Abstract
The metabolic conversion of cholesterol into bile acids in liver is initiated by the rate-limiting cholesterol 7α-hydroxylase (CYP7A1), whereas the bile salt export pump (BSEP) is responsible for the canalicular secretion of bile acids. Liver receptor homolog 1 (LRH-1) is a key transcriptional factor required for the hepatic expression of CYP7A1. We hypothesized that LRH-1 was also involved in the transcriptional regulation of BSEP. In support of our hypothesis, we found that overexpression of LRH-1 induced, whereas knockdown of LRH-1 decreased, BSEP expression. Consistent with its role in transcriptional regulation, LRH-1 dose-dependently transactivated the BSEP promoter. In addition, such transactivation by LRH-1 was required for maximal induction of BSEP expression through the bile acid/farnesoid X receptor (FXR) activation pathway. Bioinformatic and mutational analysis led to the identification of a functional liver receptor homolog 1-responsive element (LRHRE) in the BSEP promoter. Specific binding of LRH-1 to the LRHRE and recruitment of LRH-1 to the BSEP promoter were demonstrated by electrophoretic mobility shift assay and chromatin immunoprecipitation assay, respectively. In conclusion, LRH-1 transcriptionally activated the BSEP promoter and functioned as a modulator in bile acid/FXR-mediated BSEP regulation. These results suggest that LRH-1 plays a supporting role to FXR in maintaining hepatic bile acid levels by coordinately regulating CYP7A1 and BSEP for bile acid synthesis and elimination, respectively.
Keywords: bile acids, bile acid synthesis and elimination, cholesterol, cholesterol metabolism, farnesoid X receptor, small heterodimer partner, nuclear receptors
Cholesterol metabolizing into bile acids and subsequent elimination through bile secretion is a major pathway for removing excessive cholesterol from the body. The metabolic conversion of cholesterol to bile acids in the liver is initiated by the rate-limiting enzyme cholesterol 7α-hydroxylase (CYP7A1) (1, 2), whereas the bile salt export pump (BSEP) is responsible for the canalicular secretion of bile acids (3–6). Therefore, CYP7A1 and BSEP are two key players in maintaining cholesterol and bile acid homeostasis. After being synthesized in the liver by a cascade of enzymes including CYP7A1, bile acids are secreted into the bile through BSEP, reabsorbed in the small intestines via apical sodium-dependent bile acid transporter (ASBT) (7, 8) and basolateral organic solute transporters α and β (Ostα and Ostβ) (9), returned to the liver, and resecreted into bile, completing the enterohepatic circulation of bile acids. The canalicular secretion of bile acids through BSEP is the rate-limiting step in the enterohepatic circulation (5, 6). Through each circulation, ∼5% of bile acids are eliminated via fecal excretion.
The expression of CYP7A1 and BSEP is coordinately regulated by multiple transactivation pathways, notably the bile acid/farnesoid X receptor (FXR) signaling pathway (10–16). Bile acids, as FXR agonists, strongly repress CYP7A1 expression through a negative feedback circuit (10–14), whereas bile acids markedly induce BSEP expression by activating FXR (15, 16). Such coordinated feedback and feed-forward regulation of CYP7A1 and BSEP by bile acids represents an excellent mechanism for preventing the excessive accumulation of toxic bile acids in hepatocytes. On the other hand, enhancing bile acid elimination through increasing BSEP expression favors cholesterol conversion into bile acids and is a possible mechanism for the hypolipidemic agent guggulsterone to exert its cholesterol-lowering effect (17, 18).
The orphan nuclear receptor liver receptor homolog 1 (LRH-1) is a transcriptional factor expressed in multiple organs and tissues, including liver and intestine. Studies have demonstrated that LRH-1 regulates a set of target genes encoding proteins important for the biosynthesis and transport of cholesterol and bile acids, including CYP7A1 (19, 20), ASBT (21, 22), and Ostα and Ostβ (23). In the liver, LRH-1 is required for the maximal hepatic expression of CYP7A1 (19, 20). In the small intestine, LRH-1 regulates ASBT (21, 22) and Ostα and Ostβ (23). In addition, LRH-1 transactivates scavenger receptor class B type I (24), which mediates the uptake of circulating cholesterol into liver hepatocytes; ABCG5/G8, which is responsible for the canalicular secretion of cholesterol (25); cholesteryl ester transfer protein, which transfers cholesteryl esters from high density lipoproteins to triglyceride-rich lipoproteins (26); and apolipoprotein A-I, which functions as an acceptor for phospholipid and cholesterol (27). Therefore, LRH-1 is a master transcriptional regulator in maintaining cholesterol and bile acid homeostasis.
