Abstract
The phenolic methyl ether 3,5-dimethoxytoluene (DMT) is a major scent compound of many modern rose varieties, and its fragrance participates in the characteristic “tea scent” that gave their name to Tea and Hybrid Tea roses. Among wild roses, phenolic methyl ether (PME) biosynthesis is restricted to Chinese rose species, but the progenitors of modern roses included both European and Chinese species (e.g., Rosa chinensis cv Old Blush), so this trait was transmitted to their hybrid progeny. The last steps of the biosynthetic pathways leading to DMT involve two methylation reactions catalyzed by the highly similar orcinol O-methyltransferases (OOMT) 1 and 2. OOMT1 and OOMT2 enzymes exhibit different substrate specificities that are consistent with their operating sequentially in DMT biosynthesis. Here, we show that these different substrate specificities are mostly due to a single amino acid polymorphism in the phenolic substrate binding site of OOMTs. An analysis of the OOMT gene family in 18 species representing the diversity of the genus Rosa indicated that only Chinese roses possess both the OOMT2 and the OOMT1 genes. In addition, we provide evidence that the Chinese-rose-specific OOMT1 genes most probably evolved from an OOMT2-like gene that has homologues in the genomes of all extant roses. We propose that the emergence of the OOMT1 gene may have been a critical step in the evolution of scent production in Chinese roses.
Keywords: flower, volatiles, hybrid tea, dimethoxytoluene
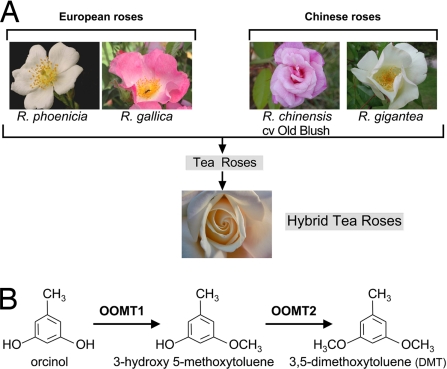
The extraordinary diversity of modern rose varieties has been obtained by intensive breeding from a relatively small number of progenitors, including both European varieties, such as Rosa gallica and Rosa moschata, and Chinese varieties, such as Rosa chinensis cv Old Blush and Rosa gigantea (1) (Fig. 1A). The Chinese roses have been grown in East Asia for thousands of years and finally reached Western Europe around 1780. Unlike European roses, they possessed the ability to bloom repeatedly throughout the summer until late autumn, and, as a result of this unique characteristic, Chinese roses were highly popular with rose breeders in the early 1800s. In this breeding process, European varieties contributed cold and disease resistance characters, and Chinese varieties contributed recurrent flowering. In addition to these traits, the European and Chinese progenitors each contributed distinctive scent characteristics (1). The major scent components of European roses include 2-phenylethanol and monoterpenes, whereas the volatiles emitted by Chinese roses contain high amounts of phenolic methyl ethers (PME) such as 3,5-dimethoxytoluene (DMT) or 1,3,5-trimethoxybenzene (TMB) (2, 3). Although PME biosynthesis was originally restricted to Chinese roses, DMT is nowadays a major scent compound of many rose varieties. Its light fragrance, with earthy and spicy notes reminiscent of black tea, is responsible, in combination with other molecules, for the characteristic “tea scent” of Tea and Hybrid Tea roses, where DMT can represent up to 90% of total flower volatiles (2–5).
Fig. 1.
Origin of Tea scent in roses. (A) Simplified genealogical tree of modern Hybrid Tea roses. Progenitors of modern roses included European varieties such as R. gallica or R. phoenicia and Chinese roses like R. chinensis cv Old Blush and R. gigantea. (B) Proposed pathway for biosynthesis of DMT (9). The reactions catalyzed by OOMT1 and OOMT2 are indicated.
Genomic approaches have recently been used to identify genes involved in rose scent biosynthesis, starting with the establishment of EST databases representing the transcriptomes of petals from different varieties (5, 6). Analysis of these petal EST databases coupled to microarray approaches allowed the identification of several genes potentially involved in scent production. Functional characterization of some of these genes led to the identification of several scent-related genes, including a sesquiterpene synthase involved in the production of germacrene D (5), an alcohol acetyltransferase catalyzing the formation of geranyl acetate (7), a phenylacetaldehyde synthase involved in the biosynthesis of 2-phenylethanol (8) and two orcinol O-methyltransferases (OOMTs) involved in the biosynthesis of PMEs (9, 10). The orcinol O-methyltransferases OOMT1 and OOMT2, which catalyze the two final methylation reactions of DMT biosynthesis (Fig. 1B), have been characterized in both modern Hybrid Tea roses and their progenitor R. chinensis cv Old Blush. Although OOMT1 and OOMT2 are very similar (96.5% identity at the amino acid level), they exhibit different substrate specificities. Based on these different catalytic properties, these two enzymes were proposed to operate sequentially to catalyze the first and the second methylation step of orcinol, respectively (9).
In this work, we first investigated the molecular basis for OOMT1 and OOMT2 substrate specificities, using molecular modeling of the three-dimensional structure of these two proteins and site-directed mutagenesis. Then, following our previous report of the presence of OOMT homologues in the genomes of two European rose varieties (11), we investigated the OOMT gene family in 18 rose species representing the genus Rosa. Finally, by combining the phylogenetic reconstruction of this gene family with functional characterization of some of its members, we investigated the role of OOMT genes in the evolution of rose scent.
