Abstract
Cell fate and organismal lifespan are controlled by a complex signaling network whose dysfunction can cause a variety of aging-related diseases. An important protection against these failures is cellular apoptosis, which can be induced by p66Shc in response to cellular stress. The precise mechanisms of p66Shc action and regulation and the function of the p66Shc-specific N terminus remain to be identified. Here, we show that the p66Shc N terminus forms a redox module responsible for apoptosis initiation, and that this module can be activated through reversible tetramerization by forming two disulfide bonds. Glutathione and thioredoxins can reduce and inactivate p66Shc, resulting in a thiol-based redox sensor system that initiates apoptosis once cellular protection systems cannot cope anymore with cellular stress.
Keywords: apoptosis, mitochondria, redox regulation
Apoptosis is an essential mechanism in cellular homeostasis and molecular stress management (1, 2). It is regulated by a network of signaling proteins that also contribute to lifespan regulation and cellular aging processes (1, 3, 4). p66Shc (Shc: Src homologous and collagen) belongs to this network and acts as a molecular sentinel that controls cellular stress responses and mammalian lifespan (5–7). p66Shc−/− mice exhibit a 30% extended lifespan, reduced H2O2 levels, and an enhanced resistance against oxidative stress (5). p66Shc activity has been implicated in the development of aging-related diseases such as early artherogenesis (8). Upon induction by stress factors, such as oxidants or UV irradiation, expression of the p66Shc protein increases (9) and it translocates to the mitochondrial intermembrane space where it induces apoptosis (10, 11). It was proposed that p66Shc oxidizes cytochrome c (Cyt c), thereby generating H2O2, which is assumed to activate the permeability transition pore (PTP), ultimately causing rupture of mitochondria and release of apoptotic factors (11). Because of its central role in lifespan regulation and aging-related diseases (6, 12, 13), p66Shc is an attractive drug target (14), and a deeper understanding of the mechanisms regulating its apoptosis-inducing activity would enable the development of novel p66Shc-targeted therapies.
p66Shc is the largest of three protein variants expressed from the shcA gene. It has the characteristic Shc domain structure consisting of an N-terminal phosphor-tyrosine-binding domain (PTB), a central collagen homology (CH) region (CH1), and a C-terminal Src homology 2 (SH2) domain. Compared with the shorter isoforms, p46Shc and p52Shc, known inductors of the Ras signaling pathway (15, 16), p66Shc exhibits an N-terminal extention comprising a second CH domain (CH2). A short sequence between the p66Shc-specific CH2 domain and the PTB was reported to act as Cyt c binding region (CB) (11) and constitutes the N terminus of p52Shc. In p52Shc, the CB domain is unstructured (17). Furthermore, the new Shc family member, RalP, has the characteristic Shc domain structure (18) with pronounced homology to p66Shc except for the CH2 and the CB domains (amino acids 1–150). The CB domain may thus be a structural and functional part of the CH2 domain. Therefore, we used here a p66Shc fragment covering both domains (p66CH2-CB) to analyze the function of the p66Shc N terminus in the mitochondrial apoptosis initiation process.
Results and Discussion
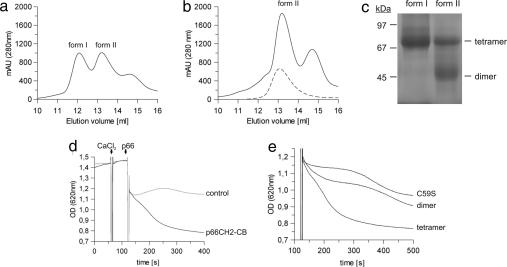
p66CH2-CB cloned from mouse colon cDNA was overexpressed in Escherichia coli and purified by affinity chromatography followed by size exclusion chromatography (SEC). Depending on the presence of a reducing agent (2.5 mM DTT), one or two major elution peaks (forms 1 and 2) appeared (Fig. 1 a and b). SDS/PAGE and MS identified both peaks to be p66CH2-CB; the third peak (14.6 ml) is a contaminating protein from the bacterial expression system (catabolite gene activator protein). No higher-order oligomers or aggregates of p66CH2-CB were observed, indicating a redox-dependent, specific oligomerization of the protein. The elution volumes of the two forms indicate that they are p66CH2-CB dimers and tetramers, respectively. Form 1 runs between the 67- and 97-kDa marker proteins in blue-native PAGE (BN-PAGE) (Fig. 1c), consistent with a tetramer of the 19.5-kDa p66CH2-CB monomer, and form 2 behaves as a dimer of ≈40 kDa. Thus, p66CH2-CB forms dimers that tetramerize in a redox-dependent manner.