In this study, we demonstrate that LRH-1 transcriptionally regulates BSEP expression through a functional liver receptor homolog 1-responsive element (LRHRE) in the BSEP promoter and functions as a modulator in bile acid/FXR-mediated BSEP regulation. Thus, LRH-1 plays a supporting role to FXR in maintaining hepatic bile acid levels by coordinately regulating CYP7A1 and BSEP for bile acid synthesis and elimination, respectively.
MATERIALS AND METHODS
Chemicals and supplies
Chenodeoxycholic acid (CDCA) and DMSO were purchased from Sigma (St. Louis, MO). DMEM, LipofectAMINE, and Plus Reagent were from Invitrogen (Carlsbad, CA). Kits for luciferase detection and the null-Renilla luciferase plasmid were from Promega (Madison, WI). Fetal bovine serum and 100× nonessential amino acids were from HyClone (Logan, UT). Unless specified otherwise, all other reagents were purchased from Fisher Scientific (Suwanee, GA). Oligonucleotides for PCR amplification, site-directed mutagenesis, cloning, and gel-shift assays were chemically synthesized by Invitrogen. Polyclonal antibodies against human LRH-1 (sc-25389X) and Jun-B (sc-73X) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA).
Plasmid constructs
The preparation of the human BSEP promoter reporter pBSEP(−2.6kb) was described elsewhere (28). The mouse promoter reporter pmBSEP(−2.6kb) was constructed by cloning a 2.6-kb fragment upstream of the transcription start site of mouse BSEP into the pGL4.10 vector (Promega) at the NheI and XhoI sites. The rat BSEP promoter reporter prBSEP(−2kb) was constructed by cloning a 2-kb fragment upstream of the transcription start site of rat BSEP into the pGL4.10 vector at the KpnI and SacI sites. All of the reporter constructs were sequence-verified before use in the experiments. The sequences of the PCR primers are listed in Table 1. Mouse LRH-1 and small heterodimer partner (SHP) expression constructs, pcDNA3-LRH-1 and pCD8-mSHP, were kindly provided by Dr. Wang Li (University of Kansas Medical Center, Kansas). Expression plasmid for the human nuclear receptor FXR were kindly provided by Dr. David Mangelsdorf (University of Texas Southwestern Medical Center, Dallas).
TABLE 1.
Sequences of oligonucleotides used in these studies
| Oligonucleotides | Sequences (5′–3′) |
|---|---|
| pmBSEP(−2.6kb)-sense | TGCCATTTCTTGAGGTTGGCTAGCTCTTAACC |
| pmBSEP(−2.6kb)-antisense | GAGAGACGCCACTGTGGAAATCAGGGTTGTACACC |
| prBSEP(−2kb)-sense | TAGTTAGGGTTTCCTTTCCTGCACAACCATCATGACC |
| prBSEP(−2kb)-antisense | GAGAGACCCCACTGTGGAAAGTCAGGATTGTACACC |
| LRHRE1 Mut-sensea | GAATCAGCAATTTCCTCGAGCTGTTGACACCCTC |
| LRHRE1 Mut-antisense | GAGGGTGTCAACAGCTCGAGGAAATTGCTGATTC |
| LRHRE2 Mut-sense | CAATTTGCCTCTCGTTCCTCGAGGAATCAGCAATTTCC |
| LRHRE2 Mut-antisense | GGAAATTGCTGATTCCTCGAGGAACGAGAGGCAAATTG |
| LRHRE3 Mut-sense | GCTTTTCTTTGCTGCTCGAGATATATTTGAGATTTG |
| LRHRE3 Mut-antisense | CAAATCTCAAATATATCTCGAGCAGCAAAGAAAAGC |