Results
Structural Basis for Substrate Discrimination in OOMTs.
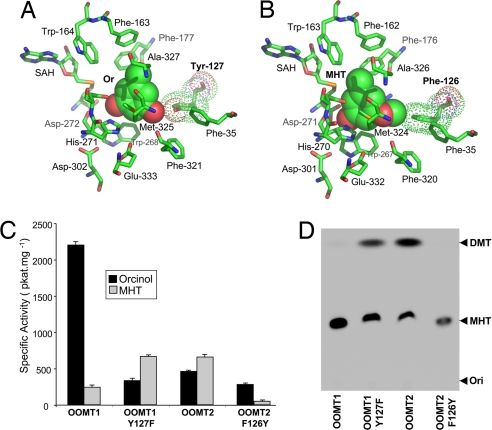
In rose petals, two closely related enzymes, OOMT1 and OOMT2, were shown to catalyze the last steps of DMT biosynthesis (9, 10). Although the substrate specificities of OOMT1 and OOMT2 were similar, they were not identical. Both enzymes were able to methylate orcinol and orcinol monomethyl ether (MHT); however, orcinol was the preferred substrate of OOMT1, and MHT was the preferred substrate of OOMT2 (9). Starting from these observations, we decided to look more closely at the biochemical basis of DMT biosynthesis using a molecular modeling approach. The three-dimensional crystal structures of several plant O-methyltransferases (OMTs) have been reported (12–15) and this information has allowed the molecular modeling of other members of this enzyme family, based on the known structures (14, 16, 17). Using this strategy, Gang et al. (16) obtained molecular models of basil chavicol-OMT and eugenol-OMT and showed that their substrate specificities could be exchanged by exchanging a single amino acid at the active site. To investigate the structural bases for their different substrate specificities, rose OOMT1 and OOMT2 structures were modeled based on the structure of IOMT from alfalfa (12). Because recombinant OOMTs were shown to behave as dimers in solution (9), OOMT1 and OOMT2 were modeled as homodimers. Before minimization cycles, the initial position of substrates was modeled based on the position of isoformononetin in the IOMT structure, orcinol replacing the A ring of isoformononetin (the ring bearing the methylated hydroxyl group) (12). According to these models, the general organization of the phenolic substrate binding site of both OOMTs is very similar (Fig. 2 A and B), and also similar to that of the IOMT substrate binding site. However, the main difference is that OOMT1 Tyr-127, which is in proximity to the second hydroxyl group of orcinol, is replaced by Phe-126 in OOMT2. In the OOMT1 model, the calculated distance between the oxygen atom of the hydroxyl group of Tyr-127 and the hydroxyl group of orcinol in position 5 is 3.05 Å. The steric hindrance of this hydroxyl group may be responsible for the fact that the monomethylated compound 3-methoxy 5-hydroxytoluene (MHT) is not methylated efficiently by OOMT1 (9, 10). In contrast, Phe-126 of OOMT2 is predicted to provide additional space and a more hydrophobic environment, favoring efficient methylation of MHT.
Fig. 2.
Molecular modeling of OOMTs. Shown are three-dimentional structure models of the active sites of OOMT1 (A) and OOMT2 (B). The models incorporate S-adenosylhomocysteine (SAH) plus orcinol (Or, OOMT1) or 3-methoxy,5-hydroxytoluene (MHT, OOMT2) substrates. (C) Substrate specificity of mutant OOMTs. The specific activities of native and mutant recombinant enzymes are compared, using orcinol (black) and MHT (gray) as substrates (500 μM); bars indicate ±SEs. (D) TLC analysis of reaction products produced after incubation of native and mutant OOMTs with orcinol (200 μM) in the presence of S-adenosyl-l-[methyl-14C]methionine (50 μM). The positions of the origin (Ori) and the reaction products MHT and DMT are indicated. The identity of the reaction products was confirmed by GC-MS analysis (Fig. S1).
Construction and Characterization of Mutant OOMTs.
To determine the contribution of Tyr-127 and Phe-126 to substrate discrimination by OOMT1 and OOMT2, respectively, these residues were exchanged by using site-directed mutagenesis. Tyr-127 was replaced by a phenylalanine in OOMT1, and Phe-126 was replaced by a tyrosine in OOMT2. Both mutant proteins were expressed in Escherichia coli, purified, and assayed for substrate preference with orcinol and MHT (Fig. 2 C and D). Orcinol was the preferred substrate of the OOMT2 F126Y mutant, and this protein showed poor activity with MHT and, thus, exhibited a similar substrate specificity to native OOMT1. On the other hand, the OOMT1 Y127F mutant behaved like the native OOMT2, exhibiting a 2-fold higher relative specific activity toward MHT than toward orcinol. As a consequence, when OOMT1 and OOMT2 F126Y were incubated with orcinol, the only accumulated product was the monomethylated MHT, but when OOMT2 and OOMT1 Y127F were incubated with orcinol, significant amounts of DMT were produced (Fig. 2D), showing that, with orcinol as substrate, OOMT2 F126Y and OOMT1 Y127F were functionally equivalent to OOMT1 and OOMT2, respectively [a detailed characterization of these enzymes is provided in supporting information (SI) Table S1]. It should be noted, however, that, although the OOMT2 F126Y and OOMT1 Y127F mutants showed the same substrate preference as the native OOMT1 and OOMT2, respectively, kinetic parameters indicated that they were not as efficient as the native proteins (Fig. 2C).
