Abstract
The evolution of new catalytic activities and specificities within an enzyme superfamily requires the exploration of sequence space for adaptation to a new substrate with retention of those elements required to stabilize key intermediates/transition states. Here, we propose that core residues in the large enzyme family, the haloalkanoic acid dehalogenase enzyme superfamily (HADSF) form a “mold” in which the trigonal bipyramidal transition states formed during phosphoryl transfer are stabilized by electrostatic forces. The vanadate complex of the hexose phosphate phosphatase BT4131 from Bacteroides thetaiotaomicron VPI-5482 (HPP) determined at 1.00 Å resolution via X-ray crystallography assumes a trigonal bipyramidal coordination geometry with the nucleophilic Asp-8 and one oxygen ligand at the apical position. Remarkably, the tungstate in the complex determined to 1.03 Å resolution assumes the same coordination geometry. The contribution of the general acid/base residue Asp-10 in the stabilization of the trigonal bipyramidal species via hydrogen-bond formation with the apical oxygen atom is evidenced by the 1.52 Å structure of the D10A mutant bound to vanadate. This structure shows a collapse of the trigonal bipyramidal geometry with displacement of the water molecule formerly occupying the apical position. Furthermore, the 1.07 Å resolution structure of the D10A mutant complexed with tungstate shows the tungstate to be in a typical “phosphate-like” tetrahedral configuration. The analysis of 12 liganded HADSF structures deposited in the protein data bank (PDB) identified stringently conserved elements that stabilize the trigonal bipyramidal transition states by engaging in favorable electrostatic interactions with the axial and equatorial atoms of the transferring phosphoryl group.
Keywords: phosphatase, phosphoryl transfer, transition state stabilization, trigonal bipyramidal phosphorane
Through the use of covalent, general acid, and/or general base catalysis, enzymes are able to provide an alternate, more energetically favorable chemical pathway from reactant to product than is possible in aqueous solvent. Equally important is the ability of the enzyme to employ electrostatic interactions to minimize the energies of the transition states formed along this chemical pathway. The library of enzyme catalysts that exists in nature was formed over time by the duplication of genes encoding enzymes that catalyze life-sustaining reactions, followed by the divergent evolution of the gene copies via random mutation and selection. Each newly evolved enzyme inherits the catalytic site of its predecessor with strategic alterations for binding a novel reactant and processing it along the same chemical pathway or along a unique pathway. Understanding this process, in chemical terms, is an important goal shared by many mechanistic enzymologists.
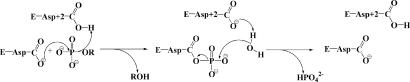
Here, we examine the conservation of structural elements for transition-state stabilization that has taken place during the evolution of a large enzyme family, the haloalkanoic acid dehalogenase enzyme superfamily (HADSF). This family is dominated by phosphotransferases; the majority of which are hydrolases (phosphatases and ATPases) and the minority of which are hexose phosphate mutases (1) (Fig. 1). The HADSF phosphotransferases have in common a core domain that binds the Mg2+ cofactor and the organophosphate reactant in an orientation that allows attack by the Asp nucleophile and general acid/base catalysis by the Asp positioned two residues upstream (the Asp+2 residue) (Fig. 1) (1, 2). The aspartylphosphate intermediate then undergoes phosphoryl-group transfer to a water molecule (in the phosphatases) or to a sugar hydroxyl group (in the mutases).
Fig. 1.
The general catalytic mechanism for phosphohydrolase members of the HAD superfamily. Catalysis proceeds through an aspartylphosphate intermediate.
The extensive biochemical range of the HADSF indicates that the catalytic scaffold is easily adapted for catalysis of phosphoryl transfer in novel substrates (1, 3). Earlier work showed that substrate-recognition elements and chemical-catalysis elements are separately located (4–9). Specifically, substrate recognition is delegated to residues located outside of the core domain active site cleft, whereas phosphoryl transfer is mediated by residues located deep inside this cleft. Whereas the substrate recognition residues are divergent among family members, the catalytic residues are conserved; thus, the catalytic residues are referred to as the “core residues” because they comprise the catalytic unit common to all HADSF phosphotransferases. Here, we propose that these core residues form a “mold” in which the trigonal bipyramidal transition-states leading from the enzyme-substrate complex to the aspartylphosphate intermediate and, hence, to the enzyme-product complex are stabilized by electrostatic forces.