Fig. 1.
Dimer–tetramer transition of p66CH2-CB regulating its apoptosis-inducing activity. (a) SEC elution profile of p66CH2-CB in the absence of reducing agent. (b) SEC elution profile of p66CH2-CB in the presence of 2.5 mM DTT, overlaid with the elution profile obtained with the mutant p66CH2-CB C59S in the absence of DTT (dashed line). (c) Stoichiometry of the two p66CH2-CB forms determined by BN-PAGE. (d) Standard experiment of the mitochondria swelling assay in the presence of 5 mM succinate. Rupture of mitochondria was observed after sequential addition of 7 μM CaCl2 and 20 μM p66CH2-CB instead of buffer (control). (e) Comparison of the apoptosis-inducing activity of dimeric and tetrameric p66CH2-CB and p66CH2-CB C59S. The initial sensitization with CaCl2 has been omitted for clarity. The decrease in OD620 observed for the dimeric p66CH2-CB and the p66CH2-CB C59S mutant beyond ≈400 s was also observed in buffer controls and is likely caused by sedimentation of the mitochondria during extended incubation.
CH2 is exclusive to p66Shc, and we therefore assumed it is responsible for the apoptosis-inducing activity of p66Shc. We tested the ability of p66CH2-CB to induce swelling and rupture of mitochondria, an initial step in mitochondria-mediated apoptosis (11, 19). Purified rat liver mitochondria were sensitized with 7 μM CaCl2, followed by addition of 20 μM p66CH2-CB and observation of their OD620 as described for full-length p66Shc (11) (Fig. 1d). Tetrameric p66CH2-CB caused a significant decrease of the OD620 because of the rupture of the mitochondria; adding instead the same amount of dimer (20 μM) had only a small effect on OD620 (Fig. 1e). Even comparing a 3-fold higher concentration of dimer (30 μM) with 10 μM tetramer (data not shown) resulted in significantly higher activity for the tetramer. Hence, the isolated p66Shc-specific N terminus is able to induce mitochondrial rupture, which inevitably leads to apoptosis (19), and it is activated through an oxidative dimer–tetramer transition.
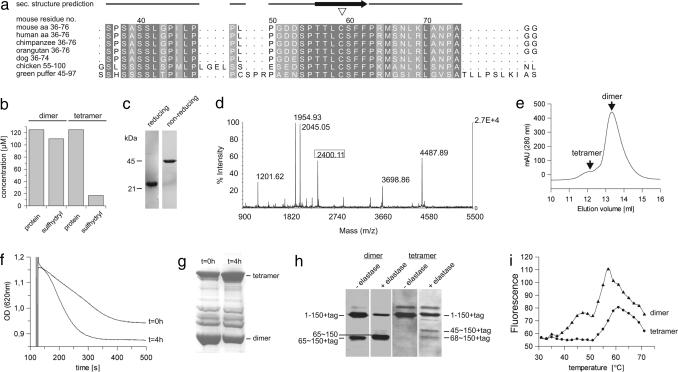
p66CH2-CB contains a single cysteine at position 59, which is conserved in p66Shc proteins from mammals and other organisms (Fig. 2a) and in the Shc family protein RalP. We hypothesized that the oxidative tetramerization of p66CH2-CB is mediated by disulfide bridging through this conserved cysteine. Testing for sulfhydryl groups with Ellman's reagent indeed identified one free cysteine per monomer exclusively in the dimer form (Fig. 2b). Consistently, the p66CH2-CB tetramer generated the dimer at ≈45 kDa in nonreducing SDS/PAGE and not the monomer observed under reducing conditions (Fig. 2c). Furthermore, through MALDI-TOF-MS of a chymotryptic digest of the tetramer we detected the disulfide-linked peptide Leu-68–Cys-59–S–S–Cys-59′–Leu-68′ (Fig. 2d) (expected: 2,400.08 Da; measured: 2,400.11 Da); this peptide signal was not detected in a digest of the dimer. The peptide's sequence was further validated by using electrospray ionization (ESI)-ion-trap-MS/MS with a high coverage of y- and b-ion series [supporting information (SI) Fig. S1]. In addition, fragments of the disulfide-linked peptide, including one complete peptide disulfide-bridged to one fragmented peptide, were identified in the ESI-ion-trap-MS/MS spectrum (e.g., Leu-68–Cys-59–S–S–Cys-59′–Phe-62′; expected: 1,683.7 Da; measured: 1,683.6 Da). Our results thus show that the noncovalent dimer has two accessible cysteines, one per monomer, which are used for two disulfide bridges in the tetramer. Tetramerization of p66CH2-CB is stable but reversible, as treatment with DTT and subsequent SEC generated the reduced dimer (Fig. 2e). Finally, we mutated Cys-59 to serine (p66CH2-CB C59S), yielding exclusively the dimer in SEC (Fig. 1b) and nonreducing SDS/PAGE (data not shown), which proofs the disulfide-mediated tetramerization and provides evidence that p66Shc can adopt two different oligomeric states, a dimer and a tetramer. The mitochondria swelling activity of p66CH2-CB C59S was comparable to the WT dimer (Fig. 1e), but only the activity of the WT increased over time (Fig. 2f). This increase in activity correlated with the sulfhydryl oxidation status (Fig. 2g), again linking the dimer–tetramer transition to the apoptosis-inducing activity of p66Shc.