Presence of OOMT-Like Genes in a Selection of Rose Species Representative of the Genus Rosa.
The similarity between OOMT1 and OOMT2, plus our previous report of the presence of OOMT homologues in European roses (11), prompted us to investigate the evolution of this gene family in a selection of 18 rose species representing the different sections of the genus Rosa. Two kinds of species were selected: on the one hand, 5 Chinese roses [R. chinensis cv Old Blush, R. chinensis spontanea, R. gigantea, R. odorata (Hume's Blush), and R. odorata ochroleuca (Park's Yellow)] grouped under the Indicae section of the genus Rosa, all of them producing PME (3, 4), and on the other hand, 13 Eurasiatic rose species representing the main sections of the genus Rosa (Banksianae, Bracteatae, Caninae, Cinnamomeae, Gallicanae, and Synstylae). Analyses of scent compounds in petals and stamens confirmed that, among the selected roses, PME production was restricted to Chinese roses; no other rose species produced significant amounts of these compounds (data not shown).
When PCR amplifications were carried out by using genomic DNA and OOMT-specific primers, DNA fragments could be amplified from all of the rose species studied. After cloning and sequencing the amplified DNA fragments, we obtained a total of 96 different OOMT sequences, 73 being potentially functional, the others corresponding to pseudogenes affected by deletion (Table S2). The number of OOMT sequences recovered from each rose species ranged from one in R. phoenicia to at least six in Chinese roses like R. chinensis spontanea or R. gigantea. The general structure of the different OOMT genes was conserved, with two exons (801 bp and 303 bp, respectively), separated by a single intron, whose size varied from 200 to 1,000 bp. Among the 73 potentially functional OOMT homologues, we identified 14 OOMT1-like genes, based on the presence of a tyrosine residue at the position corresponding to residue 127 in Old Blush OOMT1. Interestingly, OOMT1-like genes were present only in the genomes of Chinese roses, and all of the Chinese species analyzed possessed at least one OOMT1-like gene together with various numbers of OOMT2-like sequences (with a phenylalanine residue at the position corresponding to residue 127 in Old Blush OOMT1). On the other hand, the 64 different genes and pseudogenes cloned from the non-Chinese species were exclusively OOMT2-like sequences.
Phylogenetic Analysis of the OOMT Gene Family and Characterization of the Activity of OOMT-Like Enzymes.
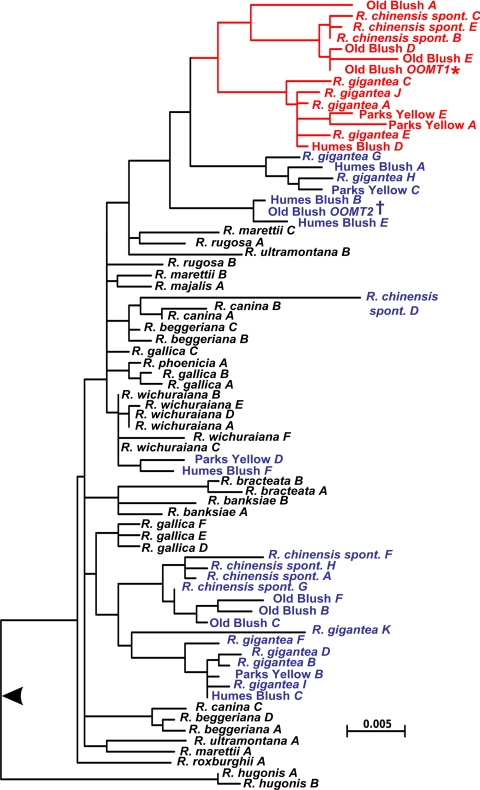
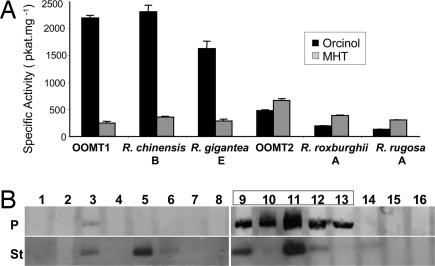
Gene duplication, followed by evolutionary divergence and acquisition of novel function, is thought to play a major role in the recruitment of genes for secondary metabolism (18). However, few studies have combined phylogenetic analyses with functional characterization, to identify the mutations responsible for novel functions in newly evolved genes (19). To understand the origin and evolution of the Chinese-rose-specific OOMT1 genes, we subjected the 73 potentially functional OOMT genes to Bayesian (Fig. 3) and maximum likelihood (data not shown) phylogenetic analyses. The tree topologies obtained using these methods were congruent, OOMT1-like sequences forming a clade in both optimal topologies. However, because the classification of the enzymes as either OOMT1-like or OOMT2-like enzymes was based solely on the presence of a tyrosine or a phenylalanine residue at the position corresponding to residue 127 in Old Blush OOMT1, it was necessary to verify that that characteristic correlated consistently with substrate specificity. We therefore tested the substrate preferences of several OOMT-like enzymes from both Chinese and non-Chinese rose species. The enzymes selected included OOMT2-like enzymes from R. roxburghii and R. rugosa because the former is only distantly related to Old Blush and the latter is a member of the Cinnamomeae, which is the largest section of the genus Rosa. We also selected OOMT1-like enzymes from the wild Chinese rose species R. chinensis spontanea and R. gigantea. The selected OOMTs were expressed in E. coli, and the specific activities of the purified recombinant OOMTs with orcinol and MHT as substrates were determined (Fig. 4A). The kinetic properties of OOMT2-like enzymes from R. roxburghii and R. rugosa were similar to those of the original OOMT2, with a preference for the monomethylated substrate MHT over orcinol. Conversely, the specific activities of OOMT1-like enzymes from R. chinensis spontanea and R. gigantea with orcinol were 10-fold higher than with MHT, as in the case of Old Blush OOMT1 (a detailed characterization of these OOMTs is provided in Table S1). Thus, the selected OOMT1-like and OOMT2-like enzymes exhibited the characteristic features of the OOMT1 and OOMT2 enzymes from Old Blush, respectively. These results indicate that the OOMT1/OOMT2 classification based on the nature of the 126/127 amino acid residue is most likely to reflect the functional characteristics of these two classes of OOMTs.
