Abstract
Bacteriocins represent a large family of ribosomally produced peptide antibiotics. Here we describe the discovery of a widely conserved biosynthetic gene cluster for the synthesis of thiazole and oxazole heterocycles on ribosomally produced peptides. These clusters encode a toxin precursor and all necessary proteins for toxin maturation and export. Using the toxin precursor peptide and heterocycle-forming synthetase proteins from the human pathogen Streptococcus pyogenes, we demonstrate the in vitro reconstitution of streptolysin S activity. We provide evidence that the synthetase enzymes, as predicted from our bioinformatics analysis, introduce heterocycles onto precursor peptides, thereby providing molecular insight into the chemical structure of streptolysin S. Furthermore, our studies reveal that the synthetase exhibits relaxed substrate specificity and modifies toxin precursors from both related and distant species. Given our findings, it is likely that the discovery of similar peptidic toxins will rapidly expand to existing and emerging genomes.
Keywords: antibiotics, bacteriocin, bioinformatics, hemolytic, streptolysin
Streptolysin S (SLS), from the human pathogen Streptococcus pyogenes, is a ribosomally synthesized, secreted toxin responsible for the classical β-hemolytic phenotype of bacterial colonies grown on blood agar media (1, 2). S. pyogenes is associated with a wide spectrum of diseases ranging from simple pharyngitis to life-threatening necrotizing fasciitis. The expression of SLS promotes virulence in animal models of invasive infection (1, 3). The molecular structure of SLS has remained elusive for more than a century; however, SLS is known to be an oxygen-stable, nonimmunogenic cytolysin (4). The gene locus responsible for SLS biosynthesis (streptolysin S-associated genes, sag) was recently identified through transposon and targeted mutagenesis coupled with heterologous expression (2, 5–7). The sag locus consists of nine genes (A–I), and allelic exchange mutagenesis has shown sagA–G to be essential for the cytolytic phenotype of S. pyogenes. The first gene, sagA, encodes a 53-aa protoxin (Fig. 1A). SagA is followed by a set of three genes (sagBCD) exhibiting low sequence identity to three genes found in the Escherichia coli mcb gene cluster that produces the DNA gyrase inhibitor microcin B17 (MccB17) [supporting information (SI) Fig. S1] (5, 8). SagBCD encode a cyclodehydratase (SagC; 13% identical to McbB), dehydrogenase (SagB; 22% identical to McbC), and a “docking” protein (SagD; 18% identical to McbD) (Figs. S2–S5). The final genes in each cluster are dedicated ATP-binding cassette (ABC) transporters.
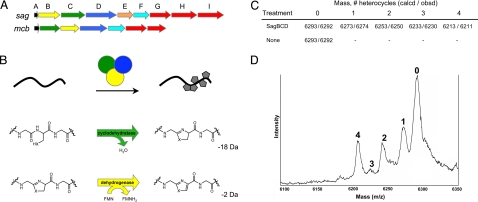
Fig. 1.
Conservation of toxin biosynthesis operons in S. pyogenes and E. coli. (A) Genetic organization of the streptolysin S-associated gene cluster (sagA–I) from S. pyogenes and the E. coli microcin B17 gene cluster (mcbA–G). (B) Through the action of a trimeric synthetase complex, oxazole and thiazole heterocycles are incorporated into a peptidic protoxin scaffold (black) and are active in vitro. Chemical transformations carried out by SagC/McbB (green, cyclodehydratase) and SagB/McbC (yellow, dehydrogenase) orthologs are shown. Molecular mass change for each reaction is shown in daltons. SagD/McbD (blue) serves as an enzymatic scaffold and facilitates substrate binding. (C) Heterocycle formation on E. coli McbA by S. pyogenes SagBCD. Shown are calculated (calcd) and observed (obsd) molecular masses for MBP-McbA after reaction with SagBCD and thrombinolysis. Formation of a single thiazole/oxazole leads to the loss of 20 Da from the mass of the peptide. (D) Linear mode MALDI-TOF mass spectrum of McbA treated with SagBCD. The number of heterocycles on the protoxin peptide is indicated above the respective mass (singly charged species).