Fig. 2.
Disulfide bridging through the conserved residue Cys-59 and accompanied structural rearrangements in p66CH2-CB. (a) Alignment of p66CH2-CB from various organisms around the conserved Cys-59 residue (▿). (b) Detection of free sulfhydryl groups in the dimer but not in the tetramer form by using Ellman's reagent. (c) Tetrameric p66CH2-CB subjected to reducing and nonreducing SDS/PAGE. (d) MALDI-TOF-MS spectrum of the chymotryptic digest of the p66CH2-CB tetramer including the disulfide-linked peptide Leu-68–Cys-59–S–S–Cys-59′–Leu-68′ (framed). (e) SEC elution profile of the tetramer after treatment with 2.5 mM DTT. (f and g) Time-dependent increase in p66CH2-CB-induced rupture of mitochondria (f) and its correlation with the protein's oxidation level determined by nonreducing SDS/PAGE (g). Dimeric p66CH2-CB was analyzed (t = 0), subsequently incubated on ice for 4 h, and then tested again (t = 4 h). (h) Limited proteolysis of p66CH2-CH dimer and tetramer with elastase. Even before the addition of elastase, fragmentation of the dimer but not the tetramer was observed, caused by proteases of the expression host. (i) Thermal denaturation of p66CH2-CH dimer and tetramer.
Secondary structure predictions with Jpred2 (20) (Fig. S2a) indicated that p66CH2-CB does not have a coiled coil structure although the CH2 domain is collagen-like, but sequence analyses with Prot-Scale (21) (Fig. S2 b and c) predict Cys-59 to be localized in a rather hydrophobic environment with low flexibility. However, limited proteolysis of the dimer but not of the tetramer (Fig. 2h) generated an N-terminally truncated fragment starting at Met-65, indicating flexibility around Cys-59 in the dimer and structural rearrangements in this tetramerization region during oxidative dimerization of the noncovalent dimers. Further, the dimer showed a biphasic thermal denaturation transition, whereas the tetramer lacks the first transition step at lower temperature (Fig. 2i), indicating that during tetramerization a more flexible subdomain gets rearranged and/or stabilized. We speculate that the activating tetramerization of p66Shc and the associated stabilizing rearrangements may be the mechanism behind how the GTPase Rac1 and the apoptosis-inducing protein p53 increase p66Shc activity, because both were reported to act by increasing the stability of p66Shc (22, 23). This mechanism resembles the activation of other proteins in response to oxidative stress (24, 25), e.g., the peroxiredoxine PrxI, which acts as dimeric peroxidase but transforms into a decameric chaperone upon its oxidation at specific cysteine residues (26).