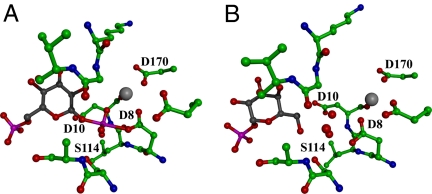
The idea of a mold for casting the trigonal bipyramidal transition state was prompted by snap shots of the β-phosphoglucomutase (βPGM) active site derived from crystal and solution structure determinations. First, the structure of the βPGM(Mg2+)(glucose-6-phosphate-1-aspartylphosphorane) complex comprises the trigonal bipyramidal phosphorane intermediate formed by attack of the Asp nucleophile at the C(1)phosphoryl group of the β-glucose-1,6-(bis)phosphate reactant [Fig. 2A and supporting information (SI) Fig. S1] (2, 10–13, §). The comparison of this structure to that reported for the βPGM(Mg2+)aspartylphosphate intermediate (2), revealed the active role that the enzyme core residues play in stabilization of the trigonal bipyramidal phosphorane intermediate. Specifically, the electrostatic interactions with electropositive groups occur at shorter distances in the βPGM(Mg2+)(glucose-6-phosphate-1-aspartylphosphorane) complex and the number of participating electropositive residues are increased (10). The propensity for the catalytic site to assume the conformation that complements the trigonal bipyramidal phosphorane became evident when the structure of the βPGM (α-galactose-1-phosphate) complex was solved (Fig. 2B and Fig. S1) (11). α-Galactose-1-phosphate is a tight-binding competitive inhibitor of βPGM rather than a substrate (6, 11) (SI Text). The structure of the crystalline βPGM(Mg2+)(α-galactose-1-phosphate) complex revealed that the α-galactose-1-phosphate is oriented in the active site with the C(1)phosphate in the region of the active site that binds the “nontransferring” phosphoryl group and the α-galactose-1-phosphate C(6)OH is in the region of the active site that binds the “transferring” phosphoryl group. Three highly ordered water molecules are observed to fill the space between the ligand C(6)OH and the Asp nucleophile. The orientation of the water molecules is striking because it coincides with the three equatorial oxygen atoms of the phosphorane observed in the βPGM(Mg2+)(glucose-6-phosphate-1-aspartylphosphorane) complex (Fig. 2B and Fig. S1). The α-galactose-1-phosphate C(6)OH and the Asp nucleophile assume the two apical positions of this mock trigonal bipyramid complex. Notably, the water molecules are held in position by the same hydrogen-bond donors used to bind the three equatorial oxygen atoms of the phosphorane intermediate.
Fig. 2.
The active site of βPGM in active and inhibited complexes. (A) The βPGM(Mg2+)(glucose-6-phosphate-1-aspartylphosphorane intermediate (black backbone). (B) The inhibitor α-galactose-1-phosphate (black backbone) binds with three water molecules (red spheres) in the transferring phosphoryl binding site. The Mg2+ cofactor is depicted as a gray sphere.
The work described here was carried out to test the hypothesis that the trigonal bipyramidal phosphorane mold observed in βPGM is conserved in the phosphatase branch of the HADSF. Previously, the x-ray structure of phosphoserine phosphatase crystallized from an equilibrium mixture of AlF3 and AlF4− identified an Al3+ coordination complex of trigonal bipyramidal geometry (9). The apical positions are occupied by the Asp nucleophile and a water ligand, whereas F− ligands fill the three equatorial positions. This structure is representative of the structure of the transition state for the aspartylphosphate hydrolysis step catalyzed by phosphoserine phosphatase. The structure of the sarcoplasmic reticulum H+, Ca2+/ATPase bound to AlF3, and ADP provided evidence that the transition state formed in the first partial reaction (leading to the aspartylphosphate intermediate) has trigonal bipyramidal geometry (17). In this structure, the Asp nucleophile and the β-phosphoryl group of the ADP ligand fill the apical positions and the F− ions fill the three equatorial positions. These two structures are consistent with the presence of a mold for the trigonal bipyramidal phosphorane in these two HADSF phosphatases, but they fall short of providing proof of concept. The reason for this is that the observed complex is an energetically favorable one for AlF3, and, therefore, the enzyme need not play an active role in its formation and stabilization other than by supplying the oxygen ligands at the two apical positions. A true test for the operation of the trigonal bipyramidal phosphorane mold is the ability of the phosphatase to form and stabilize a high energy trigonal bipyramidal complex.
The HADSF phosphatase selected for the test case is hexose phosphate phosphatase (HPP, BT4131 from Bacteroides thetaiotaomicron VPI-5482). Previous work carried out in our laboratories showed that this enzyme consists of a core domain and a type C2B cap domain with the active site at the domain-domain interface (4). The catalysis of hexose phosphate hydrolysis occurs with modest specificity and catalytic efficiency (kcat/Km ≈ 1 × 104 M−1·s−1). Here, we report the high-resolution structures of wild-type and mutant HPP bound with Mg2+ and vanadate or tungstate. The key finding is that tungstate is observed to assume a rare pentavalent trigonal coordination geometry in which the Asp-8 and an oxygen atom occupy the apical positions.
Results and Discussion
Comparison of Valency Expansion in Phosphate, Tungstate, and Vanadate.
The present investigation was carried out to test the hypothesis that the HADSF catalytic site presents a sterically confined, electrostatically defined mold for stabilization of a trigonal bipyramidal phosphorane-like transition state. A structure-activity-based approach (18) was used, in which the reactant phosphorus atom, which is resistant to expansion of its valency in the IV oxidation state beyond four unless further stabilized (19), is replaced with an element that is more amenable to valency expansion. For this purpose, we selected the elements vanadium and tungsten. Vanadium in the [V] oxidation state, namely VO43− (vanadate), is commonly used as a structural probe of phosphotransferase active sites (20). Vanadate readily accepts ligands to form a pentacovalent coordination complex with trigonal bipyramidal geometry (21). The x-ray structures of phosphotransferases complexed with vanadate provide a visual guide to the interactions that occur between active-site residues and the transition state(s) formed along the reaction coordinate that structures of phosphotransferases complexed with phosphate or phosphate ester substrates have, with only a few exceptions (10, 22–24), failed to do. To demonstrate the existence of a conserved trigonal bipyramidal mold in the HADSF phosphatase subfamily, a phosphate analog is needed that is more resistant to valency expansion than is vandate yet more pliable than orthophosphate (19, 25). Tungstate proved to be the ideal probe.