Analogous to the sag locus, the first ORF of the MccB17-producing gene cluster mcbA encodes a 69-aa precursor protein (9, 10). Walsh and colleagues (5) have shown that McbA is extensively posttranslationally modified by the McbBCD synthetase complex. These modifications convert four serines and four cysteines into oxazole and thiazole heterocycles, respectively (Fig. 1B and Fig. S1). Unmodified McbA exhibits no effect on DNA gyrase (11).
Results and Discussion
Heterocycle Formation by SagBCD.
Given similarities in organization of the mcb and sag gene clusters and the observed homology of SagBCD to McbBCD, we hypothesized that the SagBCD synthetase complex would serve to posttranslationally modify a toxin precursor in the same manner as McbBCD. The sagBCD genes were cloned individually and purified as fusions to maltose-binding protein (MBP). Because numerous attempts to observe SagA by mass spectrometry failed, we used McbA, which is amenable to mass spectrometry, to detect and confirm heterocycle formation by SagBCD (12). After removal of the MBP tag, recombinant McbA has a calculated molecular mass of 6,293 Da due to addition of Gly-Ser-His to the N terminus. For each heterocycle formed, a loss of 18 Da (oxazoline/thiazoline) or 20 Da (oxazole/thiazole) is expected from the parent peptide (Fig. 1B). Treatment of McbA with recombinant SagBCD resulted in the formation of four new masses differing from the precursor peptide by multiples of 20 Da (Fig. 1 C and D). These masses are within error for linear mode MALDI mass spectrometry and correspond to heterocycle formation at four residues of McbA. The fourth heterocycle was not visible when the synthetase concentration was reduced (data not shown), and only unmodified McbA was observed when SagBCD was omitted from the reaction. These results provide experimental evidence that SagBCD functions in a manner analogous to McbBCD.
SagBCD Converts SagA into a Cytolysin.
Because recombinant SagBCD was active in vitro, we next sought to confirm that SagA could be transformed into an active cytolysin in a SagBCD-dependent manner. SagA was produced as an MBP fusion protein and then subjected to modification by the addition of SagBCD in an in vitro synthetase reaction. After this reaction, samples were tested for lytic activity against sheep erythrocytes. In this assay, SLS extracted from S. pyogenes cultures caused rapid hemolysis (data not shown). The addition of SagA alone failed to induce lysis, but robust hemolysis was observed after treatment with SagBCD (Fig. 2A). All three synthetase proteins were required for the lytic phenotype, indicating a cooperative role in toxin maturation.
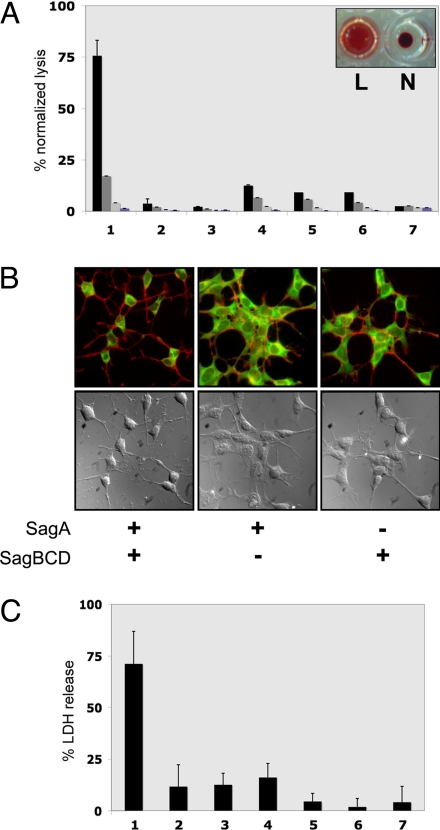
Fig. 2.
Cytolytic activity of in vitro synthetase reactions. (A) Hemolytic assays of SagA plus SagBCD synthetase reactions in microtiter wells containing defibrinated sheep blood. Bars indicate lysis normalized to a positive control (Triton X-100). Levels indicated are 1:1, 3:4, 1:2, and 1:4 ratios of synthetase reaction to blood (left to right) of 16-h reactions (n = 3). Lane 1, SagA plus SagBCD; lane 2, SagA alone; lane 3, SagBCD alone; lane 4, SagA plus SagBC; lane 5, SagA plus SagCD; lane 6, SagA plus SagBD; lane 7, vehicle. Inset demonstrates typical appearance of lytic (L) and nonlytic (N) reactions. (B) Fluorescence microscopy and DIC images of HEK293a cells treated as indicated. Actin filaments (red) and cytoplasm (green) are merged in Upper, and DIC images are in Lower. (C) LDH release assay of HEK293a cells treated with SagA plus SagBCD. Lysis is measured as A490 normalized to positive control (Triton X-100). Lane 1, SagA plus SagBCD; lane 2, SagA alone; lane 3, SagA plus SagBC; lane 4, SagA plus SagCD; lane 5, SagA plus SagBD; lane 6, SagBCD alone; lane 7, SagA-panC/S plus SagBCD.