| LRHRE4 Mut-sense | GAAGGAAAGTCTTAGGACTCGAGGACCTGTGAGCAGAT |
| LRHRE4 Mut-antisense | ATCTGCTCACAGGTCCTCGAGTCCTAAGACTTTCCTTC |
| LRH-1 siRNA-sense | AAGGATCCATCTTCCTGGTTA |
| LRHRE1 probeb | AATCAGCAATTTCCAAGGCCTGTTGACA |
| LRHRE1 Mut | AATCAGCAATTTCTCGAGCCTGTTGACA |
| CYP7A1 LRHRE | ACTTAGTTCAAGGCCAGTTACTA |
| pBSEP(+35b) | TTCACAACCTTTTCCAACCTCGG |
| pBSEP(−260b) | GGGTTGGGATAGCCTGAATTCCAGGGCTC |
| pBSEP(−805b) | TTTGGGCCTCTCAACAGCCCTAGGAGTTGG |
| pBSEP(−995b) | CACTGGCCCATCAATTGCATTTCAGAGC |
| pBSEP(−1132b) | AACCCAAAATTATAACATATAATTTTATATG |
| pBSEP(−1305b) | GGGTCCTAATTGGTGCCAAATCCAATATTAC |
| pBSEP(−1410b) | ACCTTTGCTGTGCTGAAGTTATTGACAGCA |
| pBSEP(−1629b) | TCTATAAGATGAAGTTGTTGGATGATATATC |
Mutated nucleotides are shown in boldface.
The core sequences of the liver receptor homolog 1-responsive element are underlined.
Site-directed mutagenesis
Mutagenesis was performed using the QuickChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). Three or four nucleotide substitutions in the LRHRE core sequence were introduced into pBSEP(−2.6kb), resulting in LRHRE1 Mut, LRHRE2 Mut, LRHRE3 Mut, and LRHRE4 Mut. The sequences of the mutagenic oligonucleotides are listed in Table 1. The mutagenesis reactions were performed essentially according to the manufacturer's manual. The resulting mutants were subjected to sequence analysis to confirm that the desired substitutions were made without introducing errors.
Reporter luciferase assay
Huh 7 cells were plated on 24-well plates in DMEM supplemented with 10% fetal bovine serum at a density of 8 × 104 cells per well and cultured overnight. Transient transfection was conducted with LipofectAMINE and Plus Reagent (Invitrogen) as described previously (28). For most of the transfections, standard amounts of plasmid DNA used per well were 100 ng of promoter reporter or empty pGL4.10 vector, 50–100 ng of nuclear receptor expression plasmid (FXR, LRH-1, and SHP), and 10 ng of the null-Renilla luciferase plasmid as an internal control. The luciferase activities were assayed at 36 h after transfection with the Dual-Luciferase Reporter Assay System as described previously (28). Briefly, transfected cells were washed once with PBS and lysed by adding 100 μl of passive lysis buffer (Promega) with gentle rocking for 30 min. Cell lysates (10 μl) were transferred to a 96-well reader plate, and luciferase activities were measured with an EG&G Berthold Microplate Luminometer (Perkin-Elmer, Boston, MA). The firefly luminescence was normalized based on the Renilla luminescence signal, and the ratio of treatment over control values served as fold activation. Data are presented as means ± SD of at least three separate experiments. For experiments with FXR agonist treatments, 16 h after transfection, cells were treated with 10 μM bile acid CDCA or 0.1% DMSO as a negative control for 30 h, followed by the detection of luciferase activity.