Inspection of the small-molecule and protein structure databases provided an empirical framework that could be used to evaluate the tendency of tungstate to form a pentavalent, trigonal bipyramidal complex. The analysis of the entries of the Cambridge small-molecule database (www.ccdc.cam.ac.uk/products/csd) provided evidence that the pentacoordinate trigonal bipyrimidal geometry is not a common coordination state for tungsten bearing oxygen ligands (SI Text). The analysis of the entries within the Protein Data Bank (PDB) identified 19 enzyme structures that include a four-coordinate, tetrahedral tungsten molecule bound in the active site [for example, see PDB ID codes 1UJC (26) and 2AKC (27)]. Above pH 4 in aqueous solution, this tetrahedral [WO4]2− coordination state predominates (25). There are only four reported enzyme structures in which the bound tungsten is at a higher valency state. The structures of phospholipase D and tyrosyl-DNA phosphodiesterase [PDB ID codes 1V0R (22) and 1MU7 (28)] reveal six ligands to tungsten in an octahedral geometry (the preferred state in solution at pH <4). One ligand is derived from an enzyme active-site residue, and the others are derived from either water or cryoprotectant molecules. Tungsten in a pentavalent trigonal bipyramidal geometry is observed in the structures of two enzymes from the same fold-family (HIT): the Fragile histidine triad protein [PDB ID code 6FIT (29)] and protein kinase C-interacting protein [PDB ID code 1KPE (29)], which both use a His nucleophile to displace the β-phosphoryl group from the α-phosphoryl of a nucleoside diphosphate containing substrate (SI Text) Thus, although the pentavalent trigonal bipyramidal geometry is common for phosphotransferase bound vandate, it is rare for phosphotransferase bound tungstate and has not been observed for phosphotransferase bound orthophosphate.
Structure Analysis of Wild-type HPP Complexes.
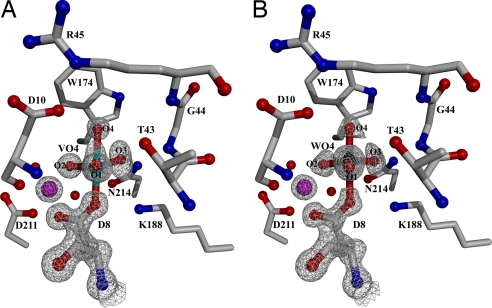
The x-ray structures of the HPP complexes bound with vanadate (Fig. 3A) and tungstate (Fig. 3B) were solved at 1.00 and 1.03 Å resolution, respectively. Notably, the vanadium atom is covalently bonded to the nucleophilic Asp-8 carboxylate oxygen atom and to four oxygen atoms (a diagram showing the covalent and hydrogen bond lengths for all pertinent atoms is shown in Fig. S2). This structure mirrors the apical attack of the water nucleophile at the phosphorus atom of the aspartylphosphate intermediate. Collectively, the three equatorial oxygen atoms form hydrogen bonds with the backbone amide NH of Asp-10, the side chain of Thr-43, the backbone amide NH of Ile-9, the side-chain amide of Asn-214, the backbone amide NH of Gly-44, and the amino group of Lys 188. In addition, one of the equatorial oxygen atoms coordinates to the Mg2+ cofactor. This network of interactions is important to the stabilization of the trigonal–bipyramidal transition states formed during catalytic turnover of the phosphate-ester substrate. Notably, the apical oxygen atom, in-line with the apical Asp-8 oxygen atom, engages in hydrogen bond formation (2.7 Å) with the carboxylic acid group of Asp-10 (Fig. S2). The key observation is that the structure of the crystalline wild-type HPP(Mg2+) (tungstate) complex (1.03 Å resolution) reveals the same pentavalent trigonal bipyramidal geometry and stabilizing hydrogen-bond interactions (Fig. 3B and Fig. S2) as are observed for the vanadate complex. It should be noted that the large number of scattering electrons in tungsten makes the identification of tungstate unique and that this density cannot be assigned to any other small molecule (or combination of small molecules) present in the crystallization milieu. Therefore, this structure is unequivocal evidence that the HPP catalytic site catalyzes the addition of Asp-8 to tetrahedral tungstate to form a stabilized pentacovalent trigonal bipyamidal complex analogous to that formed by the attack of Asp-10 activated water at the phosphorus of the HPP aspartylphosphate intermediate.
Fig. 3.
The wild-type HPP active site in the presence of phosphate mimics and the cofactor Mg2+ (magenta sphere). The 2Fo-Fc composite omit electron density maps (gray cages) are contoured at 1.5σ. (A) The 1.00 Å resolution structure of HPP complexed with vanadate. Bond angles are O1-V-O2, 120.4°; O2-V-O3, 110.4°; O1-V-O3, 124.7°; O1-V-O4, 79.0°; O2-V-O4, 93.1°; O3-V-O4, and 77.8°. (B) The 1.02 Å resolution structure of HPP complexed with tungstate. Bond angles are O1-W-O2, 118.0°; O2-W-O3, 119.2°; O1-W-O3, 119.9°; O1-W-O4, 93.8°; O2-W-O4, 101.2°; O3-W-O4, and 92.2°. Waters are depicted as red spheres.
The competitive inhibition constants of phosphate, vanadate, and tungstate reflect the amount of binding energy that is expended on transforming these ligands to the pentacovalent trigonal bipyamidal coordination geometry. Whereas phosphate binds weakly (Ki = 5.2 mM), tungstate binds with a moderate affinity (Ki = 65 μM) and the more malleable vanadate binds with high affinity (Ki = 510 nM). The ability of vanadate to bind to phosphotransferases much more tightly than does substrate has been attributed to its ability to adopt the pentavalent trigonal–bipyramidal geometry recognized as the transition state geometry by the enzyme (30).