To determine whether SagBCD-treated SagA exhibited a broader cytolytic phenotype characteristic of the native SLS toxin, reaction mixtures were applied to HEK293a cells. Cells treated with samples containing both SagA and all of the synthetase components progressively lost their focal contacts and eventually detached from the tissue culture wells (Fig. 2B). Actin staining revealed massive cytoskeletal collapse consistent with severe membrane damage. HEK cells incubated with solutions missing any component of the synthetase complex, or the protoxin precursor, were indistinguishable from untreated cells (Fig. 2B). Lactate dehydrogenase (LDH) release into the media provided quantitative confirmation that cytotoxicity required both the SagA substrate and the SagBCD complex (Fig. 2C). SagA contains seven cysteines that serve as potential sites for thiazole formation by SagBCD. Mutation of all seven cysteines to serine (SagA-panC/S) completely abolished cytolytic activity, indicating either that SagA-panC/S is not recognized as a substrate by SagBCD or that the membrane-damaging phenotype requires cysteine modification. Taken together, these experiments describe the first in vitro reconstitution of SLS activity. Furthermore, we demonstrate that SagBCD functions similarly to McbBCD by installing heterocycles on a peptidic toxin precursor.
The sagBCD Gene Clusters Are Widely Distributed Among Prokaryotes.
The similarities between the SLS and MccB17 biosynthetic operons were intriguing, given that (i) SLS is produced by a Gram-positive organism whereas MccB17 is from a Gram-negative organism and (ii) microcins have heretofore been classified as unique to Gram-negative bacteria (13). This suggested that other prokaryotes could use related machinery to introduce Ser/Thr/Cys-derived heterocycles into a wide variety of ribosomally produced peptides. We therefore initiated a comprehensive survey of the public genomic databases to identify related biosynthetic gene clusters (14). This search revealed that similar gene clusters are widespread among prokaryotes, most of which are not associated with a function (Fig. 3A). The genes encoding the SagB-like dehydrogenase (Fig. S2), SagC-like cyclodehydratase (Fig. S3), and SagD-like protein (Fig. S4) were present as adjacent ORFs in a diverse group of prokaryotic organisms spanning six phyla. The SagC and SagD orthologs were also sometimes found fused as a single ORF (Fig. S5).
Fig. 3.
The biosynthetic operon for producing thiazole/oxazole-containing toxins is widely distributed. (A) Gene clusters from organisms containing SLS- and MccB17-like bacteriocins. Members are sorted by prokaryotic phylum. Relative gene length and directionality are shown (scale for actinobacteria and cyanobacteria is reduced by 50%). Each gene cluster contains a protoxin (A, black), dehydrogenase (B, yellow), cyclodehydratase (C, green), and a docking scaffold (D, blue). These genes produce both single-domain and fusion proteins. Numerous ancillary enzymes are included and increase the chemical diversity of the toxin family. (B) Select protoxin amino acid sequences. Predicted leader peptide cleavage sites are denoted with an asterisk. Hyphens indicate a known cleavage site. Potential sites of heterocyclization are indicated in blue, and known sites of heterocycle formation are indicated in red. Green text signifies conversion to dehydroalanine. Toxins similar to SagA (top) are predicted cytolysins.
