently obtained by mating and sporulation. The abundance of individual double mutants in the query population relative to a reference mutant population is analyzed on glass slide microarrays that detect and quantify the tags marking each deletion of the library (8). Besides the strong synthetic growth defects that could be observed in some of the screens, GIM's high sensitivity allowed for the detection of a larger number of subtle, yet specific, synthetic and epistatic interactions. These subtle effects had a major impact in defining genetic interaction profiles when considered over the whole range of experiments. These profiles were specific for classes of functionally related genes and allowed functional modules to be defined. Because GIM screens are easy to scale-up, the method now makes possible the drawing of a complete quantitative genetic interaction map for yeast gene deletions.
Results
Description of the Method.
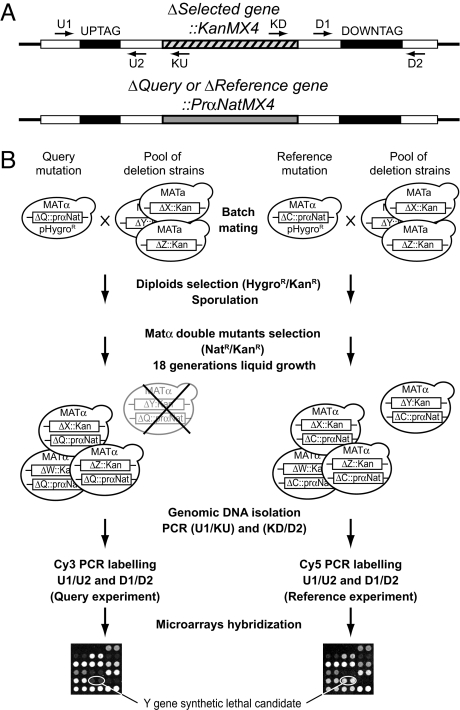
Our GIM strategy is described in Fig. 1. It combines features from both the SGA (1) and SLAM (2) approaches. In the SGA method, individual mutants of the deletion library are individually combined with the query mutation by a mating and sporulation strategy. In the GIM method, the double mutants are also generated by mating and sporulation but in a single pool combining all nonessential gene deletions of the collection. In addition, the selection of the haploid double mutants is performed in rich liquid medium in a single step, using a novel haploid-specific marker that allows for the simultaneous selection of the query mutation and of haploid cells. This marker was developed after testing several promoters of genes specifically expressed in haploid cells. We observed that strong promoters, such as those of the MFA1/YDR461W or MFα1/YPL187W genes, conferred very poor selectivity for haploid cells in association with the nourseothricin antibiotic resistance gene, NatR (nat1, GenBank gi:300195; data not shown). In contrast, the weak MF(ALPHA)2/YGL089C promoter (prMFα2) allowed excellent selectivity for MATα haploid cells when associated with NatR [see supporting information (SI) Fig. S1A]. The query mutation strains can be easily obtained by homologous recombination replacement of the KanR cassette with the new prMFα2-NatR cassette in any MATα strain from the systematic deletion collection (see Fig. S1B). After selection of the diploids, sporulation is performed in batch. Haploid double mutants are selected and grown in a single pool for a controlled number of generations in rich liquid medium containing nourseothricin and geneticin. The SLAM and dSLAM methods also work with double mutant pools but require high-efficiency transformation/recombination to introduce the query mutation in the library and double mutants are selected on plates, not allowing a strict control of the number of cell divisions. To detect subtle fitness defects, we chose to perform competitive growth of the double mutants for a relatively large number of generations (18 doublings). Although the cultures of the pools can be conducted by successive dilutions, better reproducibility and sensitivity were obtained when the cultures were performed at a constant cell density by continuous dilution using a multiple microfermenter battery (9).
Fig. 1.
The genetic interaction mapping (GIM) method. (A) The generated double-mutant haploid strains contain the resistance markers and the oligonucleotide tags for the query gene deletion and for a gene deletion from the collection. (B) Schematics of the GIM method. For every screen, two experiments are run in parallel, one with the query gene deletion strain and another with a reference deletion. Reference deletions were chosen to affect dubious ORFs, for which no significant effect on growth has been observed, either alone or in combination with any other deletion (see Fig. S1C). The query and reference deletions are tagged with a haploid-specific nourseothricin resistance marker. The strains are transformed with a plasmid that bears a hygromycin resistance cassette for diploids selection. Each of these strains is then mated with a pool of strains containing all of the viable strains from the MATa haploid gene deletion collection. The diploids are selected by using hygromycin and kanamycin resistance. Following sporulation of the heterozygous diploids, the MATα double mutant spores are selected and grown for ≈18 generations in rich liquid medium (YPD) containing nourseothricin and geneticin. Cells are collected and the tags marking the deletions of the pool are amplified by PCR and labeled with Cy3 or Cy5. Microarrays are used to measure the relative abundance of double mutants within the query versus the reference populations.