The Asp+2 Residue Asp-10 Is an Essential Component of the HADSF Trigonal–Bipyramidal Active Site Mold.
The vanadate and tungstate complex structures show that Asp-10 engages in a hydrogen-bond interaction with the oxygen ligand. This is clear evidence of its role as a general base in the catalysis of the second partial reaction (as a hydrogen bond acceptor from the water nucleophile) and as a general acid in the catalysis of the first partial reaction (as a hydrogen bond donor to the leaving group oxygen atom). The trigonal–bipyramidal transition states are thus stabilized in part through interaction with Asp-10. To test whether Asp-10 makes a significant contribution to the HADSF trigonalbipyramidal active site mold, the catalytic efficiency and structure of the Asp10Ala (D10A) mutant were determined.
HPP, like most HADSF phosphatases, displays a broad phosphate-ester substrate range. This allowed us to examine its contribution to the rate of hydrolysis of a substrate [glucose 6-phosphate (G6P)] that contains a high-energy leaving group [glucose C(6)O− alkoxide anion] and its contribution to the rate of hydrolysis of a substrate [p-nitrophenylphosphate (PNPP)] that possesses a low energy leaving group (p-nitrophenoxide anion). The kcat for the hydrolysis of the more demanding substrate is reduced 10,000-fold in the mutant (Table 1). We assume that the first partial reaction is rate limiting and that the observed rate reduction reflects the increase in energy of the trigonal–bipyramidal transition state in which the Asp nucleophile (Asp-8) and the glucose C(6)O− alkoxide anion occupy the apical positions. The kcat for hydrolysis of the PNPP is reduced 200-fold in the mutant. In this case, the rate-limiting transition state might be associated with the second partial reaction, the hydrolysis of the aspartylphosphate intermediate. The contribution of Asp-10 is consistent with the rate contribution of a general base to the rate of hydrolysis of a covalent enzyme ester intermediate (31). Thus, we conclude that the Asp-10 contributes to the trigonal–bipyramidal mold that frames the transition states of both partial reactions, and that this contribution is most pronounced in the stabilization of the transition state of the first partial reaction.
Table 1.
Kinetics constants for wild-type and mutant HPP
| Substrate | Constant | Wild-type | R45A mutant | R45K mutant | D10A mutant |
|---|---|---|---|---|---|
| PNPP | kcat, s−1 | 8.3 (± 0.2) × 10−2 | ND | 2 × 10−2 | 5 × 10−4 |
| Km, mM | 7.7 (± 0.5) × 10−1 | ND | 5 | 1 × 10−1 | |
| G6P | kcat, s−1 | 3.6 ± 0.2 | ND | 5 × 10−3 | 2 × 10−4 |
| Km, mM | 11 ± 2 | ND | 7 | 14 |
Competitive inhibition constants for wild-type HPP are: vandate Ki = 510 nM, tungstate Ki = 65 μM, phosphate Ki = 5.2 mM. Competitive inhibition constants for HPP-D10A are: vandate Ki = 700 μM, tungstate Ki = 26 μM. ND = kcat < 1 × 10−5·s−1 (detection limit).
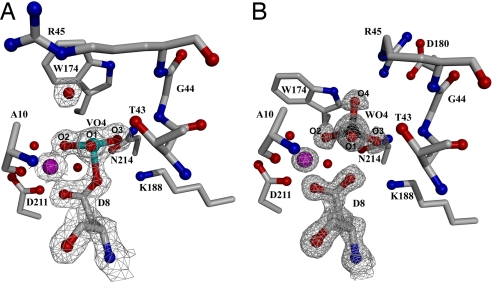
The impact of the D10A mutation on the ability of HPP to stabilize the trigonal–bipyramidal transition state is evident from the differences that exist between the structures of the HPP-D10A(Mg2+) (vanadate) and HPP-D10A(Mg2+) (tungstate) complexes and the corresponding complexes formed by the wild-type enzyme. The 1.52 Å structure of the D10A mutant bound to vanadate (Fig. 4A) reveals a tetravalent complex. A water molecule is located nearby, but it is not in-line with the Asp-8 carboxylate, nor is it within covalent bond distance to the vanadium atom (see SI Text). Overall, the vanadate assumes distorted tetrahedral geometry, in which the bond angles to the nonbridging oxygen atoms are greater than the ideal angle. Analysis of the anisotropic temperature factors for the structures resolved at <1.2 Å suggests the movement of the nonbridging oxygens in a direction parallel to the axial plane (Fig. S3), which may be reflected in the distorted geometry observed in this structure. The HPP-D10A active site stabilizes the tetravalent Asp-8-vanadate complex and binds the water molecule via hydrogen bond interactions with the Arg-45 side chain and a vanadate oxygen atom. The change in the structure of the active-site complex is reflected in the 1,400-fold reduction in vanadate binding affinity to the D10A mutant vs. wild-type HPP (Table 1). The tetravalent Asp-8-vanadate adduct is a mimic of the Asp-8-phosphate intermediate and not the pentavalent trigonal–bipyramidal transition state. Thus, the inability of the active site of the HPP-D10A mutant to stabilize the pentavalent trigonal–bipyramidal vanadate complex demonstrates the crucial role that Asp-10 plays in creating the active-site mold.
Fig. 4.