RNA interference
Small interfering RNAs (siRNAs) against human LRH-1 and enhanced green fluorescent protein (eGFP) were chemically synthesized by Qiagen (Valencia, CA). The sequences for LRH-1 siRNA and eGFP siRNA are given in Table 1. The selected LRH-1 siRNA exhibited potent specific silencing on LRH-1 expression (27). For the experiments to determine the effect of LRH-1 knockdown on endogenous BSEP expression, Huh 7 cells seeded on 12-well plates at 70% confluence were transfected with 5 or 10 nM LRH-1 siRNA using HiPerFect Transfection Reagent (Qiagen) according to the instructions of the manufacturer. Cells were also transfected with siRNA vehicle buffer or 10 nM eGFP siRNA as a negative control. Forty-eight hours after transfection, transfected cells were lysed for RNA isolation. Quantification of BSEP and LRH-1 mRNA was carried out by the TaqMan real-time PCR assay as described below. For experiments to determine the effect of LRH-1 silencing on BSEP promoter activity, Huh 7 cells seeded on 24-well plates were cotransfected with pBSEP(−2.6 kb) reporter (100 ng), 5 or 10 nM LRH-1 siRNA, and 10 ng of the null-Renilla luciferase plasmid with TransMessenger Transfection Reagent (Qiagen) as instructed. Promoter activity was assayed with the dual luciferase detection system at 48 h after transfection.
Quantitative real-time PCR
Total RNA isolation from transfected or chemically treated cells and subsequent real-time PCR assays were performed essentially as described (17). Briefly, cDNA was synthesized with 2 μg of total RNA and random primers in a total volume of 25 μl and subjected to real-time PCR using the TaqMan Gene Expression Assay (Applied Biosystems, Foster City, CA). The TaqMan assay probes for BSEP (assay identifier: Hs00184824_m1), LRH-1 (assay identifier: Hs00187067_m1), FXR (assay identifier: Hs00231968_m1), SHP (assay identifier: Hs00222677_m1), and GAPDH (assay identifier: 4352934E) were purchased from Applied Biosystems. Real-time PCR was performed using the TaqMan Universal PCR Master Mix (Applied Biosystems) in a total volume of 20 μl containing 10 μl of Universal PCR Master Mix, 1 μl of gene-specific TaqMan probe mixture, and 5 μl of cDNA templates. The cycling profile was as follows: 50°C for 2 min, 95°C for 10 min, followed by 40 cycles of 15 s at 95°C and 1 min at 60°C, as recommended by the manufacturer. Amplification and quantification were done with the Applied Biosystems 7500 Real-Time PCR System.
Nuclear extract preparation and electrophoretic mobility shift assays
Nuclear extracts of Huh 7 cells transfected with LRH-1 were prepared using the NE-PER Nuclear and Cytoplasmic Extraction kit (Pierce, Rockford, IL) according to the instructions of the manufacturer. All of the preparation procedures were carried out at 4°C. The total protein concentrations were determined by the BCA protein assay (Pierce). The prepared nuclear extracts were stored at −80°C until use. The electrophoretic mobility shift assays (EMSAs) were performed using the LightShift Chemiluminescent EMSA kit (Pierce). The sense and antisense oligonucleotide probes containing the functional LRHRE (LRHRE1) were biotin-labeled at the 5′ end (Invitrogen) and annealed by heating at 94°C for 5 min followed by gradually cooling to room temperature. Nuclear proteins (10 μg) were incubated with biotin-labeled probe (20 fM) in a final volume of 20 μl in 1× DNA binding buffer containing 2.5% glycerol, 50 ng/μl poly(dI.dC), and 0.05% Nonidet P-40. To establish the specificity of the binding, competition assays were included using various competing oligonucleotides, including unlabeled LRHRE1 (10× and 50×), mutated LRHRE1 (50×), and the LRHRE from human CYP7A1 (50×). The sequences of those oligonucleotides are listed in Table 1. For the competition experiments, nuclear extracts were first incubated with excess competing oligonucleotides (10–50×) and then mixed with the biotin-labeled probe. To determine whether LRH-1 is a component of the protein complex binding to the probe, supershift assays were performed using polyclonal antibodies against human LRH-1 (sc-25389; Santa Cruz Biotechnology). Antibodies against Jun-B were included as a negative control. For the supershift assays, antibodies (2 μg) were incubated with nuclear extracts at 4°C for 30 min before adding the biotin-labeled probe. The protein-DNA complexes were resolved by 6% PAGE, transferred to nylon membranes, and detected by chemiluminescence according to the protocol provided by the manufacturer (Pierce).