Fig. 3.
Molecular phylogenetic tree of the OOMT gene family in roses. Branch lengths are drawn proportional to the amount of inferred character change (scale in substitution per site). Topology was rooted at the base of R. hugonis sequences (black arrow) because this location was the favored rooting point when three divergent OOMT2-like sequences recovered from Prunus dulcis were included in the analysis. The red subtree highlights OOMT1-like sequences. This clade was supported by maximal (i.e., 1.00) posterior probability values but received only weak bootstrap support (45%). However, this bootstrap value might be partly explained by the low number of substitutions observed. OOMT2-like genes from Chinese roses are indicated in blue. * and †, previously characterized OOMT1 (AJ786302) and OOMT2 (AJ786303) sequences (10).
Fig. 4.
Characterization of OOMT-like proteins. (A) Specific activities of OOMT-like enzymes with orcinol and MHT. Purified recombinant OOMT1, OOMT2, OOMT1-like (R. chinensis spontanea OOMT-B and R. gigantea OOMT-E), and OOMT2-like (R. roxburghii OOMT-A and R. rugosa OOMT-A) enzymes were incubated with orcinol or MHT (500 μM) in the presence of S-adenosyl-l-[methyl-14C]methionine (35 μM). (B) Western blot analysis of OOMT expression in the selected Rosa species. Total proteins (15 μg) from petals (P) and stamens (St) were subjected to SDS/PAGE and analyzed by Western blot using anti-OOMT antibody (11). Lane 1, R. banksiae; 2, R. bracteata; 3, R. canina; 4, R. beggeriana; 5, R. majalis; 6, R. marettii; 7, R. rugosa; 8, R. gallica; 9, R. chinensis cv Old Blush; 10, R. chinensis spontanea; 11, R. gigantea; 12, R. odorata cv Hume's Blush; 13, R. odorata cv Park's Yellow; 14, R. hugonis; 15, R. wichuraiana; 16, R. roxburghii. Lanes corresponding to Chinese roses are boxed.
Analysis of OOMT Protein Accumulation in Petals.
In some cases, modifications of flower scent during evolution have been shown to occur as a result of an increase in the expression of a scent-related gene in petals (20). Previous work on R. gallica and Damask rose showed that, although these European roses possessed OOMT homologs encoding potentially active proteins, these genes were not expressed in flower tissues (11). Extending these investigations to the rose species selected in this study, we used anti-OOMT antibodies (11) to monitor OOMT protein accumulation in stamens and petals. Western blot analyses (Fig. 4B) showed that, although OOMT proteins were detected in the stamens of some Eurasiatic roses, they did not accumulate in petals of these species (except for a low amount in the petals of R. canina). On the other hand, OOMT proteins accumulated to high levels in petals of Chinese roses, indicating that PME production correlates with OOMT gene expression in petals.
Discussion
The Substrate Specificities of OOMT Proteins Are Determined Principally by a Single Amino Acid Polymorphism in Their Phenolic Substrate Binding Site.
The characterization of the three-dimensional structures of alfalfa chalcone and isoflavone-OMTs identified residues important for the function of these enzymes, including catalytic residues and residues involved in substrate binding (12). It has subsequently been shown that this information can be used to identify functionally important residues in other OMTs (16). In rose petals, the closely related OOMT1 and OOMT2 enzymes have been proposed to operate sequentially to catalyze the first and the second methylation step of orcinol, respectively, because of their different substrate specificities (9). To investigate the structural bases for these differences, we modeled the structures of rose OOMT1 and OOMT2 based on the crystal structure of IOMT from alfalfa. The molecular models suggested that the difference in substrate specificity could be explained by a single amino acid polymorphism in the phenolic substrate binding site (Tyr-127 in OOMT1 and the equivalent Phe-126 in OOMT2), which was predicted to alter the steric and hydrophobicity characteristics of the active site. This predicted major role of Tyr-127 and Phe-126 in substrate discrimination was confirmed by using site-directed mutagenesis; OOMT2 F126Y and OOMT1 Y127F mutants showed the same substrate preference as the native OOMT1 and OOMT2, respectively (Fig. 2). Apart from the Phe/Tyr difference, OOMT1 and OOMT2 genes are remarkably similar (97.5% overall nucleotide identity). Within the plant OMT family, several studies have provided detailed information about the structural basis for substrate discrimination (21). In particular, studies on isoeugenol-OMT from Clarkia breweri (22), phenylpropene-OMTs (16), and cinnamate/p-coumarate carboxyl methyltransferase (23) from basil have shown that a small number of amino acid changes may have been responsible for the evolution of novel substrate specificities. In a few cases, such amino acid changes have been shown to be favored by positive selection mechanisms (24, 25). Thus, as has been observed with a number of other OMT enzymes involved in plant secondary metabolite biosynthesis (16, 18, 21), rose OOMTs provide additional evidence that minor changes, sometimes a single amino acid substitution, may be sufficient to produce an OMT with a significantly modified substrate specificity over a short period of evolutionary time.