Of particular interest is the fact that gene clusters that highly resemble sagA–I are present in major mammalian pathogens such as Clostridium botulinum, Listeria monocytogenes, and Staphylococcus aureus RF122 (Fig. 3A). Similar gene clusters are also found in distantly related prokaryotes, such as cyanobacteria (15, 16) and archaea (e.g., Pyrococcus furiosus DSM 3638). Other family members are found throughout proteobacteria. For instance, a gene cluster identical in arrangement to E. coli mcbA–G is present in the plant symbiont Pseudomonas putida KT2440 (13). P. putida colonizes the nutrient-rich rhizosphere of plants and induces plant growth while secreting antibiotics to limit the growth of competing soil bacteria (17, 18). Furthermore, two other plant symbionts, Bacillus amyloliquefaciens FZB42 (Bam) and Bradyrhizobium japonicum USDA110, encode analogous synthetase proteins. Bacillus thuringiensis harbors a similar operon; although not a plant symbiont, B. thuringiensis is of agricultural interest owing to its ability to secrete toxins that are effective against a variety of pests, such as moths, butterflies, flies, mosquitoes, and beetles (19). The conservation of this synthetase gene cluster across prokaryotes—in particular, among both pathogenic and symbiotic bacteria—suggests that these gene products play an important role in the establishment of a survival niche for these organisms.
Presence of Protoxins Across Prokaryotic Lineages.
McbA and SagA encode protoxins of 69 and 53 residues, respectively (Fig. 3B). These are extremely short ORFs, which many gene-identification algorithms do not always recognize. For this reason, manual ORF searches were performed in the intergenic regions for each biosynthetic cluster identified. Short ORFs encoding proteins of 50–70 residues that are rich in Cys/Ser/Thr were present in all organisms containing sagBCD-like genes as adjacent ORFs (Fig. 3B). Although the functions of these potential protoxins are not known, based on similarity to McbA and SagA, we speculate that some will function as membrane-damaging agents (e.g., S. aureus and L. monocytogenes) or DNA gyrase poisons (P. putida). Secondary metabolites produced by biosynthetic clusters such as these have historically been an abundant source of pharmaceuticals. The bacterial products of these gene clusters are potential candidates for novel antibiotic design as well as promising targets for vaccine development.
Most of the gene clusters identified contain ORFs encoding additional modifying enzymes, suggesting that, in addition to heterocycle formation, these protoxins will undergo further modification. For example, clusters containing acetyltransferases and lantibiotic dehydratases, such as the goadsporin-producing organism Streptomyces sp. TP-A0584 (20), will likely produce toxins with acetylated and dehydrated amino acids (Fig. S1). Furthermore, methylated toxins are expected from gene clusters with methyltransferases. There are also gene clusters containing radical S-adenosylmethionine (SAM)-like proteins, which catalyze the formation of many types of chemical linkages in complex natural products (21). Inclusion of these ancillary enzymes is expected to dramatically increase the chemical diversity of the toxins, perhaps for niche purposes. The cyanobacteria Prochloron didemni (15) and Trichodesmium erythraeum IMS101 (16) have been previously identified as sources of cyclic peptides with Ser/Thr/Cys-derived heterocycles (Fig. S1). The sag-like biosynthetic clusters of T. erythraeum and P. didemni harbor an additional gene encoding a putative serine protease (Fig. 3A). Although the exact function of this protein is unclear, it is proposed to catalyze a head-to-tail macrocyclization reaction by using the N terminus of a peptide, instead of water, to hydrolyze the acyl-enzyme intermediate. After this reaction, the volume occupied by the toxin is much smaller and may eliminate the need for an elaborate export system. The lack of ABC transporters in these gene clusters supports this notion (Fig. 3A). Based on a similar arrangement of genes, we hypothesize that Rhodobacter sphaeroides, Lyngbya sp. PCC 8106, and Microcystis aeruginosa will also form macrocyclized peptides.
The Heterocycle Synthetase Complex Displays Functional Promiscuity.