Similarly to the SLAM approach, the estimation of the individual double mutants' relative growth rates was performed by using bar-code DNA microarrays. DNA regions encompassing the deletion-specific tags were amplified by PCR from the cultures and quantified in comparison with the tags amplified from a reference population (see Fig. 1 and SI Methods). The intensity of the signal measured on microarrays for each double mutant in the query population, when compared with a reference population, allowed an estimation of the relative proportion of cells of a given double mutant in each population after 18 doublings of competitive growth. Normalized results are expressed as log2(Q/R) where Q represents the signal intensities of the tags marking a given mutant when combined with the query mutation and R the signal from the same mutant when introduced in a reference strain (see SI Methods). In each screen, two reference experiments with one or two different control deletions (ymr326cΔ, yfr057wΔ, or yel068cΔ) were generated in parallel to the query populations by submitting reference strains to an identical procedure. To validate their neutrality, we used each reference deletion as a query mutation in GIM screens and verified that the log2(Q/R) value was never >1 (Fig. S1C).
Identification of Novel Factors Interfering with mRNA Decapping.
To illustrate the sensitivity and reproducibility of the GIM screens, the results of a typical experiment, performed by using the deletion of EDC3 (an enhancer of mRNA decapping) as a query mutation, are shown in Fig. 2. The deletion of EDC3 was previously shown to be synthetic lethal at 30°C with double mutants of genes involved in mRNA decapping and degradation (dcp1-2 ski3Δ or dcp2-7 ski8Δ). Critically diminished decapping activity in these mutant combinations correlates with a cellular growth defect (10). The presence of EDC3 is also essential for decapping mediated autoregulation mechanisms that control the levels of the RPS28B mRNA (11) and YRA1 pre-mRNA (12).
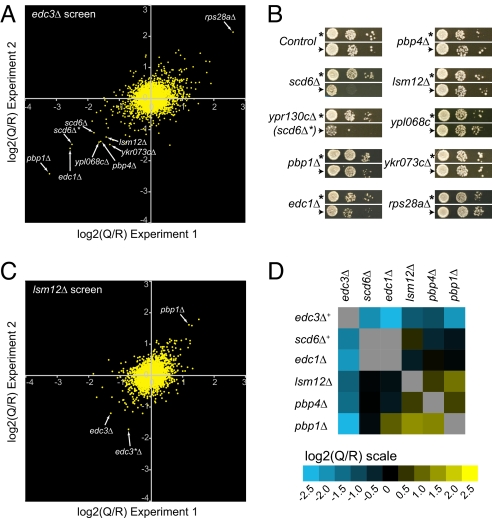
Fig. 2.
Reproducibility and sensitivity of GIM. (A) Comparison of two independent screens with edc3Δ as a query mutation and ymr326cΔ as a reference. Arrows point to the values obtained for mutants that were subsequently tested on plates. (B) Plate assays for double deletions with edc3Δ. Dilution series of sporulated cultures of double mutants obtained by combining either the query mutation edc3Δ::prMFα2NatR (arrowhead) or the control mutation ymr326cΔ::prMFα2NatR (asterisk) and the indicated deletions (“Control” represents the deletion of YEL068C, another reference). Cultures were grown at 23°C on rich medium containing geneticin and nourseothricin. The deletion of the dubious ORF YPR130C was called “scd6Δ*” because it overlaps the scd6Δ deletion and thus constitutes an independent mutation of SCD6. (C) As in A except that lsm12Δ was used as the query mutation. (D) Matrix of genetic interactions between genes whose deletions showed a synthetic growth defect with edc3Δ. The values are means of at least two independent screens. The plus sign after edc3Δ and scd6Δ indicates that the shown values were averaged from edc3Δ and yel014cΔ and from scd6Δ and ypr130cΔ, respectively.