Chromatin immunoprecipitation assays
Chromatin immunoprecipitation (ChIP) assay was performed with the ChIP-IT Express Kit (Active Motif, Carlsbad, CA). Chromatins were prepared from the following sources: Huh 7 cells transfected with LRH-1 with or without CDCA treatment and normal human liver tissue. For the preparation of chromatin from Huh 7 cells, cells seeded on six-well plates were transfected with 2 μg of LRH-1 plasmid, followed by treatment of transfected cells with 10 μM CDCA or vehicle DMSO (0.1%) for 24 h. Cells were fixed with 1% formaldehyde-containing DMEM at room temperature for 10 min and then treated with Glycine Stop-Fix solution. The cells were washed with PBS, harvested, and centrifuged at 720 g for 10 min at 4°C. Pellets were resuspended in 1 ml of lysis buffer and incubated on ice for 30 min, then processed with a Dounce homogenizer for 10 strokes and centrifuged at 2,400 g for 10 min at 4°C. Nuclei were suspended in 0.5 ml of Shearing Buffer and sonicated with the Branson Sonifier 150 (Branson, Danbury, CT) by three strokes of 20 s pulse at power level 5 with a 30 s interval on ice to achieve optimal chromatin sizes ranging from 400 to 800 bp. The sheared chromatins were collected as the supernatant after centrifuging at 12,000 g for 12 min. The preparation of chromatins from human liver tissues was carried out as described (29). Briefly, normal human liver tissues from two individual donors were obtained from the Liver Tissues Procurement and Distribution System (University of Minnesota). Upon arrival, the liver tissues were frozen in liquid nitrogen and pulverized into powder, followed by cross-linking with 1% formaldehyde for 15 min at room temperature and subsequent termination of cross-linking with the addition of Glycine Stop-Fix solution. The soluble chromatins were prepared by following the same procedure described above. For ChIP experiments, 50 μl aliquots of sheared chromatins were used for immunoprecipitation with the addition of 25 μl of protein G magnetic bead suspension and 2 μg of LRH-1 antibodies (sc-25389; Santa Cruz Biotechnology) or 2 μg of IgG antibodies as a negative control (Active Motif). The chromatins were eluted from the magnetic beads after washing with ChIP buffers, followed by reversing the cross-link with the addition of Reverse Cross-link Buffer and heating at 94°C for 15 min. The proteins in the solution were digested by proteinase K at 37°C for 1 h, followed by the addition of proteinase K stop solution to terminate the reaction. The final volume of the eluted chromatin DNA was 100 μl. For the preparation of input DNA, 10 μl of sheared chromatins was diluted up to 100 μl to reverse cross-link and proteinase K digestion, followed by 10-fold dilution with water.
Four sets of primers were used for PCR amplification. The sequences of those primers are listed in Table 1. The primer set flanking the LRHRE1 site at position −175 bp consisted of the forward primer pBSEP(−260b) and the reverse primer pBSEP(+35b). The expected PCR product is a 295 bp fragment encompassing sequence from nucleotides +35 to −260 in the BSEP promoter. The negative control primer set consisted of the forward primer pBSEP(−995b) and the reverse primer pBSEP(−805b) and amplified a 190 bp fragment encompassing the sequence from nucleotides −995 to −805 in the BSEP promoter, a region 630 bp upstream the LRHRE1 site. The two primer sets detecting the recruitment of LRH-1 to LRHRE3 and LRHRE4 were pBSEP(−1305b)/pBSEP(−1132b) and pBSEP(−1629b)/pBSEP(−1410b), respectively. PCR amplification was carried out in a final volume of 25 μl with 2 μl of eluted chromatin DNA, input DNA, or water (mock) as template and 1 unit of high-fidelity Taq DNA polymerase (Invitrogen) with the following cycling parameters: 94°C for 3 min, 40 cycles of 94°C for 45 s, 53°C for 45 s, and 68°C for 45 s, followed by an extension at 68°C for 5 min. PCR products were resolved on 2% agarose gels and scanned with the Typhoon 9410 Image Scanner (GE Healthcare, Piscataway, NJ).