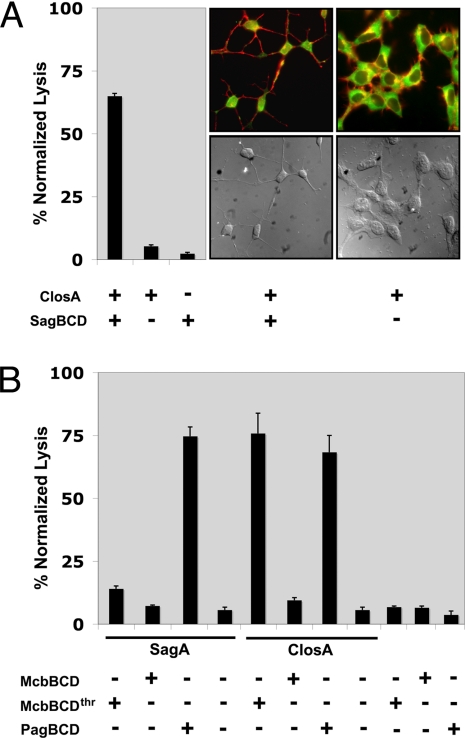
A gene cluster that is highly similar to the sag cluster was found in the pathogenic bacterium C. botulinum (Fig. 3A). The organization and amino acid sequence of proteins present in the C. botulinum cluster closA–I displays the highest similarity to sagA–I. The protoxin, which we have designated ClosA, shares many characteristics with SagA (39% identical) including the presence of numerous adjacent cysteines, other conserved heterocyclizable residues, and a putative protease site for removal of a leader peptide (Fig. 3B). To explore the possibility that closA encodes an SLS-like cytolysin, recombinant MBP-ClosA was prepared and subjected to in vitro synthetase assays with SagBCD. Hemolysis was observed only in samples containing both ClosA and the SagBCD synthetase complex (Fig. 4A). Microscopy revealed that cytotoxicity in HEK cells treated with ClosA plus SagBCD was identical to cells treated with SagA plus SagBCD (Figs. 2B and 4A). These data demonstrate that SagBCD accepts substrates beyond its cognate peptide, SagA.
Fig. 4.
Disparate heterocycle synthetases accept substrates from related and distant prokaryotes. (A) The S. pyogenes synthetase SagBCD accepts the C. botulinum protoxin and produces a cytolytic toxin. (Left) Hemolytic assay. (Center and Right) Fluorescence microscopy and DIC images of HEK293a cells treated as indicated. (B) Hemolytic assay of SagA and ClosA treated with the synthetase complex from E. coli (McbBCD) and the hyperthermophile P. furiosus (PagBCD). Recombinant MBP-McbD must be first treated with thrombin (thr) before synthetase activity is observed.
Given the ability of SagBCD to accept McbA and ClosA as alternative substrates, we assessed whether other synthetases were permissive in their substrate selectivity. To this end, the heterocycle-forming synthetases from E. coli (McbBCD) and the thermophilic archaeon P. furiosus (PagBCD, identity to corresponding Sag: 17–24%) were prepared as MBP fusions. SagA and ClosA were then tested for erythrocyte lytic activity after reaction with McbBCD and PagBCD. The PagBCD complex efficiently converted both SagA and ClosA into a hemolysin (Fig. 4B). In contrast, the McbBCD complex converted only ClosA into a potent lytic species. It is intriguing that McbBCD accepted and converted an alternate substrate into a cytolytic factor given that its biological function is to produce a DNA gyrase inhibitor (22). Therefore, we conclude that the mechanism of toxin action is governed by the protoxin amino acid sequence and that the synthetase proteins are functionally redundant. The extent of substrate tolerance and the kinetics and selectivity of ring formation require further investigation.
In sum, our bioinformatic survey has uncovered the presence of sag-like gene clusters in a myriad of prokaryotes, leading us to define a new class of bacteriocin. This class is characterized by a biosynthetic gene cluster that encodes a small protoxin and three adjacent synthetase proteins that endow an organism with the ability to cyclize Ser/Thr/Cys residues from a ribosomally synthesized protoxin scaffold. The finding that the heterocycle synthetase genes are exclusively found as adjacent ORFs will facilitate the identification of orthologous biosynthetic clusters in emerging genomes. Using the protoxin and synthetase proteins from the human pathogen S. pyogenes, we demonstrate in vitro reconstitution of SLS activity. Furthermore, we show that the synthetase enzymes are functionally redundant and catalyze heterocycle formation on alternative substrates despite their significantly distinct evolutionary lineages. Our data suggest that all of the newly identified gene clusters are responsible for the production of bacteriocins containing at least one Ser/Thr/Cys-derived heterocycle. Many of the gene clusters encode ancillary modifying enzymes that will install further modifications onto the bacteriocin, thus increasing the chemical diversity of this family. Further insights into this bacteriocin family will lead to the identification of novel targets for antibiotic and vaccine development.
Materials and Methods
Additional Details.
See SI Text for additional methods.
Bioinformatics.