The HPP-D10A active site in the presence of phosphate mimics and the cofactor Mg2+ (magenta sphere). The 2Fo-Fc composite-omit electron density maps (gray cages) are contoured at 1.5σ. (A) The 1.52 Å resolution structure of HPP-D10A complexed with vanadate. Bond angles are O1-V-O2, 111.6°; O2-V-O3, 122.9°; O1-V-O3, 117.7°; O1-V-OAsp8, 95.2°; O2-V-OAsp8, 100.1°; O3-V-OAsp8, 102.3°. (B) The 1.07 Å resolution structure of HPP-D10A complexed with tungstate. Bond angles are O1-W-O2, 112.8°; O2-W-O3, 118.8°; O1-W-O3, 112.3°; O1-W-O4, 103.8°; O2-W-O4, 104.1°; O3-W-O4, 102.9°{ideal bond angles tungstate [PDB accession code 1FR3 (32)] O1-W-O2, 111.1°; O2-W-O3, 111.1°; O1-W-O3, 108.2°; O1-W-O4, 107.7°; O2-W-O4, 111.3°; O3-W-O4, 107.6°}. The water molecules are depicted as red spheres.
The contribution of Asp-10 to the stabilization of the pentavalent trigonal–bipyramidal tungstate is also evidenced by the structure of the HPP-D10A mutant complexed with tungstate (Fig. 4b). In this complex, the tungstate assumes a sterically and energetically favored tetrahedral configuration [see Fig. S2 and Fig. 4 for comparison of bond lengths and angles to an “ideal” tetrahedral tungstate (PDB ID code 1FR3 (32)]. This “ground-state” complex resembles those observed for HADSF phosphotransferases bound to phosphate or phosphate analogs. Specifically, three of the oxygen atoms interact with the Mg2+ cofactor and the enzyme residues Ala-10, Ile-9, Gly-44, Thr-43, Lys-188, and Asn-214. The oxygen atom positioned “in line” with the Asp-8 nucleophile forms a hydrogen bond with Arg-45. The absence of the side chain of Asp-10 allows the side chain of Arg-45 to form a salt bridge with the cap domain residue Glu-180. There is no water molecule observed in the space vacated by substitution of the Asp-10 side chain with the methyl group of Ala in the D10A mutant. The absence of a water molecule replacing the carboxylate moiety in the mutant protein is significant, indicating that the mutated Asp is part of a preorganized environment supported in its exact position by the protein folding energy (33). The inhibitory activity of tungstate toward HPP-D10A is comparable with that observed with wild-type HPP (Table 1). This result suggests that the favorable energy gained from the coordination of the Asp-10 carboxylate oxygen to the tungstate (and the increased interaction of the trigonal–bipyramidal adduct with the active-site residues) is approximately equal to the energy required to distort the geometry of tungstate. Unfortunately, the ΔG of stabilization of the trigonal bipyramidal form of tungstate cannot be estimated at this time because the corresponding equilibrium in aqueous solution for monomeric species is not known.
Conservation of the Trigonal–Bipyramidal Mold Among HADSF Phosphotransferases.
The essential nature of the role of Asp-10 in transition-state stabilization is supported by the finding that in all HADSF subfamilies, the equivalent residue (the second D of the DXD motif conserved among HAD phosphatases and phosphomutases) is itself positioned by a salt-bridge or hydrogen bond formed with a residue that is conserved in role, but not in identity (4, 34). In HPP this residue, Arg-45 forms a salt bridge with Asp-10. Mutation of Arg-45 to Ala produces a diminution in activity (kcat < 1 × 10−5·s−1) similar in magnitude to that resulting from the mutation of Asp-10 (Table 1). Analysis of HADSF phosphohydrolases from all members for that liganded structures are available (the current structures and PDB ID codes 1L7N, 1TA0, 2C4N, 1U7O, 2HLL, 1Q91, and 1WZC), and encompassing all three subfamilies reveals that the Asp+2 general base is held in position by a hydrogen bond formed with the side chain of a residue (Thr, Tyr, Arg, Trp, or Lys) that is located at the same active site position as the Arg-45 in HPP.
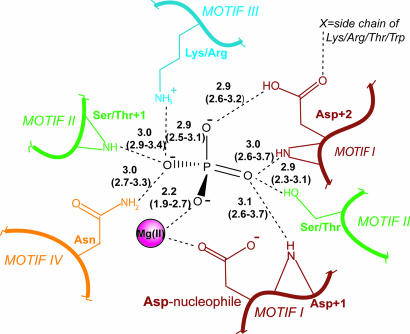
A composite trigonal–bipyramidal mold for all HADSF phosphotransferases was also derived from this analysis (Fig. 5). The mold is formed in part by amino acid side chains and in part by backbone amide groups. The use of backbone amide groups in the stabilization of the trigonal–bipyramidal transition state evidences the direct connection between fold and catalytic function.
Fig. 5.
Composite scheme of the residues involved in forming the pentavalent trigonal–bipyramidal mold in the HADSF. Hydrogen bonds are depicted as dashed lines labeled with average bond lengths in Å (range of bond lengths in parenthesis). The notation +1 denotes the next amino acid in sequence. Note that, in addition to the strictly conserved elements, the side-chain of an Asn from Motif IV participates in a hydrogen bond with the phosphoryl oxygen in ≈30% of the structures.
Materials and Methods
Procedures used for protein preparation and kinetic analysis were performed as described in ref. 4 (see SI Text).
Crystallization and Diffraction Data Collection.