To have a global view of the results obtained in an edc3Δ screen, we plotted the log2(Q/R) of every double mutant in relation to the genomic position of the deleted prey ORFs (Fig. S1D). For a robust estimation of the values, two reference deletions were used in independent hybridizations for most experiments. In addition, for most of the query mutations, the experiments were performed twice independently. The values obtained in these two independent experiments were highly correlated, indicating good reproducibility of the GIM screens (see Fig. 2A for an example).
To estimate how the log2(Q/R) correlate with visible growth defects, the double mutants showing the strongest synthetic effects in our screens with edc3Δ were tested individually on plates (Fig. 2B). Only the double mutants with the strongest log2(Q/R) exhibited a visible phenotype. We also tested the edc3Δ rps28aΔ double mutant that exhibited the lowest log2(Q/R) value (≈2.5) and found that edc3Δ partially suppressed the slow growth phenotype induced by rps28aΔ, as expected (11).
Synthetic growth defects such as those measured for edc3Δ lsm12Δ or edc3Δ pbp4Δ were thus too weak to be readily discernible on plates, yet they appeared to be reproducible. To test reciprocity, we performed screens with the newly identified gene deletions as query mutations. Fig. 2C shows the results of the experiment performed with an lsm12Δ query strain. The overall amplitude of genetic interactions observed with lsm12Δ was weak when compared with the edc3Δ screens. Strikingly, edc3Δ and the deletion of the adjacent dubious ORF YEL014W (called edc3Δ* because it consists of a deletion of the 3′ UTR of EDC3 and always behaved similarly to edc3Δ in the screens) exhibited the strongest synthetic growth defects [lowest log2(Q/R) values] with lsm12Δ. Interestingly, pbp1Δ, which exhibited a synthetic growth defect when combined with edc3Δ, showed a strong positive log2(Q/R) value in combination with lsm12Δ. Lsm12 and Pbp1 were recently shown to physically interact (13). The positive genetic interaction observed between pbp1Δ and lsm12Δ is thus likely to illustrate the epistatic interactions observed between mutants affecting proteins of the same complex (5). Indeed, the growth defect induced by a mutation can be masked by the growth defect of the query mutation (or vice versa). Because the query and reference populations are not grown for the same period but for the same number of doublings, epistatic (buffering) interactions result in positive values in the screens (for an example and explanations of this effect, see Fig. S2). Positive values thus correspond to either epistatic (buffering) or suppressive (alleviating) interactions.
The results of screens performed with other deletions that showed synthetic growth defects with edc3Δ (scd6Δ, edc1Δ, pbp1Δ, and pbp4Δ) as query mutations are summarized in a matrix (Fig. 2D; see also Fig. S3), which is symmetrical, demonstrating the reciprocity of the interactions. From all of these observations, we conclude that our approach is simple, is very sensitive, and can measure not only strong synthetic growth defects but also weak genetic interactions.
Functional Validation of Synthetic Interactions.
The above results identified new genetic interactions between EDC3 and SCD6, PBP1, LSM12, PBP4, and EDC1. With the exception of Pbp4, the other proteins, Scd6/Lsm13, Lsm12, Pbp1, and Edc3/Lsm16, share a common Lsm (like Sm) domain (14) thought to mark their involvement in RNA processing (15). Edc1 was previously described as an enhancer of decapping, like Edc3 (16).
To identify protein associated with Scd6, we performed both a two-hybrid screen and a tandem affinity purification (17) experiment. In agreement with the newly identified genetic interactions, Scd6 was found to be associated with different factors involved in mRNA decapping: Edc3, Dcp1, Dcp2, and Dhh1 (Fig. S4A and Table S1; for a summary of the interactions from this work and the literature, see Fig. 3A). Scd6 also colocalizes with the decapping factor Dcp2 within P bodies (18). These observations suggest the appealing hypothesis that the wild-type growth of the edc3Δ strain is due to the redundant function of Scd6 and not to a minor role of Edc3 in mRNA decay, as previously suggested (10).
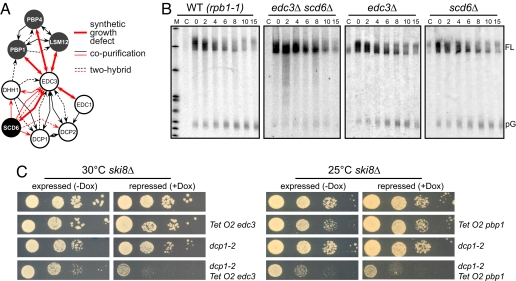
Fig. 3.