The amino acid sequences of SagBCD from S. pyogenes (2) and McbBCD from E. coli (23) were initially used as a BLAST query against the nonredundant protein sequence database of the National Center for Biotechnology Information (14). Highly similar proteins were identifiable in the firmicutes phylum, specifically in C. botulinum, S. aureus RF122, and L. monocytogenes. The surrounding ORFs for each identified gene were analyzed for the presence of all three synthetase enzymes by using the Genomic Context feature of Entrez Gene and the SEED tool from The Fellowship for the Interpretation of Genomes. Gene clusters that contained all three synthetase enzymes were also found to contain other bacteriocin-type operon components, such as a probable protease, immunity protein, and ABC transporters. In cases where the automatic annotation algorithms do not identify the smaller toxin structural genes (e.g., Listeria, ListA), manual inspection of all predicted ORFs in the local intergene region was performed, and these were subsequently scored by using ClustalW alignment to known toxin structural genes from S. pyogenes and E. coli. Streptococcus iniae has been shown to contain a gene cluster nearly identical to S. pyogenes and was therefore not used for bioinformatics searching (24, 25). Furthermore, another Streptococcus species, Streptococcus dysgalactiae sp. equisimilis, contains a SagA and SagB homolog (3). Because of an incomplete genome sequence, we cannot confirm the presence of SagC–I at this time.
Wider subsequent searches of the nonredundant protein sequence database (National Center for Biotechnology Information) were initiated using all proteins confirmed to have a sag-like gene locus (with bcd genes located directly adjacent to or within a few ORFs of each another). A lower limit of 30% amino acid similarity was chosen as a threshold to assign a candidate protein as being homologous (BLOSUM30 matrix). This second search identified gene clusters from three other organisms that produce a heterocycle-containing bacteriocin with a known structure (Fig. S1), Streptomyces sp. TP-A0584 (goadsporin) (26), P. didemni (patellamide A/C) (15), and T. erythraeum IMS101 (trichamide) (16) and from additional organisms.
Mass Spectrometry.
Sample preparation.
Recombinant MBP-McbA (110 μl at 40 μM) was proteolytically digested with 50 NIH units of thrombin (Sigma–Aldrich) for 4 h at room temperature in thrombin cleavage buffer: 50 mM Tris (pH 7.8) and 10 mM CaCl2. During the course of the reaction, cut McbA precipitates (pI ≈ 7) as a white solid. Precipitated McbA was subsequently harvested by centrifugation and washed two to three times with 100 μl of deionized water to remove the majority of buffer, salts, detergent, cut MBP, and uncut MBP-McbA. This precipitate was then resolubilized in 60% MeCN/2% formic acid (40 μl). Samples of MBP-McbA that were treated with BCD synthetase complex (Sag and Pag) were first reacted, as described in In Vitro Synthetase Assay, before thrombinolysis.
The MALDI target (stainless steel, Applied Biosystems) was prepared by allowing a saturated solution of sinapic acid (Sigma–Aldrich) in 50% MeCN/0.1% TFA to fully dry on the target (1.5 μl per sample, in duplicate). The precipitated McbA sample was then diluted (1:1) in the saturated matrix solution before spotting on top of the dried spot (1 μl in duplicate).
Instrument settings.
MALDI-TOF mass spectra were acquired on a Voyager DE-STR instrument (Applied Biosystems) in linear positive mode. Specific settings were typically as follows: 2,000–2,100 laser power, 93% grid, 0.12% guide wire, 400 nsec delay, mass window 3,000–18,000 Da.
In Vitro Synthetase Assay.
Synthetase reactions using MBP-tagged SagA and SagBCD and other fusion proteins were performed in a manner described earlier (27, 28). Reaction mixtures consisted of MBP-SagA (10 μM) and MBP-SagBCD (2 μM each) in a total volume of 100 μl of synthetase buffer (50 mM Tris·HCl, pH 7.5/125 mM NaCl/20 mM MgCl2/2 mM ATP/10 mM DTT). Reactions were allowed to proceed at 37°C for 16 h unless otherwise stated. These reactions were used for hemolytic assays and LDH release assays to quantify membrane damage. Omission of either ATP or DTT resulted in loss of hemolytic activity (data not shown).
Hemolytic Assay.
Fresh defibrinated whole sheep blood was obtained from Hemostat Laboratories. Whole blood was washed three times in PBS and diluted to a final concentration of 1:25 vol/vol in PBS. Prepared whole blood (100 μl) was then placed into individual wells of a flat-bottom 96-well microtiter plate (Costar, Corning). Reactions from in vitro synthetase assays were then added directly to the wells, and the mixture was reacted for 16 h at 37°C unless otherwise noted. After incubation, plates were centrifuged at 500 × g for 10 min and an aliquot of supernatant (100 μl) was placed in a separate microtiter plate for measuring hemoglobin absorbance at 450 nm on a Victor3 microplate reader (Perkin–Elmer).