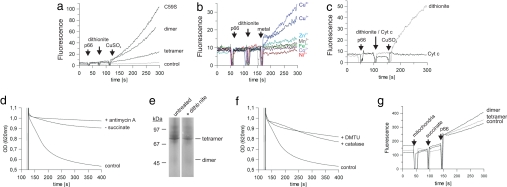
It has been reported that p66Shc generates reactive oxygen species (ROS) and it was supposed that these ROS then mediate p66Shc-induced apoptosis (11). For testing our protein for ROS generation we used the dye H2DDFDA, which becomes fluorescent after oxidation by ROS, as described (11). Fluorescence increased significantly after addition of p66CH2-CB in the presence of CuSO4 and Na-dithionite (Fig. 3a). Adding only Na-dithionite and CuSO4 without protein did not show any effect on the fluorescence (Fig. 3a, control). Hence, the p66Shc-specific N terminus is the ROS-generating part of p66Shc. Consistent with findings for the full-length protein (11), only the addition of copper ions lead to p66CH2-CB-dependent generation of ROS, contrary to other metal ions tested (CuCl2, FeCl3, NiSO4, ZnCl2, CoCl2, MnCl2) (Fig. 3b), possibly hinting at a copper-specific metal center in p66CH2-CB. No difference between Cu1+ and Cu2+ was observed for the p66CH2-CB ROS-generating activity, possibly indicating that both copper redox states might be involved in catalysis. However, we cannot exclude the possibility that Na-dithionite reduces dissolved Cu2+ to Cu1+, which then acts as active ion.
Fig. 3.
Copper-dependent ROS formation by p66CH2-CB and contribution of Cyt c to the apoptosis-inducing process. (a–c) Changes in fluorescence of H2DDFDA after sequential addition of the p66CH2-CB forms (20 μM) or buffer (control), 85 μM Na-dithionite (a–c) or 20 μM Cyt c (c), and 50 μM CuSO4 (a and c) or 50 μM of other metal ions (b). (d) Rupture of mitochondria after addition of 7 μM CaCl2 followed by buffer (control) or 0.3 μg/ml antimycin A, respectively, followed by addition of tetrameric p66CH2-CB. The experiment was done in the presence (control and + antimycin A) or absence (− succinate) of 5 mM succinate. (e) BN-PAGE of tetrameric p66CH2-CB treated with 85 μM Na-dithionite. (f) Rupture of mitochondria after addition of 7 μM CaCl2 followed by buffer (control), 280 units of catalase or 20 mM DMTU, and 20 μM tetrameric p66CH2-CB in the presence of 5 mM succinate. (g) Changes in fluorescence of H2DDFDA after sequential addition of mitochondria (100 μg total mitochondrial protein), succinate (5 mM), and dimeric or tetrameric p66CH2-CB (40 μM); as a control, buffer was added instead of protein.
To identify a physiological electron donor for ROS formation we added reduced Cyt c instead of Na-dithionite to the assay, but no ROS generation was observed (Fig. 3c) in contrast to results published for full-length p66Shc (11). However, consistent with the results on full-length protein (11) p66CH2-CB-dependent mitochondrial rupture required the presence of succinate as substrate for the respiratory chain, and addition of the cytochrome reductase inhibitor antimycin A prevented rupture of mitochondria in the swelling assay (Fig. 3d). Thus, the respiratory chain contributes to the apoptosis-inducing process, but the potential interaction between Cyt c and p66CH2-CB might, e.g., be weakened because of missing domains of p66Shc or other factors necessary for interaction but not present in the in vitro assay.
We next compared the ROS-generating activity of the different p66CH2-CB forms. Surprisingly, the tetramer generated less ROS than the dimer (Fig. 3a), in contrast to the activating effect of tetramerization on the apoptosis-inducing activity of p66CH2-CB. p66CH2-CB C59S (Fig. 3a) and p66CH2-CB with N-ethylmaleimide-modified cysteine (data not shown) also generated more ROS than the tetramer, which further shows that the dimer–tetramer transition is not part of the catalytic cycle for ROS formation but instead has a regulatory role. Consistently, Na-dithionite is a potent electron donor for p66CH2-CB-dependent ROS formation but it is not able to reduce the disulfide bridges of the tetramer (Fig. 3e). We next tested whether ROS indeed act downstream from p66Shc or if ROS generation is a side product of a different p66Shc-dependent apoptosis-initiating mechanism. Quenching ROS through addition of catalase or dimethyl-thiourea (DMTU), respectively, inhibited rupture of mitochondria in our swelling assay (Fig. 3f), which shows that ROS indeed mediate p66Shc-initiated mitochondrial rupture. To exclude that the lower ROS-generating activity of the tetramer compared with the dimer might be caused by the artificial electron donor Na-dithionite we added instead isolated mitochondria into the ROS assay to provide physiological interaction partners. Again, dimeric p66CH2-CB generated more ROS than the tetrameric form (Fig. 3g), validating the lower ROS-generating activity of the tetramer. We therefore assume that the activation of p66CH2-CB might not result in a general increase of ROS generation but maybe results in an elevated affinity for an interaction partner such as the PTP and thereby in a locally increased ROS level.