Functional validation of novel genes identified by GIM as linked to RNA degradation. (A) Overview of biochemical and genetic interactions shared by EDC3, EDC1, SCD6, PBP1, PBP4, and LSM12. Red lines correspond to interactions uncovered during this work (Fig. 2, Table S1, Dataset S3, and Fig. S4A), whereas black lines correspond to published data compiled in the BioGRID database (24). (B) MFA2pG mRNA degradation time courses in edc3Δ, scd6Δ, or edc3Δ scd6Δ strains compared with a wild type (WT). All strains carried the temperature-sensitive RNA-pol II mutation rpb1-1 and the MFA2pG reporter mRNA under the control of a pGAL1 promoter. Cultures were grown at 25°C in raffinose medium, shifted for 1 h at 25°C in galactose medium to induce MFA2pG transcription. At time 0, the cells were transferred in glucose-containing medium at 37°C, and aliquots were taken at the indicated time points. Lane C corresponds to a noninduced culture as control. Northern blots from total RNAs separated on 5% polyacrylamide gels were probed with a radiolabeled oligo-C specific for the poly(G) of the MFA2pG mRNA. FL and pG indicate the positions of full-length mRNAs and of the polyG degradation intermediate, respectively. (C) Genetic interactions between EDC3 (Left) or PBP1 (Right) and the decapping enzyme DCP2 in a cytoplasmic exosome mutant background (ski8Δ). EDC3 and PBP1 were placed under the control of the doxycycline repressible TetO2 promoter (25) in ski8Δ or ski8Δ, dcp1-2 double mutants (10). After growth at 25°C, 10-fold dilution series of cultures at the same optical density were spotted on YPD complete medium without or with doxycycline (4 μg/ml; −Dox/+Dox) and incubated at 25°C or 30°C as indicated.
A prediction of this hypothesis is that the growth impairment observed in the edc3Δ scd6Δ double mutant is accompanied by a strong defect in mRNA decapping. To test this prediction, we used the unstable MFA2 mRNA marked with an oligo(G) tract (MFA2pG) under the control of a galactose inducible promoter (19). The absence of the small pG intermediate RNA in the experiment performed with the edc3Δ scd6Δ strain compared with the wild type or the corresponding single mutants (Fig. 3B) indicated that the 5′ to 3′ degradation pathway was impaired in this double mutant. Analysis of the full-length transcripts indicated that deadenylation proceeded normally but the deadenylated form accumulated in the edc3Δ scd6Δ double mutant (Fig. S4 B and C), excluding the hypothesis of an enhanced 3′ to 5′ decay. Altogether, these results are fully consistent with the hypothesis that Scd6 affects, directly or indirectly, the 5′ to 3′ mRNA degradation pathway.
As for Scd6, the Pbp1, Lsm12, or Pbp4 factors had not been previously linked to mRNA decapping. Pbp1, Pbp4, and Lsm12 were recently found to be part of the same complex (13). We focused our analysis on PBP1, which showed the strongest synthetic growth defect with edc3Δ. In contrast to edc3Δ scd6Δ, the synthetic growth defect observed when combining edc3Δ with pbp1Δ was not correlated with strong changes in MFA2pG mRNA degradation (Fig. S4D). In addition, Pbp1-eGFP did not accumulate in cytoplasmic foci upon glucose deprivation (data not shown). To investigate whether the observed genetic interaction between edc3Δ and pbp1Δ is nevertheless linked to the decapping function of Edc3, we tested whether the depletion of Pbp1 exhibited a synthetic growth defect with a dcp1-2 mutant. Such an effect is expected to be enhanced in a ski8Δ background [impairment of the cytoplasmic exosome activity (20)] in which a compromised decapping activity becomes rate-limiting for growth. As a control, and in agreement with previous reports (10), repression of EDC3 in this genetic context resulted in growth impairment (Fig. 3C Left). Pbp1 was toxic when overexpressed. Nevertheless, when the weak TetO2 promoter was used at 25°C, repression of PBP1 upon addition of doxycycline resulted in further growth inhibition in the presence of the dcp1-2 mutation (Fig. 3C Right). Thus, the depletion of Pbp1 led to a synthetic growth inhibition when combined with two independent mutants compromised in decapping, edc3Δ or dcp1-2. Pbp1 thus genetically interacts with mRNA decapping factors and therefore may have some role, probably in association with Lsm12 and Pbp4, in mRNA degradation. In conclusion, all of the factors identified in the edc3Δ screens seem to interfere, directly or indirectly, with mRNA turnover.