Supplementary Material
Acknowledgments.
We thank Joyce Limm for technical assistance and Christopher Walsh (Harvard Medical School, Boston) for the mcbA–D clones. We also thank Elizabeth Komives and Michael VanNieuwenhze for their helpful suggestions. Seema Mattoo critically reviewed the manuscript, for which we are grateful. This research was supported by grants from the National Institutes of Health, the Walther Cancer Institute, the Ellison Foundation, and the Howard Hughes Medical Institute (to J.E.D.). D.A.M. is supported by a Hartwell Foundation postdoctoral fellowship. S.W.L. is supported by a University of California at San Diego Heme and Blood Proteins postdoctoral training grant.
Footnotes
The authors declare no conflict of interest.
See Commentary on page 5655.
This article contains supporting information online at www.pnas.org/cgi/content/full/0801338105/DCSupplemental.
References
- 1.Datta V, et al. Mutational analysis of the group A streptococcal operon encoding streptolysin S and its virulence role in invasive infection. Mol Microbiol. 2005;56:681–695. doi: 10.1111/j.1365-2958.2005.04583.x. [DOI] [PubMed] [Google Scholar]
- 2.Nizet V, et al. Genetic locus for streptolysin S production by group A streptococcus. Infect Immun. 2000;68:4245–4254. doi: 10.1128/iai.68.7.4245-4254.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Humar D, et al. Streptolysin S and necrotising infections produced by group G streptococcus. Lancet. 2002;359:124–129. doi: 10.1016/S0140-6736(02)07371-3. [DOI] [PubMed] [Google Scholar]
- 4.Nizet V. Streptococcal beta-hemolysins: Genetics and role in disease pathogenesis. Trends Microbiol. 2002;10:575–580. doi: 10.1016/s0966-842x(02)02473-3. [DOI] [PubMed] [Google Scholar]
- 5.Li YM, Milne JC, Madison LL, Kolter R, Walsh CT. From peptide precursors to oxazole and thiazole-containing peptide antibiotics: Microcin B17 synthase. Science. 1996;274:1188–1193. doi: 10.1126/science.274.5290.1188. [DOI] [PubMed] [Google Scholar]
- 6.Yorgey P, Davagnino J, Kolter R. The maturation pathway of microcin B17, a peptide inhibitor of DNA gyrase. Mol Microbiol. 1993;9:897–905. doi: 10.1111/j.1365-2958.1993.tb01747.x. [DOI] [PubMed] [Google Scholar]
- 7.Betschel SD, et al. Reduced virulence of group A streptococcal Tn916 mutants that do not produce streptolysin S. Infect Immun. 1998;66:1671–1679. doi: 10.1128/iai.66.4.1671-1679.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.San Millan JL, Kolter R, Moreno F. Plasmid genes required for microcin B17 production. J Bacteriol. 1985;163:1016–1020. doi: 10.1128/jb.163.3.1016-1020.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davagnino J, et al. The DNA replication inhibitor microcin B17 is a forty-three-amino-acid protein containing sixty percent glycine. Proteins. 1986;1:230–238. doi: 10.1002/prot.340010305. [DOI] [PubMed] [Google Scholar]
- 10.Yorgey P, et al. Posttranslational modifications in microcin B17 define an additional class of DNA gyrase inhibitor. Proc Natl Acad Sci USA. 1994;91:4519–4523. doi: 10.1073/pnas.91.10.4519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Milne JC, et al. Cofactor requirements and reconstitution of microcin B17 synthetase: A multienzyme complex that catalyzes the formation of oxazoles and thiazoles in the antibiotic microcin B17. Biochemistry. 1999;38:4768–4781. doi: 10.1021/bi982975q. [DOI] [PubMed] [Google Scholar]
- 12.Kelleher NL, Belshaw PJ, Walsh CT. Regioselectivity and chemoselectivity analysis of oxazole and thiazole ring formation by the peptide-heterocyclizing microcin B17 synthetase using high-resolution MS/MS. J Am Chem Soc. 1998;120:9716–9717. [Google Scholar]