Statistical analysis
Student's t-test was applied to pair-wise comparisons to determine statistical significance. Values of 0.05 or lower were considered significant.
RESULTS
Overexpression of LRH-1 induced BSEP expression
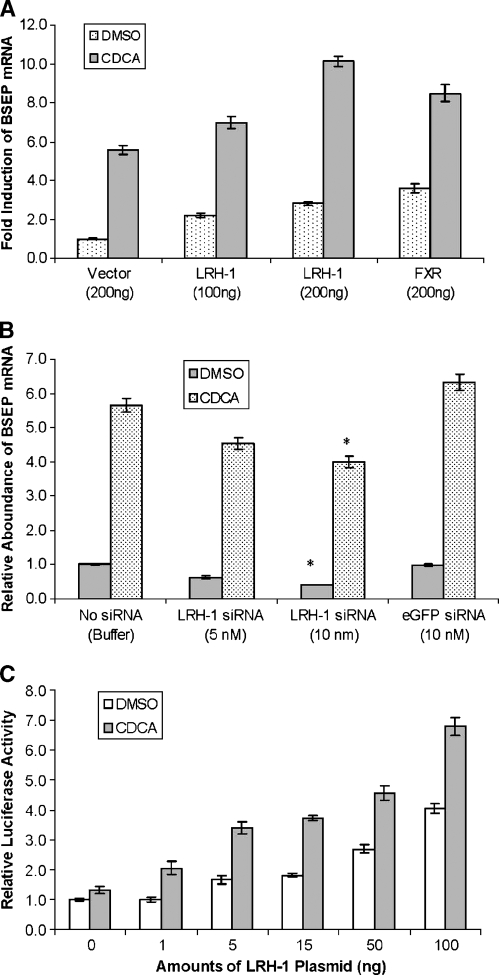
To determine whether LRH-1 is involved in the regulation of BSEP expression, human hepatoma Huh 7 cells were transfected with LRH-1 expression plasmid, followed by detection of BSEP mRNA by real-time PCR. As shown in Fig. 1A, cells transfected with 100 or 200 ng of LRH-1 plasmid exhibited an increase in BSEP mRNA level, ∼2.5- to 3-fold induction compared with the cells transfected with empty expression vector. Such increased levels were comparable to those in cells transfected with 200 ng of FXR plasmid DNA. To eliminate the possibility that LRH-1 may modulate FXR expression and in turn upregulate BSEP expression, FXR mRNA levels were measured by real-time PCR after LRH-1 transfection. As shown in Fig. 1B, LRH-1 had a minimal effect on FXR expression, suggesting that LRH-1-mediated induction of BSEP expression was independent of FXR. On the other hand, overexpression of FXR also had no effect on endogenous LRH-1 expression (Fig. 1B), suggesting that LRH-1 is not an FXR target gene. Together, these results demonstrate that LRH-1 induced BSEP expression in an FXR-independent manner.
Fig. 1.
Liver receptor homolog 1 (LRH-1) upregulated bile salt export pump (BSEP) expression. A: Huh 7 cells seeded on 12-well plates were transfected with LRH-1 expression plasmid, empty vector as a negative control, or farnesoid X receptor (FXR) expression plasmid as a positive control. Forty-eight hours after transfection, total RNA was isolated and subjected to cDNA synthesis and real-time PCR using the TaqMan Gene Expression Assay with a BSEP-specific probe. The mRNA levels for GAPDH were detected as an internal control. The PCR cycle times (Ct) value in cells transfected with vector control is given at the top of the column. B: After the same treatment of Huh 7 cells as in A, the mRNA levels of LRH-1 and FXR were measured by real-time PCR using LRH-1- and FXR-specific probes. The Ct values for FXR and LRH-1 in cells transfected with vector control are given at the top of the columns. C: Huh 7 cells seeded on 12-well plates were transfected with 5 or 10 nM LRH-1 small interfering RNA (siRNA). Cells were also transfected with vehicle buffer or 10 nM enhanced green fluorescent protein (eGFP) siRNA as a negative control. Forty-eight hours after transfection, cells were lysed for RNA isolation and quantification by real-time PCR. LRH-1- and BSEP-specific probes were used to detect LRH-1 and BSEP mRNA expression levels, respectively. Student's t-test was applied to determine the significance of the differences between siRNA- and vehicle-treated groups. The asterisks indicate significant differences (P < 0.05). All data are presented as means ± SD of at least three separate experiments.