The purified wild-type and D10A mutant HPP proteins (≈17–34 mg/ml) in 1 mM Hepes (pH 7.0) were crystallized using the hanging drop method. Equal volumes (2 μl) of protein solution were mixed with a well solution consisting of 30% wt/vol polyethelene glycol (PEG) 1,500, 10 mM MgCl2, and 5 mM either sodium tungstate or sodium orthovanadate at room temperature (≈25°C). The purified HPP wild-type protein (17 mg/ml) was also crystallized in 10 mM MgCl2, 0.2 M NH4OAc, 0.1 M Bis-Tris (pH 5.5), and 25% PEG 3350 at room temperature (≈25°C). Large orthorhombic crystals appeared within several days. The crystals obtained from the PEG 3350 solution were soaked with mother liquor plus 2 mM sodium orthovanadate for 2 h before data collection. Orthovanadate solutions were prepared fresh by boiling sodium orthovanadate in water for 30 min.
Crystals were frozen for data collection by passing them through 100% Paratone-N (Hampton Research) and then placing them directly in a stream of nitrogen gas cooled by liquid nitrogen. Diffraction data were collected at −180°C, using CuKα radiation from a Rigaku RU-300 generator equipped with an R-Axis IV2+ image plate located at Boston University School of Medicine (D10A mutant/VO4) or the X-12C beamline equipped with ADSC Q210 CCD detector (wild type/WO4 and D10A mutant/WO4) or the X-12B beamline equipped with ADSC Q4 CCD detector (wild type/VO4) at Brookhaven National Laboratory. Data were indexed and integrated by using DENZO and scaled by using SCALEPACK (35). Data collection statistics are summarized in Table 2. Both crystal forms are orthorhombic and belong to space group P212121.
Table 2.
X-ray data collection and structure refinement statistics
| Parameter | Wild-type |
HPP-D10A |
||
|---|---|---|---|---|
| HPP/WO42− | HPP/ VO43− | /WO42− | /VO43− | |
| Data Collection | ||||
| Space group | P212121 | P212121 | P212121 | P212121 |
| Unit cell dimensions a/b/c, Å | 49.387/56.467/94.447 | 49.533/ 56.356/94.026 | 49.526/56.339/93.770 | 49.363/56.476/94.421 |
| Wavelength, Å | 0.9 | 0.9 | 0.9 | 1.54 |
| Resolution, Å | 50–1.03 (1.07–1.03) | 50–1.0 (1.04–1.00) | 50–1.07(1.11–1.07) | 50–1.52 (1.57–1.52) |
| I/σ | 32.0(3.6)a | 15.0(2.0) | 21.2(2.8) | 32.0(3.8) |
| Total/unique reflections observed, no. | 1,657,856/125,015 | 1,270,695/136,055 | 915,102/114,002 | 238,416/40,407 |
| Completeness, % | 95.5 (75.3) | 95.5(71.4) | 98.0(91.6) | 97.1 (78.8) |
| Rmerge, % | 7.5 (47.9) | 12.2 (42.3) | 8.1 (50.1) | 3.6 (23.6) |
| Structure refinement | ||||
| Resolution, Å | 10–1.03 | 10–1.00 | 10–1.07 | 50–1.52 |
| Rwork/Rfree, % | 11.1/13.6 | 12.3/14.8 | 11.7/14.9 | 16.7/19.0 |
Numbers in parentheses are related to the highest-resolution shell. Rmerge = Σ(I − 〈I〉)2/ΣI2.
Phase Determination, Refinement, and the Final Model.
The phases were solved via the molecular replacement method, using the published HPP structure (PDB ID code 1YMQ) excluding any ligand as the search model. The program MOLREP (36) in the CCP4 program suite was used to solve the rotation and translation functions and the models were further refined by using CNS (37) and SHELX (except for the D10A mutant/VO4 structure) (38).
Successive rounds of manual rebuilding were performed by using the molecular graphics program Coot (39) alternating with rounds of minimization and simulated annealing in CNS (37). High-resolution (<1.5 Å) structures were further refined by using SHELX (38). Refinement statistics are summarized in Table 2 and Table S1.
Supplementary Material
Acknowledgments.
We thank Drs. Jan Florian and Deborah Crans for critical reading of the manuscript and valuable discussions. This work was supported by National Institutes of Health Grant GM61099 to (to K.N.A. and D.D.-M.). Data for this study were measured at Beamline X12B and X12C of the National Synchrotron Light Source, Brookhaven National Laboratory. Financial support comes principally from the Offices of Biological and Environmental Research and of Basic Energy Sciences of the US Department of Energy and the National Center for Research Resources of the National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The coordinates have been deposited in the Protein Data Bank, www.pdb.org [PDB ID codes 2RB5, 2RBK, 2RAV, and 2RAR corresponding, respectively, to the tungstate complex and vanadate complex of wild-type hexose phosphate phosphatase (HPP) and the tungstate complex and vanadate complex of HPP D10A mutant].
This article contains supporting information online at www.pnas.org/cgi/content/full/0710800105/DCSupplemental.
An alternative interpretation of the x-ray coordinates was given in a recent report of the solution NMR structure determination of the complex formed from an equilibrium mixture of βPGM, MgCl2, NH4F and glucose-6-phosphate at neutral pH (14). The authors claim that their NMR data evidence a βPGM(Mg2+)(glucose-6-phosphate) (MgF3−) complex in which the Mg(F)3− species assumes the position of the transferring phosphoryl group exchanged between the aspartylphosphate and the glucose-6-phosphate C(1)OH during normal catalytic turnover and that because the crystals of βPGM were grown in the presence of MgCl2 and NH4F that the x-ray structure depicts the βPGM(Mg2+)(glucose-6-phosphate) (MgF3−) complex and not the βPGM(Mg2+)(glucose-6-phosphate-1-aspartylphosphorane) complex. Earlier publications have supported the assignment of the x-ray structure as the βPGM(Mg2+)(glucose-6-phosphate-1-aspartylphosphorane) complex (3, 4, 10–13, 15, 16).