GIM as a Genome-Wide Approach.
We conducted 73 screens performed with 41 different query mutations (32 duplicates) chosen within genes involved in several RNA metabolism pathways. Of the 4,821 strains initially present in the pool, 3,812 exhibited a signal-to-noise ratio above background in more than half of the experiments (see Dataset S1 and SI Methods). We improved the direct genetic interaction results by introducing a synthetic growth defect (SGD) score that introduces, in addition to the direct log2(Q/R) value, parameters reflecting the specificity of the interaction, both within a single experiment (kurtosis of the log2 values and rank within a given screen) and within the whole dataset [median of the log2(Q/R) for a given target deletion in all screens]. The SGD score was computed as the linear combination of these values, where their respective weights were optimized based on a logistic regression model with a manually selected list of specific interactions as target (Dataset S2; and see SI Methods). We arbitrarily defined three threshold values, 6.5, 5, and 3.5 times the standard deviation of the SGD scores, corresponding to high (176 pairs; 182 pairs including the reciprocal interactions—176/182), medium (258/265), and lower confidence (622/634) categories (Dataset S3). The performance of the SGD scores was compared with the similar measures from a recent extensive quantitative study of epistasis conducted in matrix (E-MAP) (5). We tested 41 query mutations against 3,812 target mutations, giving rise to ≈140,000 reproducible measurements, whereas in the largest E-MAP published set, 745 mutants were tested, generating data for ≈150,000 double mutants. Thus, although they have different structures, both series of data are of comparable size. 1,972 pairs of deletions were tested in both datasets and had both an SGD and E-MAP score (Fig. 4A). The two sets of data were found to be very highly correlated (P < 10−15; Kendall correlation test), thus providing independent validation of our method on a large scale. For a few cases with strong or medium SGD scores but not selected by the E-MAP screens, an aggravating phenotype could be observed for the mutant combinations on plates (see Fig. S5A for examples). Note, however, that some discrepancies between the results obtained with the two methods can result from differences between growth conditions (mid-log liquid medium versus growth on plates). In addition, pairs with weaker SGD scores might not exhibit a detectable phenotype on plates, as shown in Fig. 2 A and B. Reproducibility of the screens was also assessed by the good correlation of values obtained for reciprocal pairs, when available (Fig. S5B and Dataset S1). We conclude that large-scale GIM screens give reliable and sensitive estimates of synthetic growth effects.
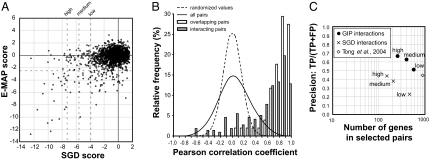
Fig. 4.
GIM provides robust, specific genetic interaction profiles that correlate with function. (A) SGD scores, centered on 0, and E-MAP scores (5) for the 1,972 pairs of deletions for which the synthetic growth defects were measured by both methods were plotted, with dotted lines indicating the arbitrarily defined strong, medium, and weak SGD scores as well as the E-MAP threshold value for significant genetic interactions. (B) To evaluate the reproducibility of genetic interaction profiles and their correlation with protein–protein interactions, we calculated the Pearson correlation coefficient for all of the possible 598,965 pairs of combinations of the 1,095 selected target gene deletions. The frequency distribution of the correlation coefficients (continuous line) was compared with the frequency distribution of a randomized set (dashed line). Subsets of pairs for known interacting proteins (21) (gray bars; “interacting pairs”) or for deletions that overlap (open bars; “overlapping pairs”) showed a highly skewed distribution with most of the values having strong positive correlation (P < 10−15; χ2 test). (C) The performance of both SGD and GIP scores in predicting functional association of genes was assessed by comparison with a Gene Ontology (GO) “gold standard” (22). To estimate both the coverage and the functional predictive value of the identified genetic interactions, we plotted, at different thresholds of the two scores, precision [number of predictions found as true in the gold standard (TP, true pairs) divided by the sum of TP and the number of predictions scored as false in the gold standard (FP)] against the number of distinct genes found in the corresponding pairs. The estimated precision and coverage of data derived from SGD scores are shown as crosses, and data derived from correlation of genetic interaction profiles (GIP scores) are shown as filled circles. Coverage and precision for data obtained in a large-scale (132 query mutations) SGA genetic screen (4) is shown by a diamond.
Genetic Interaction Profiles Provide Specific Functional Signatures.