Acquisition of PME Biosynthesis in Chinese Roses Involved both an Increase in Expression Level in Petals and Evolution of More Efficient OOMT Enzymes.
An interesting question is whether PME were produced by a common ancestor of Eurasiatic and Chinese species and subsequently lost by the Eurasiatic species or whether this trait was acquired in Chinese roses. Previous phylogenetic analyses showed that the Chinese roses, grouped under the Indicae section of the genus Rosa, evolved recently within this genus (26), and this recent emergence would be consistent with the evolution of new functions within in this group. The restricted distribution of the OOMT1 genes within the Chinese rose group, together with the clustering of all of the OOMT1 sequences in a single clade, suggests that OOMT1 arose by duplication of a preexisting OOMT2 gene, followed by functional diversification. OOMT1 and OOMT2 thus illustrate the process of gene duplication followed by functional divergence, with the OOMT1 protein now being optimized to carry out the first, and OOMT2 the second, methylation reaction leading to the biosynthesis of DMT from orcinol. However, if both enzymes are necessary to efficiently catalyze DMT biosynthesis, then the question arises as to what was the original function of the ancestral OOMT2-like gene. There is no clear answer to this question because OOMT-like genes exist in different plant species (10) and, although OOMT2-like enzymes from R. roxburghii and R. rugosa were found to exhibit OOMT2 activity in vitro, predicting the physiological substrates of OMTs is not trivial (27). It should be noted that OOMT2 from Old Blush is able to catalyze the first methylation reaction leading to DMT in vitro although with a relatively low efficiency (Fig. 2). Therefore, DMT biosynthesis would theoretically be possible with only one OOMT2-like enzyme. However, OOMT1 catalyzes this first methylation reaction (from orcinol to MHT) much more efficiently, as shown by specific activities (2,200 pkat·mg−1 for OOMT1, 460 pkat·mg−1 for OOMT2; Fig. 2), and Kcat/Km values (2,500 M−1·s−1 for OOMT1, 300 M−1·s−1 for OOMT2; Table S1), and, in this respect, the evolution of OOMT1 activity can be considered to have been a process optimization corresponding to a major transition in the evolution of the DMT biosynthetic pathway. Consistent with this hypothesis, the PME-producing wild rose species R. chinensis spontanea and R. gigantea both possess OOMT1 genes encoding functional enzymes with characteristic OOMT1 features, especially a high specific activity toward orcinol. These OOMT1 genes were also found in later Chinese hybrids of R. chinensis spontanea or R. gigantea, like Old Blush, Hume's Blush, and Parks Yellow. Finally, modern roses varieties, which produce DMT, inherited OOMT1 genes from their Chinese progenitors, and, in this respect, it is noteworthy that the OOMT1 proteins from the DMT-rich modern hybrid Golden Gate and from R. chinensis cv Old Blush are identical (9, 10).
In addition to gene duplication and evolution, changes in scent-related gene expression may allow the evolution of new scent characteristics, as shown in the genus Clarkia (20). Taken together with the observation that OOMT proteins accumulate to high levels only in petals of roses that produce PME, the data presented here indicate that Chinese roses evolved their ability to produce these scent compounds as a result of a combination of increased OOMT gene expression in petals and the evolution of more catalytically active enzymes. Because DMT has been shown to be perceived by bees (28, 29), evolution of the PME biosynthetic pathway in Chinese roses might therefore have provided distinctive olfactory cues for insect pollinators.
Among PME-producing roses, R. chinensis spontanea exhibits unique scent characteristics. Indeed, this very rare wild species, found only twice over a period of one century (30), produces large amounts of TMB (60% total volatiles) and no DMT (3). In this species, biosynthesis of the trimethylated molecule TMB involves a phloroglucinol-OMT (POMT) that catalyzes the first methylation step of phloroglucinol, which is a poor substrate of OOMTs (31). POMT and OOMTs are therefore linked metabolically, posing the question of the evolutionary relationship between these enzymes (32). Phylogenetic analyses of the plant OMT gene family revealed that POMT and OOMTs belong to different subfamilies. Indeed, POMT is related to the large caffeic acid-OMT (COMT) gene family, whereas OOMTs belong to a distinct clade, which includes chavicol and eugenol-OMT from basil (31, 32). The evolutionary distance between POMT and OOMTs suggests that they have evolved independently, from different ancestors within the OMT gene family. It is therefore possible that R. chinensis spontanea represents a further evolution of the PME metabolism leading to the biosynthesis of TMB instead of DMT. According to this hypothesis, POMT would have evolved in R. chinensis spontanea to function with existing OOMTs, allowing efficient production of TMB and giving this species its unique scent characteristics. Several additional OMTs with overlapping substrate specificities are expressed in Rosa chinensis spontanea petals. In particular, the COMT-like enzyme RcOMT2 exhibits a broad substrate specificity and is able to catalyze the same reaction as OOMT1 (33). RcOMT2 and OOMT1 may represent a case of repeated evolution, where enzymes with similar functions evolved from different progenitors (32). Additional work will be needed to understand the contribution of these different OMTs to scent biosynthesis in R. chinensis spontanea.