References
- 1.Allen KN, Dunaway-Mariano D. Phosphoryl group transfer: Evolution of a catalytic scaffold. Trends Biochem Sci. 2004;29:495–503. doi: 10.1016/j.tibs.2004.07.008. [DOI] [PubMed] [Google Scholar]
- 2.Lahiri SD, Zhang G, Dunaway-Mariano D, Allen KN. Caught in the act: The structure of phosphorylated beta-phosphoglucomutase from Lactococcus lactis. Biochemistry. 2002;41:8351–8359. doi: 10.1021/bi0202373. [DOI] [PubMed] [Google Scholar]
- 3.Burroughs AM, Allen KN, Dunaway-Mariano D, Aravind L. Evolutionary genomics of the HAD superfamily: Understanding the structural adaptations and catalytic diversity in a superfamily of phosphoesterases and allied enzymes. J Mol Biol. 2006;361:1003–1034. doi: 10.1016/j.jmb.2006.06.049. [DOI] [PubMed] [Google Scholar]
- 4.Lu Z, Dunaway-Mariano D, Allen KN. HAD superfamily phosphotransferase substrate diversification: Structure and function analysis of HAD subclass IIB sugar phosphatase BT4131. Biochemistry. 2005;44:8684–8696. doi: 10.1021/bi050009j. [DOI] [PubMed] [Google Scholar]
- 5.Lahiri SD, Zhang G, Dunaway-Mariano D, Allen KN. Diversification of function in the haloacid dehalogenase enzyme superfamily: The role of the cap domain in hydrolytic phosphoruscarbon bond cleavage. Bioorg Chem. 2006;34:394–409. doi: 10.1016/j.bioorg.2006.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lahiri SD, Zhang G, Dai J, Dunaway-Mariano D, Allen KN. Analysis of the substrate specificity loop of the HAD superfamily cap domain. Biochemistry. 2004;43:2812–2820. doi: 10.1021/bi0356810. [DOI] [PubMed] [Google Scholar]
- 7.Morais MC, et al. The crystal structure of bacillus cereus phosphonoacetaldehyde hydrolase: Insight into catalysis of phosphorus bond cleavage and catalytic diversification within the HAD enzyme superfamily. Biochemistry. 2000;39:10385–10396. doi: 10.1021/bi001171j. [DOI] [PubMed] [Google Scholar]
- 8.Zhang G, et al. Investigation of metal ion binding in phosphonoacetaldehyde hydrolase identifies sequence markers for metal-activated enzymes of the HAD enzyme superfamily. Biochemistry. 2004;43:4990–4997. doi: 10.1021/bi036309n. [DOI] [PubMed] [Google Scholar]
- 9.Wang W, et al. Structural characterization of the reaction pathway in phosphoserine phosphatase: Crystallographic “snapshots” of intermediate states. J Mol Biol. 2002;319:421–431. doi: 10.1016/S0022-2836(02)00324-8. [DOI] [PubMed] [Google Scholar]
- 10.Lahiri SD, Zhang G, Dunaway-Mariano D, Allen KN. The pentacovalent phosphorus intermediate of a phosphoryl transfer reaction. Science. 2003;299:2067–2071. doi: 10.1126/science.1082710. [DOI] [PubMed] [Google Scholar]
- 11.Tremblay LW, Zhang G, Dai J, Dunaway-Mariano D, Allen KN. Chemical confirmation of a pentavalent phosphorane in complex with beta-phosphoglucomutase. J Am Chem Soc. 2005;127:5298–5299. doi: 10.1021/ja0509073. [DOI] [PubMed] [Google Scholar]
- 12.Zhang G, et al. Catalytic cycling in beta-phosphoglucomutase: A kinetic and structural analysis. Biochemistry. 2005;44:9404–9416. doi: 10.1021/bi050558p. [DOI] [PubMed] [Google Scholar]
- 13.Dai J, Wang L, Allen KN, Radstrom P, Dunaway-Mariano D. Conformational cycling in beta-phosphoglucomutase catalysis: Reorientation of the beta-d-glucose 1,6-(Bis)phosphate intermediate. Biochemistry. 2006;45:7818–7824. doi: 10.1021/bi060136v. [DOI] [PubMed] [Google Scholar]
- 14.Baxter NJ, et al. A Trojan horse transition state analogue generated by MgF3− formation in an enzyme active site. Proc Natl Acad Sci USA. 2006;103:14732–14737. doi: 10.1073/pnas.0604448103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Allen KN, Dunaway-Mariano D. Response to Comment on “The Pentacovalent Phosphorus Intermediate of a Phosphoryl Transfer Reaction.”. Science. 2003;301:1184d. doi: 10.1126/science.1082710. [DOI] [PubMed] [Google Scholar]
- 16.Blackburn GM, Williams NH, Gamblin SJ, Smerdon SJ. Comment on “The Pentacovalent Phosphorus Intermediate of a Phosphoryl Transfer Reaction.”. Science. 2003;301:1184c. doi: 10.1126/science.1085796. [DOI] [PubMed] [Google Scholar]
- 17.Sorensen TL, Moller JV, Nissen P. Phosphoryl transfer and calcium ion occlusion in the calcium pump. Science. 2004;304:1672–1675. doi: 10.1126/science.1099366. [DOI] [PubMed] [Google Scholar]