One of the recurrent features of the screens was the presence of a relatively large number of both negative and positive log2(Q/R) values, representing aggravating and buffering (or alleviating) effects, respectively. The GIM technique is thus sensitive to positive and negative epistasis, similarly to E-MAP, but on a whole-genome scale. Because the assay is sensitive, a number of target mutations exhibited weak interactions with at least some of the query mutations, thus generating genetic interaction profiles (GIPs) for these mutants. To what extent would two independent mutations of the same genes exhibit similar GIPs in our screens? Such independent mutations are already present in the yeast deletion library because a number of deletions of “dubious” ORFs overlap deletions of verified ORFs. We filtered the set of analyzed strains to retain only those deletions that showed a log2(Q/R) with an absolute value >1 (at least 3% change in generation time) in at least one of the performed screens. One-fourth of the tested mutations, 1,095 target genes, were thus selected for further analysis. This dataset contained 51 pairs of overlapping deletions. We calculated the Pearson correlation coefficient of the series of log2(Q/R) values for each pair of mutants as a measure of similarity of GIPs. Strikingly, the correlation coefficients calculated for the overlapping deletion pairs were globally shifted to very high correlation values when compared with the complete distribution of Pearson coefficients (Fig. 4B). Likewise, we observed very high Pearson correlation coefficients between the GIPs of genes encoding interacting proteins, as compiled by Collins et al. (21), confirming that functionally connected genes have similar GIPs (Fig. 4B). We can thus conclude that, because GIM measures weak yet reproducible genetic interactions, performing the screens with 41 independent query mutations generated specific GIPs for up to one-fourth of the tested mutations.
To what extent can these profiles be used to predict gene function? We chose an evaluation method based on the Gene Ontology (GO) annotations developed by Myers et al. (22) and their “gold standard” set consisting of pairs of genes sharing a common GO term (positive set) and pairs of genes that are unlikely to share the same GO term (negative set). A relative measure of precision can be obtained as TPs/(TPs + FPs) where TPs and FPs represent the number of selected gene pairs found in the positive and negative sets, respectively (true positive and false positive pairs). To minimize the effects of some target mutations that showed large absolute log2(Q/R) values in many screens and were thus highly correlated despite a lack of obvious functional link, we introduced a specificity correction factor to generate what we call the GIP score. This score was derived from the Pearson correlation coefficient between GIPs and favored the correlations between profiles that were similar to only a small number of other profiles (see SI Methods). Based on the correlation with GO, we arbitrarily defined three threshold GIP score values that distinguished high, medium, and low confidence classes for prediction of functional association (Fig. 4C and Dataset S4). GIP scores were found to have a considerably higher predictive value for functional relationship between genes than SGD scores.
Importantly, approximately one-half of the genes in the selected GIP pairs exhibited no strong synthetic interactions in any of the screens (they were absent from selected SGD pairs). Analysis of the hierarchical clustering of the log2(Q/R) for the set of 1,095 gene deletions as defined in Fig. 4B gives some clues for explaining this observation (Fig. S6). Although some clusters of functionally related genes were based on strong direct synthetic growth defects (see Fig. S6B for an example with splicing related genes), some highly correlated gene profiles were essentially defined by epistasis [positive log2(Q/R) ratios]. Fig. S6C shows such an example with ribosomal protein genes. In addition, some clusters of strongly correlated genes do not rest on any strong genetic interactions but rather on the combination of many weak effects. These weak genetic interactions did not pass the significance threshold when taken individually yet are globally sufficiently reproducible to provide GIPs robust enough to predict functional connections. An example of such a cluster grouping genes involved in peroxisome function can be seen in Fig. S6D. This example also illustrates that genes completely unrelated to the starting query mutations are sensitive to the approach, expanding the spectrum of functional predictions that can be obtained with even a relatively modest number of screens.
Discussion
Several recent large-scale experiments demonstrated the power of genetic interaction screens for function prediction when applied at a large scale. The first approaches described, SGA (1, 4) or SLAM and dSLAM (2, 3), were applied to the complete, unbiased mutant collection, but only relatively crude quantitative fitness defects were measured. The E-MAP method (7) added precise quantitative measurements of both aggravating (negative) and alleviating (positive) interactions. If the E-MAP approach proved extremely powerful in exploring defined cellular pathways, it is intrinsically not designed for uncovering unpredicted functional gene connections because its principle imposes the limitation of the analysis to rationally selected subsets of genes.