Evolution of new functions after gene duplication has been proposed to play a fundamental role in the origin of phenotypic diversity. However, it is rarely possible to substantiate such propositions by tracing the gene duplication and specialization processes back through evolutionary time (19, 34). In conclusion, we have exploited the unique scent characteristics of Chinese roses, which gave rise to the Tea scent character in modern roses, to provide direct functional and phylogenetic evidence for the evolution of specialized OOMT enzymes in these species. We propose that this process was a key event in the acquisition of PME production by Chinese roses.
Materials and Methods
Plant Material.
All rose species were grown in open soil conditions in the Botanical Garden of the Parc de la Tête d'Or, Lyon, France. To reflect the diversity of the genus Rosa, our sampling was based on the recent phylogeny of the genus Rosa (26). The selected species cover all sections of this genus, except the Carolinae, which is a small (four species) subsection of the Cinnamomeae. The Cinnamomeae representing the largest section of the genus Rosa, accounting for about half of all roses species, we selected four rose species exhibiting very different geographic repartitions.
Chemicals and Radiochemicals.
MHT was prepared as described in ref. 10. S-adenosyl-l-[methyl-14C]methionine (55 mCi/mmol) ([14C]SAM) was from GE Healthcare–Amersham Biosciences. All other chemicals and reagents were from Sigma (St. Quentin Fallavier, France).
Molecular Modeling of OOMT1 and OOMT2.
The structures of OOMT1 and OOMT2 were modeled by homology as homodimers by using Modeller (35). Orcinol and MHT were constructed from neighbor structures found in the HIC-UP database (Hetero-Compound Information Centre, Uppsala). Topology and parameters for these molecules were generated with HETZE and XPLO2D (36) and entered into the Modeller database. The template model was the homodimer of IOMT complexed with isoformononetin (PDB ID code 1FP2) (12) where cycle A of isoformononetin was replaced by orcinol or MHT, respectively, in OOMT1 and OOMT2. The quality of final models was verified by ProsaII (37). Further refinement of the ligand position was carried out by using the docking software Autodock (38).
Cloning of OOMT-Like Genes.
OOMT genes were amplified by PCR using the upstream primer 5′-ATGGAAAGGCTAAACAGCTTTAGACACCTTAAC-3′ and the downstream primer 5′-TCAAGGATAAACCTCAATGAGAGACCTTAAACC-3′ as described in ref. 11. Amplified DNA fragments were cloned into pGEM T-easy (Promega, Madison, WI).
Phylogenetic Reconstruction of the OOMT Gene Family.
Seventy-one previously undescribed OOMT sequences and previously published OOMT1 and OOMT2 sequences were aligned by using ClustalW (39). Phylogenetic relationships were then assessed under a Bayesian framework using the most complex model implement in MrBayes v3.0B4 (40). Most probable topologies and nodal posterior probabilities were computed by using four independent chains run for 1,000,000 generations and an empirically defined “burn in” fraction. In addition to posterior probabilities, nodal support was estimated by nonparametric bootstrapping under a maximum likelihood framework using Paup* 4.0b10 (41). Paup* 4.0b10 default parameters were used, with the exception of the model of evolution that we selected using the AIC criterion (42) as implemented in Modeltest 3.6 (43).
Characterization of Recombinant OOMTs.
Old Blush OOMT1 and OOMT2 coding regions were cloned into pGEX-4T1. Site-directed mutagenesis was performed by using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). Recombinant OOMT proteins were expressed as GST fusion proteins and purified as in ref. 10, with the exception that an optimized cleavage procedure using high quality thrombin (GE Healthcare-Amersham) yielded recombinant proteins exhibiting higher specific activities. To characterize OOMT-like enzymes, OOMT-like genes were spliced by PCR-driven overlap extension. The corresponding cDNAs were amplified by PCR using the primers 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTGGTTCCGCGTGGATC C A T GGAAAGGCTAAACAGCTTT-3′ and 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTTCATCAAGGATAAACCTCAATGAGAGA-3′, cloned into pDONR207 vector (Invitrogen, Carlsbad, CA). cDNAs were sequenced and subsequently transferred into the pDEST15 destination vector (Invitrogen). OOMT-like enzymes were purified and characterized after cleavage of the GST moiety as described above. Purified recombinant OOMTs were incubated as described in ref. 11, in 25 μl final volume with 35 μM [14C]SAM and 200 μM to 1 mM phenolic substrates. Ranges of phenolic substrate concentrations of between 5 μM and 1 mM were used for Km determination. Km and Vmax values were calculated from Lineweaver–Burk plots. Reaction products were analyzed by TLC as described in ref. 10.
Western Blot Analyses.
Western blot analyses were performed as indicated in ref. 11, except that, for protein extraction, tissues were grounded in 50 mM Tris·HCl (pH 6.8), 2.5% SDS, 100 mM DTT, 10% glycerol (4 ml per g of fresh weight) and incubated at 95°C for 5 min, before centrifugation (5 min, 10,000 × g). Each lane was loaded with 15 μg of total proteins.
Supplementary Material
Acknowledgments.