- 13.Severinov K, et al. Low-molecular-weight post-translationally modified microcins. Mol Microbiol. 2007;65:1380–1394. doi: 10.1111/j.1365-2958.2007.05874.x. [DOI] [PubMed] [Google Scholar]
- 14.Altschul SF, et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schmidt EW, et al. Patellamide A and C biosynthesis by a microcin-like pathway in Prochloron didemni, the cyanobacterial symbiont of Lissoclinum patella. Proc Natl Acad Sci USA. 2005;102:7315–7320. doi: 10.1073/pnas.0501424102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sudek S, Haygood MG, Youssef DT, Schmidt EW. Structure of trichamide, a cyclic peptide from the bloom-forming cyanobacterium Trichodesmium erythraeum, predicted from the genome sequence. Appl Environ Microbiol. 2006;72:4382–4387. doi: 10.1128/AEM.00380-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dos Santos VA, et al. Insights into the genomic basis of niche specificity of Pseudomonas putida KT2440. Environ Microbiol. 2004;6:1264–1286. doi: 10.1111/j.1462-2920.2004.00734.x. [DOI] [PubMed] [Google Scholar]
- 18.Nelson KE, et al. Complete genome sequence and comparative analysis of the metabolically versatile Pseudomonas putida KT2440. Environ Microbiol. 2002;4:799–808. doi: 10.1046/j.1462-2920.2002.00366.x. [DOI] [PubMed] [Google Scholar]
- 19.Broderick NA, Raffa KF, Handelsman J. Midgut bacteria required for Bacillus thuringiensis insecticidal activity. Proc Natl Acad Sci USA. 2006;103:15196–15199. doi: 10.1073/pnas.0604865103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Onaka H, et al. Cloning and characterization of the goadsporin biosynthetic gene cluster from Streptomyces sp. TP-A0584. Microbiology. 2005;151:3923–3933. doi: 10.1099/mic.0.28420-0. [DOI] [PubMed] [Google Scholar]
- 21.Sofia HJ, et al. Radical SAM, a novel protein superfamily linking unresolved steps in familiar biosynthetic pathways with radical mechanisms: Functional characterization using new analysis and information visualization methods. Nucleic Acids Res. 2001;29:1097–1106. doi: 10.1093/nar/29.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heddle JG, et al. The antibiotic microcin B17 is a DNA gyrase poison: Characterisation of the mode of inhibition. J Mol Biol. 2001;307:1223–1234. doi: 10.1006/jmbi.2001.4562. [DOI] [PubMed] [Google Scholar]
- 23.Roy RS, et al. Thiazole and oxazole peptides: Biosynthesis and molecular machinery. Nat Prod Rep. 1999;16:249–263. doi: 10.1039/a806930a. [DOI] [PubMed] [Google Scholar]
- 24.Locke JB, et al. Streptococcus iniae beta-hemolysin streptolysin S is a virulence factor in fish infection. Dis Aquat Organ. 2007;76:17–26. doi: 10.3354/dao076017. [DOI] [PubMed] [Google Scholar]
- 25.Fuller JD, et al. Identification of a streptolysin S-associated gene cluster and its role in the pathogenesis of Streptococcus iniae disease. Infect Immun. 2002;70:5730–5739. doi: 10.1128/IAI.70.10.5730-5739.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Igarashi Y, et al. Goadsporin, a chemical substance which promotes secondary metabolism and morphogenesis in streptomycetes. II. Structure determination. J Antibiot (Tokyo) 2001;54:1045–1053. doi: 10.7164/antibiotics.54.1045. [DOI] [PubMed] [Google Scholar]
- 27.Sinha Roy R, Belshaw PJ, Walsh CT. Mutational analysis of posttranslational heterocycle biosynthesis in the gyrase inhibitor microcin B17: Distance dependence from propeptide and tolerance for substitution in a GSCG cyclizable sequence. Biochemistry. 1998;37:4125–4136. doi: 10.1021/bi9728250. [DOI] [PubMed] [Google Scholar]
- 28.Sinha Roy R, Kelleher NL, Milne JC, Walsh CT. In vivo processing and antibiotic activity of microcin B17 analogs with varying ring content and altered bisheterocyclic sites. Chem Biol. 1999;6:305–318. doi: 10.1016/s1074-5521(99)80076-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.