LRH-1 was required for maximal BSEP expression
To confirm the finding, we next performed experiments to determine the effect of LRH-1 knockdown by siRNA on BSEP expression. We have shown previously that BSEP is expressed at a relatively high level in human hepatoma Huh 7 cells (28). Therefore, the effect of LRH-1-specific siRNA on BSEP expression was evaluated in Huh 7 cells. A validated potent and specific human LRH-1 siRNA was used to silence LRH-1 expression (27), and its effect on BSEP expression was assessed by quantitative real-time PCR. As expected, transfection of Huh 7 cells with 5 or 10 nM LRH-1 siRNA for 48 h resulted in a reduction of LRH-1 expression (Fig. 1C). More importantly, and in parallel with LRH-1 reduction, BSEP mRNA levels were also decreased significantly in cells transfected with LRH-1 siRNA (Fig. 1C). Reductions of >50% in BSEP mRNA levels were detected in cells transfected with 10 nM LRH-1 siRNA. As a control, transfection of eGFP-specific siRNA had minimal effects on both LRH-1 and BSEP mRNA levels (Fig. 1C), indicating the specificity of the effect. These data indicated that LRH-1 was required for maximal expression of BSEP, consistent with the previous results from LRH-1 overexpression experiments.
LRH-1 transactivated the BSEP promoter
To determine whether the upregulation of BSEP expression by LRH-1 is mediated at the transcriptional level rather than through other posttranscriptional mechanisms, such as stabilizing BSEP mRNA, the effect of LRH-1 on human BSEP promoter activation was evaluated. Huh 7 cells were cotransfected with the BSEP promoter reporter pBSEP(−2.6kb) (28) and increasing amounts of LRH-1 expression plasmid or empty vector (0, 5, 25, 100, and 200 ng). As shown in Fig. 2A, LRH-1 dose-dependently activated the BSEP promoter, whereas such activation was not detected in cells cotransfected with increasing amounts of control vector. Increased activation was detected in cells transfected with 25 ng of LRH-1, with a further increase in cells transfected with 100 and 200 ng of LRH-1 (4- to 5-fold increase) (Fig. 2A). These data indicated that LRH-1 regulated BSEP expression at the transcriptional level, consistent with its function as a transcriptional factor in regulating other LRH-1 target genes (19–26).
Fig. 2.
LRH-1 transactivated the human BSEP promoter. A: Huh 7 cells seeded on 24-well plates were transfected with 100 ng of the human BSEP promoter reporter pBSEP(−2.6kb) (28), 10 ng of the null-Renilla luciferase plasmid as an internal control, and increasing amounts of LRH-1 plasmid DNA or empty vector as a negative control (0, 5, 25, 100, and 200 ng). The luciferase activities were assayed at 36 h after transfection with the Dual-Luciferase Reporter Assay System. The firefly luminescence was normalized based on the Renilla luminescence signal. B: Huh 7 cells were transfected with pBSEP(−2.6kb) (100 ng) and 5 or 10 nM LRH-1-specific siRNA. Treatments with vehicle buffer or eGFP-specific siRNA (10 nM) were also included in the experiment as negative controls. Forty-eight hours after transfection, promoter activities were detected using the Dual-Luciferase Reporter Assay System. The asterisks indicate significant differences between vehicle and siRNA treatment groups by Student's t-test (P < 0.05). All data are presented as means ± SD of at least three separate experiments.