We thank Isabelle Desbouchages, Alexis Lacroix, Armand Guillermin (Ecole Normale Supérieure de Lyon), Christophe Ferry (Jardin Botanique de la Ville de Lyon, Parc de la Tête d'Or), and Henriette Mourgues for help with plant material. We thank Marie-Rose Viricel (Université Lyon 1), Patricia Claudel [Institut National de la Recherche Agronomique (INRA), Colmar], Anne-Marie Thierry, Hervé Leyral, and Claudia Bardoux (Ecole Normale Supérieure de Lyon) for technical assistance. We thank Raquel Tavares, Dominique Mouchiroud (Université Lyon 1), Marc Robinson-Rechavi (University of Lausanne), Francis Karst (INRA Colmar), Emmanuelle Schmitt, and Yves Mechulam (Ecole Polytechnique, Palaiseau) for helpful comments on the manuscript. This work was supported by the Région Rhône-Alpes (France), the Institut National de la Recherche Agronomique, and the Centre National de la Recherche Scientifique.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. E.P. is a guest editor invited by the Editorial Board.
Data deposition: The sequences reported in this paper have been deposited in the GenBank/EMBL databases (accession nos. AM182763–AM182856).
This article contains supporting information online at www.pnas.org/cgi/content/full/0711551105/DCSupplemental.
References
- 1.Krüssmann G. Roses. London: B. T. Batsford; 1982. [Google Scholar]
- 2.Flament I, Debonneville C, Furrer A. Volatile constituents of roses: Characterization of cultivars based on the headspace analysis of living flower emissions. In: Teranishi R, Buttery RG, Sugisawa H, editors. Bioactive Volatile Compounds from Plants. Washington, DC: Am Chem Soc; 1993. pp. 269–281. [Google Scholar]
- 3.Joichi A, Yomogida K, Awano K, Ueda Y. Volatile components of tea-scented modern roses and ancient Chinese roses. Flavour Fragr J. 2005;20:152–157. [Google Scholar]
- 4.Nakamura S. Scent and component analysis of the hybrid tea rose. Perfum Flavor. 1987;12:43–45. [Google Scholar]
- 5.Guterman I, et al. Rose scent: Genomics approach to discovering novel floral fragrance-related genes. Plant Cell. 2002;14:2325–2338. doi: 10.1105/tpc.005207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Channelière S, et al. Analysis of gene expression in rose petals using expressed sequence tags. FEBS Lett. 2002;515:35–38. doi: 10.1016/s0014-5793(02)02413-4. [DOI] [PubMed] [Google Scholar]
- 7.Shalit M, et al. Volatile ester formation in roses: Identification of an acetyl-coenzyme A-Geraniol/Citronellol acetyltransferase in developing rose petals. Plant Physiol. 2003;131:1868–1876. doi: 10.1104/pp.102.018572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaminaga Y, et al. Plant phenylacetaldehyde synthase is a bifunctional homotetrameric enzyme that catalyzes phenylalanine decarboxylation and oxidation. J Biol Chem. 2006;281:23357–23366. doi: 10.1074/jbc.M602708200. [DOI] [PubMed] [Google Scholar]
- 9.Lavid N, et al. O-methyltransferases involved in the biosynthesis of volatile phenolic derivatives in rose petals. Plant Physiol. 2002;129:1899–1907. doi: 10.1104/pp.005330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scalliet G, et al. Biosynthesis of the major scent components 3,5-dimethoxytoluene and 1,3,5-trimethoxybenzene by novel rose O-methyltransferases. FEBS Lett. 2002;523:113–118. doi: 10.1016/s0014-5793(02)02956-3. [DOI] [PubMed] [Google Scholar]
- 11.Scalliet G, et al. Role of petal-specific orcinol O-methyltransferases in the evolution of rose scent. Plant Physiol. 2006;140:18–29. doi: 10.1104/pp.105.070961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zubieta C, He XZ, Dixon RA, Noel JP. Structures of two natural product methyltransferases reveal the basis for substrate specificity in plant O-methyltransferases. Nat Struct Biol. 2001;8:271–279. doi: 10.1038/85029. [DOI] [PubMed] [Google Scholar]
- 13.Zubieta C, Kota P, Ferrer JL, Dixon RA, Noel JP. Structural basis for the modulation of lignin monomer methylation by caffeic acid/5-hydroxyferulic acid 3/5-O-methyltransferase. Plant Cell. 2002;14:1265–1277. doi: 10.1105/tpc.001412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zubieta C, et al. Structural basis for substrate recognition in the salicylic acid carboxyl methyltransferase family. Plant Cell. 2003;15:1704–1716. doi: 10.1105/tpc.014548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferrer JL, Zubieta C, Dixon RA, Noel JP. Crystal structures of alfalfa caffeoyl coenzyme A 3-O-methyltransferase. Plant Physiol. 2005;137:1009–1017. doi: 10.1104/pp.104.048751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gang DR, et al. Characterization of phenylpropene O-methyltransferases from sweet basil: Facile change of substrate specificity and convergent evolution within a plant O-methyltransferase family. Plant Cell. 2002;14:505–519. doi: 10.1105/tpc.010327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pott MB, et al. Biochemical and structural characterization of benzenoid carboxyl methyltransferases involved in floral scent production in Stephanotis floribunda and Nicotiana suaveolens. Plant Physiol. 2004;135:1946–1955. doi: 10.1104/pp.104.041806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pichersky E, Gang DR. Genetics and biochemistry of secondary metabolites in plants: An evolutionary perspective. Trends Plant Sci. 2000;5:439–445. doi: 10.1016/s1360-1385(00)01741-6. [DOI] [PubMed] [Google Scholar]