We describe here a simple method that allows the sensitive and quantitative measure of a large number of aggravating and alleviating genetic interactions. The identified strong effects correlated very well with the results of previous screens but were restricted to the partners of the 41 query genes. In contrast, the subtle effects quantified by GIM, considered over the whole range of experiments, had a major impact in defining genetic interaction profiles specific for functionally related hit genes. Moreover, when using the GO gold standard to estimate the precision of functional predictions associated with gene pairs, we defined associations for five times more genes using the similarity of GIPs than with direct synthetic interactions. This difference is dramatically reflected by the fact that, when compared with an SGA analysis involving 132 query mutants (4), we report genetic interactions for about five times fewer mutants, yet the GIP analysis allowed us to predict functional associations for a similar number of genes at the same precision level (Fig. 4C). In conclusion, an unexpected outcome of the GIM strategy is the crucial contribution of weak interactions in defining functional links between genes involved in various cellular processes, many of them unrelated to the pathways assigned to the query genes.
When using our data in a graphical network representation, we observed groups of genes (modules) mostly formed by GIP interactions. Connections between modules mainly consisted of SGD links (Fig. S7). Based on the GO terms shared by the genes forming the modules, specific cellular processes could be assigned to many subnetworks. In addition to annotated factors, many modules contained uncharacterized genes that are likely to be connected to the same pathway. These modules are linked to processes as various as translation, chromatin modification, and vesicle-mediated or nucleocytoplasmic transport.
The coverage and precision of the functional links defined on the basis of GIM results should increase by performing additional screens and using query mutations affecting different cellular pathways. In addition, the flexibility of the GIM method will allow a large panel of different growth conditions to be tested extensively.
Materials and Methods
The query strains were generated starting with the mutants from the systematic deletion library in the MATα background (BY4742) (8), by changing the KanMX4 marker to the prMFα2NatR marker upon transformation with the restriction digest of the specifically designed plasmid pGID3 (see above, SI Methods, and Fig. S1B). The query strains were transformed with a plasmid carrying the hygromycin resistance marker (pGID1) (23) and mated in mass with the pooled MATa yeast deletion library (KanR). The diploids were selected for hygromycin and kanamycin resistance. After sporulation, the MATα double mutants haploid cells were directly selected for combined nourseothricin and kanamycin resistance in standard rich liquid medium (YPD; see also Fig. 1) and grown at constant turbidity for 18 generations. Labeling of PCR products that amplified the upstream and downstream deletion tags was performed with Cy3 or Cy5, end-labeled oligonucleotides. Yeast glass slide oligonucleotides microarrays (SLRI-Yeast-Barcode-13k; A-MEXP-714 EBI ArrayExpress) were used to estimate the proportion of each double mutant in the screen as compared with a screen performed in parallel with a “neutral” deletion (ymr326cΔ, yfr057wΔ, or yel068cΔ). A detailed description of the GIM screen protocol, data analyses, and additional protocols can be found in SI Methods and Table S2.
Supplementary Material
Acknowledgments.
We thank C. Proux and J.-Y. Coppé for help with microarrays experiments; F. Feuerbach-Fournier and G. G. Cabal for the data in Fig. S5A; M.-T. Teixeira for work on early phases of the project; C. Fairhead for critical reading of the manuscript; and J. Beggs, O. Lefebvre, D. Tollervey, M.-C. Daugeron, and B. Séraphin for stimulating discussions and strains. K.Z. was the recipient of a Ministère de la Recherche fellowship. This work was supported by grants from Institut Pasteur, Centre National de la Recherche Scientifique, and the European Union (RNOMICS QLG2-CT-2001-01554).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0710533105/DCSupplemental.