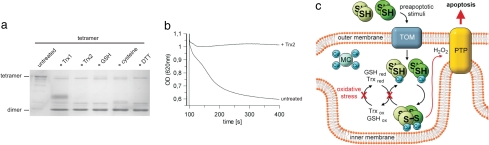
To identify potential physiological reducing agents, and thus inactivators, of tetrameric p66Shc we monitored the oxidation state of p66CH2-CB by BN-PAGE (data not shown) and nonreducing SDS/PAGE (Fig. 4a) after incubation with cytosolic thioredoxin 1 (Trx1), mitochondrial thioredoxin 2 (Trx2), glutathione (GSH), cysteine, and as control with DTT. p66CH2-CB gets completely reduced by these reducing agents (Fig. 4a), and reduction with Trx2 leads to the expected loss of activity in the mitochondrial swelling assay (Fig. 4b). Thus, the cellular redox systems are able to inhibit and to reverse the activating dimer–tetramer transition of p66CH2-CB and thereby to prevent apoptosis initiation.
Fig. 4.
Reduction of tetrameric p66CH2-CB by Trxs and GSH and model for the redox-mediated regulation of p66Shc. (a) Nonreducing SDS/PAGE of tetrameric p66CH2-CB incubated with a 2-fold molar excess of Trx1 and Trx2, 5 mM GSH, 5 mM cysteine, or 5 mM DTT. (b) Pretreatment of the p66CH2-CB tetramer with Trx2 reduces its apoptosis inducing activity. (c) Model of the redox-mediated regulation of p66Shc. After cellular stress dimeric p66Shc translocates to mitochondria. Under conditions of lower stress the Trx and GSH systems reduce tetrameric p66Shc and prevent apoptosis, whereas excessive oxidative stress leads to an overload of the redox systems and accumulation of active, tetrameric p66Shc. p66Shc-generated ROS then activate the PTP, which results in rupture of the mitochondria and release of apoptotic factors. MC, metal chaperone; TOM, translocase of the outer membrane.
Based on our results we propose the following model for the redox-mediated activation of p66Shc (Fig. 4c). Under normal physiological conditions p66Shc mainly localizes to the cytosol (11, 27) where Trx1 and GSH keep it in its inactive reduced state. Cellular stress induces translocation of p66Shc into the mitochondrial intermembrane space (IMS) (10). We speculate that the import of p66Shc might be assisted by a disulfide relay system such as Mia40 (28). Mia40 exchanges its intramolecular disulfide bridges with sulfhydryls of proteins being imported, resulting in a coupling of import and oxidative folding. Interestingly, Mia40 binds copper and zinc ions, and it was proposed to supply the imported protein with these metals (28). In mitochondria, thiols are sensitive to oxidation because of the alkaline pH (25), thus favoring tetramerization of p66Shc. However, under conditions of lower stress the mitochondrial redox systems GSH and Trx reduce tetrameric p66Shc and thereby prevent apoptosis. GSH is present in mitochondria in high concentrations (29), and components of the Trx system appear also to be present in the IMS (30). Excessive oxidative stress, however, leads to an overload of the GSH and Trx systems, which results in an accumulation of active, tetrameric p66Shc. The thereby increased local ROS levels then activate the PTP, which results in rupture of the mitochondrial membranes (11).
We have presented here the function of the N-terminal domain of p66Shc. The domain has two redox centers: (i) a copper-dependent catalytic site for ROS formation, which subsequently induce mitochondrial rupture, a key step of mitochondria-mediated apoptosis, and (ii) a regulatory disulfide/thiol site mediating a reversible dimer–tetramer transition, which increases the protein's mitochondria disintegrating activity. Our results yield insights into regulation of p66Shc activity and provide approaches for targeting this important stress and lifespan regulator protein with drugs.
Materials and Methods
Cloning and Protein Purification.