- 19.Zhang J. Evolution by gene duplication: An update. Trends Ecol Evol. 2003;18:292–298. [Google Scholar]
- 20.Dudareva N, Cseke L, Blanc VM, Pichersky E. Evolution of floral scent in Clarkia: Novel patterns of S-linalool synthase gene expression in the C. breweri flower. Plant Cell. 1996;8:1137–1148. doi: 10.1105/tpc.8.7.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Noel JD, Dixon RA, Pichersky E, Zubieta C, Ferrer J. Structural, functional and evolutionary basis for methylation of plant small molecules. In: Romeo JT, editor. Recent Advances in Phytochemistry. Vol 37. Oxford: Elsevier Science Ltd; 2003. pp. 253–283. [Google Scholar]
- 22.Wang J, Pichersky E. Identification of specific residues involved in substrate discrimination in two plant O-methyltransferases. Arch Biochem Biophys. 1999;368:172–180. doi: 10.1006/abbi.1999.1304. [DOI] [PubMed] [Google Scholar]
- 23.Kapteyn J, et al. Evolution of cinnamate/p-coumarate carboxyl methyltransferases and their role in the biosynthesis of methylcinnamate. Plant Cell. 2007;19:3212–3229. doi: 10.1105/tpc.107.054155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barkman TJ. Evidence for positive selection on the floral scent gene isoeugenol O-methyltransferase. Mol Biol Evol. 2003;20:168–172. doi: 10.1093/molbev/msg030. [DOI] [PubMed] [Google Scholar]
- 25.Barkman TJ, Martins TR, Sutton E, Stout JT. Positive selection for single amino acid change promotes substrate discrimination of a plant volatile-producing enzyme. Mol Biol Evol. 2007;24:1320–1329. doi: 10.1093/molbev/msm053. [DOI] [PubMed] [Google Scholar]
- 26.Wissemann V, Ritz CM. The genus Rosa (Rosoideae, Rosaceae) revisited: Molecular analysis of nrITS-1 and atpB-rbcL intergenic spacer (IGS) versus conventional taxonomy. Bot J Linn Soc. 2005;147:275–290. [Google Scholar]
- 27.Schröder G, Wehinger E, Schröder J. Predicting the substrates of cloned plant O-methyltransferases. Phytochemistry. 2002;59:1–8. doi: 10.1016/s0031-9422(01)00421-6. [DOI] [PubMed] [Google Scholar]
- 28.Shalit M, et al. Volatile compounds emitted by rose cultivars: Fragrance perception by man and honeybees. Isr J Plant Sci. 2004;52:245–255. [Google Scholar]
- 29.Wright GA, Lutmerding A, Dudareva N, Smith BH. Intensity and the ratios of compounds in the scent of snapdragon flowers affect scent discrimination by honeybees (Apis mellifera) J Comp Physiol A. 2005;191:105–114. doi: 10.1007/s00359-004-0576-6. [DOI] [PubMed] [Google Scholar]
- 30.Rix M. China roses. In: Foster R, editor. Historic Roses Journal No. 17. Hertfordshire, England: Royal National Rose Society, St. Albans; 1999. pp. 8–12. [Google Scholar]
- 31.Wu S, et al. The key role of phloroglucinol O-methyltransferase in the biosynthesis of Rosa chinensis volatile 1,3,5-trimethoxybenzene. Plant Physiol. 2004;135:1–8. doi: 10.1104/pp.103.037051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gang DR. Evolution of flavors and scents. Annu Rev Plant Biol. 2005;56:301–325. doi: 10.1146/annurev.arplant.56.032604.144128. [DOI] [PubMed] [Google Scholar]
- 33.Wu S, et al. Two O-methyltransferases isolated from flower petals of Rosa chinensis var. spontanea involved in scent biosynthesis. J Biosci Bioeng. 2003;96:119–128. [PubMed] [Google Scholar]
- 34.Yokoyama S. Evaluating adaptive evolution. Nat Genet. 2002;30:350–351. doi: 10.1038/ng0402-350. [DOI] [PubMed] [Google Scholar]
- 35.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 36.Kleywegt GJ, Jones TA. Databases in protein crystallography. Acta Crystallogr D. 1998;54:1119–1131. doi: 10.1107/s0907444998007100. [DOI] [PubMed] [Google Scholar]
- 37.Sippl MJ. Recognition of errors in three-dimensional structures of proteins. Proteins. 1993;17:355–362. doi: 10.1002/prot.340170404. [DOI] [PubMed] [Google Scholar]
- 38.Morris GM, et al. Automated docking using a lamarckian genetic algorithm and empirical binding free energy function. J Comput Chem. 1998;19:1639–1662. [Google Scholar]
- 39.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 41.Swofford DL. PAUP*: Phylogenetic Analysis Using Parsimony (*and Other Methods) Sunderland, MA: Sinauer Associates; 2002. Version 4. [Google Scholar]
- 42.Akaike H. A new look at the statistical model identification. IEEE Trans Autom Contr. 1974;19:716–723. [Google Scholar]
- 43.Posada D, Crandall KA. MODELTEST: Testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.