- 18.Hammett LP. The effect of structure upon the reactions of organic compounds. Benzene derivatives. J Am Chem Soc. 1937;59:96–103. [Google Scholar]
- 19.Holmes RR. Pentacoordinated Phosphorous Vol. I. Doolin Memorial Lecture. Washington, DC: American Chemical Society; 1980. [Google Scholar]
- 20.Davies DR, Hol WG. The power of vanadate in crystallographic investigations of phosphoryl transfer enzymes. FEBS Lett. 2004;577:315–321. doi: 10.1016/j.febslet.2004.10.022. [DOI] [PubMed] [Google Scholar]
- 21.Crans DC, Smee JJ, Gaidamauskas E, Yang L. The chemistry and biochemistry of vanadium and the biological activities exerted by vanadium compounds. Chem Rev. 2004;104:849–902. doi: 10.1021/cr020607t. [DOI] [PubMed] [Google Scholar]
- 22.Leiros I, McSweeney S, Hough E. The reaction mechanism of phospholipase D from Streptomyces sp. strain PMF. Snapshots along the reaction pathway reveal a pentacoordinate reaction intermediate and an unexpected final product. J Mol Biol. 2004;339:805–820. doi: 10.1016/j.jmb.2004.04.003. [DOI] [PubMed] [Google Scholar]
- 23.Wang Y, et al. Seeing the process of histidine phosphorylation in human bisphosphoglycerate mutase. J Biol Chem. 2006;281:39642–39648. doi: 10.1074/jbc.M606421200. [DOI] [PubMed] [Google Scholar]
- 24.Choe JY, et al. Inhibition of fructose-1,6-bisphosphatase by a new class of allosteric effectors. J Biol Chem. 2003;278:51176–51183. doi: 10.1074/jbc.M308396200. [DOI] [PubMed] [Google Scholar]
- 25.Cruywagen JJ. Protonation, oligomerization, and ondensation reactions of vanadate(V), molybdate(VI) and tungstate(VI) Adv Inorg Chem. 2000;49:127–182. [Google Scholar]
- 26.Hamada K, et al. Crystal structure of the protein histidine phosphatase SixA in the multistep His-Asp phosphorelay. Genes Cells. 2005;10:1–11. doi: 10.1111/j.1365-2443.2005.00817.x. [DOI] [PubMed] [Google Scholar]
- 27.Makde RD, Mahajan SK, Kumar V. Structure and mutational analysis of the PhoN protein of Salmonella typhimurium provide insight into mechanistic details. Biochemistry. 2007;46:2079–2090. doi: 10.1021/bi062180g. [DOI] [PubMed] [Google Scholar]
- 28.Davies DR, Interthal H, Champoux JJ, Hol WG. Insights into substrate binding and catalytic mechanism of human tyrosyl-DNA phosphodiesterase (Tdp1) from vanadate and tungstate-inhibited structures. J Mol Biol. 2002;324:917–932. doi: 10.1016/s0022-2836(02)01154-3. [DOI] [PubMed] [Google Scholar]
- 29.Lima CD, Klein MG, Hendrickson WA. Structure-based analysis of catalysis and substrate definition in the HIT protein family. Science. 1997;278:286–290. doi: 10.1126/science.278.5336.286. [DOI] [PubMed] [Google Scholar]
- 30.Linquist RN, Lynn JL, Jr, Lienhard GE. Possible transition-state analogs for ribonuclease. Complexes of uridine with oxovanadium(IV) ion and vanadium(V) ion. J Am Chem Soc. 1973;95:8762–8768. doi: 10.1021/ja00807a043. [DOI] [PubMed] [Google Scholar]
- 31.Zhang W, et al. Histidine 90 function in 4-chlorobenzoyl-coenzyme a dehalogenase catalysis. Biochemistry. 2001;40:13474–13482. doi: 10.1021/bi0114426. [DOI] [PubMed] [Google Scholar]
- 32.Wagner UG, Stupperich E, Kratky C. Structure of the molybdate/tungstate binding protein mop from Sporomusa ovata. Structure. 2000;8:1127–1136. doi: 10.1016/s0969-2126(00)00525-6. [DOI] [PubMed] [Google Scholar]
- 33.Warshel A, Florian J. Computer simulations of enzyme catalysis: Finding out what has been optimized by evolution. Proc Natl Acad Sci. 1998;95:5950–5955. doi: 10.1073/pnas.95.11.5950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tremblay LW, Dunaway-Mariano D, Allen KN. Structure and activity analyses of Escherichia coli K-12 NagD provide insight into the evolution of biochemical function in the haloalkanoic acid dehalogenase superfamily. Biochemistry. 2006;45:1183–1193. doi: 10.1021/bi051842j. [DOI] [PubMed] [Google Scholar]
- 35.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 36.Vagin A, Teplyakov A. A translation-function approach for heavy-atom location in macromolecular crystallography. Acta Crystallogr D. 1998;54:400–402. doi: 10.1107/s0907444997014923. [DOI] [PubMed] [Google Scholar]
- 37.Brünger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 38.Sheldrick GM, Schneider TR. SHELXL: High resolution refinement. Methods Enzymol. 1997;277:319–343. [PubMed] [Google Scholar]
- 39.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.