The p66Shc gene was PCR-amplified from mouse colon cDNA, and residues 1–150 were subcloned into pET101/D-Topo (Invitrogen) with a C-terminal His tag. An influence of the His tag on the apoptosis-inducing activity was excluded by doing control experiments with untagged protein prepared by specific proteolytic tag removal. Site-directed mutagenesis was done with the QuikChange protocol (Stratagene). Expression in E. coli BL21(DE3)Rosetta2 was induced at OD600 = 0.6 with 0.5 mM isopropyl β-D-thiogalactoside, and cell culturing continued for 18 h at 20°C. The dimer was prepared by reduction of p66CH2-CB with 5 mM tris(2-carboxyethyl)phosphine before SEC in absence of reducing agent; reducing agent for stabilization of the dimer was only added if explicitly stated. The tetramer was obtained by overexpressing p66CH2-CB in E. coli BL21(DE3)Rosetta2gami. The protein was purified by affinity chromatography on Talon resin in buffer A [20 mM Tris (pH 7.8), 150 mM NaCl] followed by SEC in buffer A with or without 2.5 mM DTT, respectively. Stoichiometry was determined by BN-PAGE according to Schaegger and von Jagow (31) and protein identity by mass spectrometry as described (32). Limited proteolysis was done as described (33).
MALDI-TOF-MS and ESI-Ion-Trap-MS/MS.
Ten micrograms of pure tetrameric p66CH2-CB in 25 mM NH4HCO3 was digested by using 125 ng of chymotrypsin at 37°C for 18 h. The reaction was stopped through addition of 0.25% trifluoroacetic acid. MALDI-TOF-MS was performed with an Applied Biosystems Voyager-DE Pro system in reflector mode as described (32) with ferulic acid as matrix. For ESI-ion-trap-MS/MS, the digested sample (5 pmol/μl in 2% acetonitrile, 0.1% formic acid) was injected by the syringe pump of the LTQ XL system (Thermo Fisher Scientific) with a flow rate of 1 μl/min. Spray needles were pulled in-house as described (34). The LTQ was operated in the positive mode to acquire a full MS scan in the enhanced mode between m/z = 400 and 2,000, followed by a full MS/MS scan of m/z = 1,200.5 ion from the preceding MS scan. The heated desolvation capillary was set to 180°C. The relative collision energy for collision-induced dissociation was set to 35%.
Thermal Denaturation and In Vitro Activity Assay.
Melting transitions were determined as described (35) with a FluoDiaT70 fluorescence plate reader (Photal Otsuka Electronics) with 12.5 μg of protein in buffer A supplemented with SYPRO (Sigma). The ROS-generating activity was measured by using H2DDFDA (Invitrogen) as described (11). Changes in fluorescence [λ(ex) = 498 nm, λ(em) = 525 nm] of 10 μM H2DDFDA in 100 μl buffer A were recorded with a PerkinElmer LS50B spectrofluorimeter. Fresh reduced rat Cyt c (Sigma) was prepared before use in the in vitro assay through treatment with a 4-fold molar excess of Na-dithionite and subsequent removal of the reducing agent by using a NAP column (GE Healthcare). The stability of the reduced form of the Cyt c stock was verified through UV spectra, and no significant reoxidation was observed over several hours.
Preparation of Mitochondria and Mitochondria Swelling Assay.
Rat liver cells in isolation buffer (2 mM Hepes, 220 mM mannite, and 70 mM sucrose) were disrupted mechanically, and mitochondria were isolated by centrifugation at 4°C at 500 × g for 3 min. The supernatant was centrifuged two more times, followed by centrifugation at 26,000 × g for 10 min. The pellet was washed and recentrifuged twice. Mitochondria were resuspended in 5 mM Tris, pH 7.4, and their outer membrane was permeabilized with 25 μM digitonin for 10 min. Rupture of mitochondria pretreated with 7 μM CaCl2 was monitored photometrically as a decrease of the OD620 as described (11).
Supplementary Material
Acknowledgments.
We thank Bing Liu (Ruhr-University Bochum) for supplying rat liver tissue.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0800691105/DCSupplemental.