References
- 1.Tong AH, et al. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science. 2001;294:2364–2368. doi: 10.1126/science.1065810. [DOI] [PubMed] [Google Scholar]
- 2.Ooi SL, Shoemaker DD, Boeke JD. DNA helicase gene interaction network defined using synthetic lethality analyzed by microarray. Nat Genet. 2003;35:277–286. doi: 10.1038/ng1258. [DOI] [PubMed] [Google Scholar]
- 3.Pan X, et al. A robust toolkit for functional profiling of the yeast genome. Mol Cell. 2004;16:487–496. doi: 10.1016/j.molcel.2004.09.035. [DOI] [PubMed] [Google Scholar]
- 4.Tong AH, et al. Global mapping of the yeast genetic interaction network. Science. 2004;303:808–813. doi: 10.1126/science.1091317. [DOI] [PubMed] [Google Scholar]
- 5.Collins SR, et al. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature. 2007;446:806–810. doi: 10.1038/nature05649. [DOI] [PubMed] [Google Scholar]
- 6.Segre D, Deluna A, Church GM, Kishony R. Modular epistasis in yeast metabolism. Nat Genet. 2005;37:77–83. doi: 10.1038/ng1489. [DOI] [PubMed] [Google Scholar]
- 7.Schuldiner M, et al. Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell. 2005;123:507–519. doi: 10.1016/j.cell.2005.08.031. [DOI] [PubMed] [Google Scholar]
- 8.Winzeler EA, et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- 9.Frachon E, Bondet V, Munier-Lehmann H, Bellalou J. Multiple microfermentor battery: A versatile tool for use with automated parallel cultures of microorganisms producing recombinant proteins and for optimization of cultivation protocols. Appl Environ Microbiol. 2006;72:5225–5231. doi: 10.1128/AEM.00239-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kshirsagar M, Parker R. Identification of Edc3p as an enhancer of mRNA decapping in Saccharomyces cerevisiae. Genetics. 2004;166:729–739. doi: 10.1534/genetics.166.2.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Badis G, Saveanu C, Fromont-Racine M, Jacquier A. Targeted mRNA degradation by deadenylation-independent decapping. Mol Cell. 2004;15:5–15. doi: 10.1016/j.molcel.2004.06.028. [DOI] [PubMed] [Google Scholar]
- 12.Dong S, et al. YRA1 autoregulation requires nuclear export and cytoplasmic Edc3p-mediated degradation of its pre-mRNA. Mol Cell. 2007;25:559–573. doi: 10.1016/j.molcel.2007.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fleischer TC, et al. Systematic identification and functional screens of uncharacterized proteins associated with eukaryotic ribosomal complexes. Genes Dev. 2006;20:1294–1307. doi: 10.1101/gad.1422006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anantharaman V, Aravind L. Novel conserved domains in proteins with predicted roles in eukaryotic cell-cycle regulation, decapping and RNA stability. BMC Genomics. 2004;5:45. doi: 10.1186/1471-2164-5-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Albrecht M, Lengauer T. Novel Sm-like proteins with long C-terminal tails and associated methyltransferases. FEBS Lett. 2004;569:18–26. doi: 10.1016/j.febslet.2004.03.126. [DOI] [PubMed] [Google Scholar]
- 16.Muhlrad D, Parker R. The yeast EDC1 mRNA undergoes deadenylation-independent decapping stimulated by Not2p, Not4p, and Not5p. EMBO J. 2005;24:1033–1045. doi: 10.1038/sj.emboj.7600560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rigaut G, et al. A generic protein purification method for protein complex characterization and proteome exploration. Nat Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 18.Barbee SA, et al. Staufen- and FMRP-containing neuronal RNPs are structurally and functionally related to somatic P bodies. Neuron. 2006;52:997–1009. doi: 10.1016/j.neuron.2006.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Decker CJ, Parker R. A turnover pathway for both stable and unstable mRNAs in yeast: evidence for a requirement for deadenylation. Genes Dev. 1993;7:1632–1643. doi: 10.1101/gad.7.8.1632. [DOI] [PubMed] [Google Scholar]
- 20.Anderson JS, Parker RP. The 3′ to 5′ degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SKI2 DEVH box protein and 3′ to 5′ exonucleases of the exosome complex. EMBO J. 1998;17:1497–1506. doi: 10.1093/emboj/17.5.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Collins SR, et al. Toward a comprehensive atlas of the physical interactome of Saccharomyces cerevisiae. Mol Cell Proteomics. 2007;6:439–450. doi: 10.1074/mcp.M600381-MCP200. [DOI] [PubMed] [Google Scholar]
- 22.Myers CL, et al. Finding function: evaluation methods for functional genomic data. BMC Genomics. 2006;7:187. doi: 10.1186/1471-2164-7-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berger AB, et al. Hmo1 is required for TOR-dependent regulation of ribosomal protein gene transcription. Mol Cell Biol. 2007;27:8015–8026. doi: 10.1128/MCB.01102-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stark C, et al. BioGRID: A general repository for interaction datasets. Nucleic Acids Res. 2006;34:D535–D539. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Belli G, et al. An activator/repressor dual system allows tight tetracycline-regulated gene expression in budding yeast. Nucleic Acids Res. 1998;26:942–947. doi: 10.1093/nar/26.4.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.