References
- 1.Balaban RS, Nemoto S, Finkel T. Mitochondria, oxidants, and aging. Cell. 2005;120:483–495. doi: 10.1016/j.cell.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 2.Ranganathan AC, Adam AP, Aguirre-Ghiso JA. Opposing roles of mitogenic and stress signaling pathways in the induction of cancer dormancy. Cell Cycle. 2006;5:1799–1807. doi: 10.4161/cc.5.16.3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee HC, Wei YH. Oxidative stress, mitochondrial DNA mutation, and apoptosis in aging. Exp Biol Med. 2007;232:592–606. [PubMed] [Google Scholar]
- 4.Hekimi S. How genetic analysis tests theories of animal aging. Nat Genet. 2006;38:985–991. doi: 10.1038/ng1881. [DOI] [PubMed] [Google Scholar]
- 5.Migliaccio E, et al. The p66shc adaptor protein controls oxidative stress response and life span in mammals. Nature. 1999;402:309–313. doi: 10.1038/46311. [DOI] [PubMed] [Google Scholar]
- 6.Migliaccio E, Giorgio M, Pelicci PG. Apoptosis and aging: Role of p66Shc redox protein. Antioxid Redox Signal. 2006;8:600–608. doi: 10.1089/ars.2006.8.600. [DOI] [PubMed] [Google Scholar]
- 7.Purdom S, Chen QM. p66Shc: At the crossroad of oxidative stress and the genetics of aging. Trends Mol Med. 2003;9:206–210. doi: 10.1016/s1471-4914(03)00048-0. [DOI] [PubMed] [Google Scholar]
- 8.Napoli C, et al. Deletion of the p66Shc longevity gene reduces systemic and tissue oxidative stress, vascular cell apoptosis, and early atherogenesis in mice fed a high-fat diet. Proc Natl Acad Sci USA. 2002;100:2112–2116. doi: 10.1073/pnas.0336359100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Favetta LA, Robert C, King WA, Betts DH. Expression profiles of p53 and p66shc during oxidative stress-induced senescence in fetal bovine fibroblasts. Exp Cell Res. 2004;299:36–48. doi: 10.1016/j.yexcr.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 10.Pinton P, et al. Protein kinase C beta and prolyl isomerase 1 regulate mitochondrial effects of the life-span determinant p66Shc. Science. 2007;315:659–663. doi: 10.1126/science.1135380. [DOI] [PubMed] [Google Scholar]
- 11.Giorgio M, et al. Electron transfer between cytochrome c and p66Shc generates reactive oxygen species that trigger mitochondrial apoptosis. Cell. 2005;122:221–233. doi: 10.1016/j.cell.2005.05.011. [DOI] [PubMed] [Google Scholar]
- 12.Jackson JG, Yoneda T, Clark GM, Yee D. Elevated levels of p66 Shc are found in breast cancer cell lines and primary tumors with high metastatic potential. Clin Cancer Res. 2000;6:1135–1139. [PubMed] [Google Scholar]
- 13.Pagnin E, et al. Diabetes induces p66shc gene expression in human peripheral blood mononuclear cells: Relationship to oxidative stress. J Clin Endocrinol Metab. 2005;90:1130–1136. doi: 10.1210/jc.2004-1283. [DOI] [PubMed] [Google Scholar]
- 14.Fantin VR, Leder P. Mitochondriotoxic compounds for cancer therapy. Oncogene. 2006;25:4787–4797. doi: 10.1038/sj.onc.1209599. [DOI] [PubMed] [Google Scholar]
- 15.Migliaccio E, et al. Opposite effects of the p52shc/p46shc and p66shc splicing isoforms on the EGF receptor-MAP kinase-fos signalling pathway. EMBO J. 1997;16:706–716. doi: 10.1093/emboj/16.4.706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bonfini L, Migliaccio E, Pelicci G, Lanfrancone L, Pelicci PG. Not all Shc's roads lead to Ras. Trends Biochem Sci. 1996;21:257–261. [PubMed] [Google Scholar]
- 17.Farooq A, Zeng L, Yan KS, Ravichandran KS, Zhou MM. Coupling of folding and binding in the PTB domain of the signaling protein Shc. Structure. 2003;11:905–913. doi: 10.1016/s0969-2126(03)00134-5. [DOI] [PubMed] [Google Scholar]
- 18.Fagiani E, et al. RaLP, a new member of the Src homology and collagen family, regulates cell migration and tumor growth of metastatic melanomas. Cancer Res. 2007;67:3064–3073. doi: 10.1158/0008-5472.CAN-06-2301. [DOI] [PubMed] [Google Scholar]
- 19.Bernardi P, Petronilli V, Di Lisa F, Forte M. A mitochondrial perspective on cell death. Trends Biochem Sci. 2001;26:112–117. doi: 10.1016/s0968-0004(00)01745-x. [DOI] [PubMed] [Google Scholar]
- 20.Cuff JA, Barton GJ. Evaluation and improvement of multiple sequence methods for protein secondary structure prediction. Proteins. 1999;34:508–519. doi: 10.1002/(sici)1097-0134(19990301)34:4<508::aid-prot10>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 21.Gasteiger E, et al. Protein identification and analysis tools on the ExPASy server. In: Walker JM, editor. The Proteomics Protocols Handbook. Clifton, NJ: Humana; 2005. pp. 571–607. [DOI] [PubMed] [Google Scholar]
- 22.Khanday FA, et al. Rac1 leads to phosphorylation-dependent increase in stability of the p66shc adaptor protein: Role in Rac1-induced oxidative stress. Mol Biol Cell. 2006;17:122–129. doi: 10.1091/mbc.E05-06-0570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trinei M, et al. A p53–p66Shc signaling pathway controls intracellular redox status, levels of oxidation-damaged DNA, and oxidative stress-induced apoptosis. Oncogene. 2002;21:3872–3878. doi: 10.1038/sj.onc.1205513. [DOI] [PubMed] [Google Scholar]
- 24.Barford D. The role of cysteine residues as redox-sensitive regulatory switches. Curr Opin Struct Biol. 2004;14:679–686. doi: 10.1016/j.sbi.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 25.Hansen JM, Go YM, Jones DP. Nuclear and mitochondrial compartmentation of oxidative stress and redox signaling. Annu Rev Pharmacol Toxicol. 2006;46:215–234. doi: 10.1146/annurev.pharmtox.46.120604.141122. [DOI] [PubMed] [Google Scholar]
- 26.Jang HH, et al. Two enzymes in one: Two yeast peroxiredoxins display oxidative stress-dependent switching from a peroxidase to a molecular chaperone function. Cell. 2004;117:625–635. doi: 10.1016/j.cell.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 27.Nemoto S, et al. The mammalian longevity-associated gene product p66shc regulates mitochondrial metabolism. J Biol Chem. 2006;281:10555–10560. doi: 10.1074/jbc.M511626200. [DOI] [PubMed] [Google Scholar]
- 28.Mesecke N, et al. A disulfide relay system in the intermembrane space of mitochondria that mediates protein import. Cell. 2005;121:1059–1069. doi: 10.1016/j.cell.2005.04.011. [DOI] [PubMed] [Google Scholar]
- 29.Koehler CM, Beverly KN, Leverich EP. Redox pathways of the mitochondrion. Antioxid Redox Signal. 2006;8:813–822. doi: 10.1089/ars.2006.8.813. [DOI] [PubMed] [Google Scholar]
- 30.Inarrea P, et al. Mitochondrial respiratory chain and thioredoxin reductase regulate intermembrane Cu,Zn-superoxide dismutase activity: Implications for mitochondrial energy metabolism and apoptosis. Biochem J. 2007;405:173–179. doi: 10.1042/BJ20061809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schaegger H, von Jagow G. Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal Bioanal Chem. 1991;199:223–231. doi: 10.1016/0003-2697(91)90094-a. [DOI] [PubMed] [Google Scholar]
- 32.Schluesener D, Fischer F, Kruip J, Rogner M, Poetsch A. Mapping the membrane proteome of Corynebacterium glutamicum. Proteomics. 2005;5:1317–1330. doi: 10.1002/pmic.200400993. [DOI] [PubMed] [Google Scholar]
- 33.Rossoll W, et al. Specific interaction of Smn, the spinal muscular atrophy determining gene product, with hnRNP-R and gry-rbp/hnRNP-Q: A role for Smn in RNA processing in motor axons? Hum Mol Genet. 2002;11:93–105. doi: 10.1093/hmg/11.1.93. [DOI] [PubMed] [Google Scholar]
- 34.Gatlin CL, Kleemann GR, Hays LG, Link AJ, Yates JR., 3rd Protein identification at the low femtomole level from silver-stained gels using a new fritless electrospray interface for liquid chromatography-microspray and nanospray mass spectrometry. Anal Biochem. 1998;263:93–101. doi: 10.1006/abio.1998.2809. [DOI] [PubMed] [Google Scholar]
- 35.Veladi M, Niesen FH, Allali-Hassani A, Fedorov OY, Finerty PJ. Chemical screening methods to identify ligands that promote protein stability, protein crystallization, and structure determination. Proc Natl Acad Sci USA. 2006;103:15835–15840. doi: 10.1073/pnas.0605224103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.