To further confirm such findings, we next tested whether knockdown of LRH-1 by siRNA has an effect on the basal activity of the BSEP promoter. It should be mentioned that endogenous LRH-1 expression in Huh 7 cells was confirmed by RT-PCR (Fig. 1B). Huh 7 cells were transfected with pBSEP(−2.6kb) and LRH-1-specific siRNA, followed by detection of reporter activity. The BSEP promoter had relatively low basal activity. However, such low basal activity was further decreased by ∼35–40% by LRH-1 siRNA (Fig. 2B). As expected, siRNA against eGFP had no effect on BSEP promoter basal activity (Fig. 2B). These results demonstrated that knockdown of LRH-1 by siRNA resulted in a decrease in BSEP promoter activation, consistent with the finding that LRH-1 knockdown led to a decrease in BSEP mRNA expression (Fig. 1C).
LRH-1 had a modulating effect on the bile acid/FXR-mediated regulation of BSEP expression
It is well established that BSEP expression is regulated by bile acids through activating FXR (15, 16). To determine whether LRH-1 has modulating effect on bile acid/FXR-mediated regulation of BSEP expression, Huh 7 cells were transfected with LRH-1, followed by treatment of transfected cells with FXR agonist CDCA or vehicle DMSO. As shown in Fig. 3A, compared with cells transfected with vector, cells transfected with LRH-1 exhibited increased basal (DMSO) as well as CDCA-induced BSEP expression. Similar effects were observed in cells transfected with FXR (Fig. 3A). These results indicated that LRH-1 modulated the bile acid/FXR-mediated induction of BSEP expression by increasing its basal expression. To confirm this finding, the effect of LRH-1 silencing by siRNA on the bile acid/FXR-mediated regulation of BSEP expression was determined. As shown in Fig. 3B, knockdown of LRH-1 with 10 nM siRNA resulted in a significant decrease in the CDCA-mediated induction of BSEP expression. Such a decrease was not observed with eGFP-specific siRNA, indicating the specificity of the action. It should be mentioned that silencing with 5 nM siRNA also resulted in decreased BSEP expression, although it was not statistically significant. Thus, we demonstrated that the bile acid/FXR-mediated regulation of BSEP expression was modulated by the levels of LRH-1.
Fig. 3.
LRH-1 modulated the bile acid/FXR-mediated regulation of BSEP expression. A: Huh 7 cells seeded on 12-well plates were transfected with LRH-1, FXR, or vector as a control, followed by treatment of transfected cells with chenodeoxycholic acid (CDCA; 10 μM) or vehicle DMSO (0.1%) for 30 h. Total RNA was isolated and subjected to cDNA synthesis and real-time PCR using the TaqMan Gene Expression Assay with a BSEP-specific probe. The mRNA levels for GAPDH were detected as an internal standard. B: Huh 7 cells were transfected with LRH-1 siRNA, vehicle buffer, or eGFP siRNA. Thirty-six hours after transfection, the cells were treated with CDCA (10 μM) or vehicle DMSO (0.1%) for 24 h. Cells were lysed for RNA isolation and BSEP mRNA quantification by real-time PCR. The asterisks indicate significant differences (P < 0.05) between vehicle and treatment groups. C: Huh 7 cells seeded on 24-well plates were cotransfected with the BSEP promoter reporter pBSEP(−2.6kb) (100 ng), FXR expression plasmid (5 ng), and the null-Renilla luciferase plasmid (10 ng) in the absence or presence of increasing amounts of LRH-1. Sixteen hours after transfection, cells were treated with CDCA (10 μM) or vehicle DMSO (0.1%) for 30 h, followed by detection of promoter activity with the Dual-Luciferase Reporter Assay System. All data are presented as means ± SD of at least three separate experiments.
To further confirm these results, the effect of LRH-1 on the bile acid/FXR-mediated transactivation of the BSEP promoter was investigated. Huh 7 cells were cotransfected with the BSEP promoter reporter and FXR in the absence or presence of increasing amounts of LRH-1. The cells were then treated with CDCA or DMSO, followed by the detection of BSEP promoter activity. As shown in Fig. 3C, LRH-1 dose-dependently increased basal (DMSO) as well as CDCA-mediated activation of the BSEP promoter, consistent with the effects of LRH-1 overexpression on BSEP mRNA expression. Thus, we conclude that LRH-1 functioned as a modulator in the bile acid/FXR-mediated regulation of BSEP expression.
Identification of functional LRHREs in the BSEP promoter
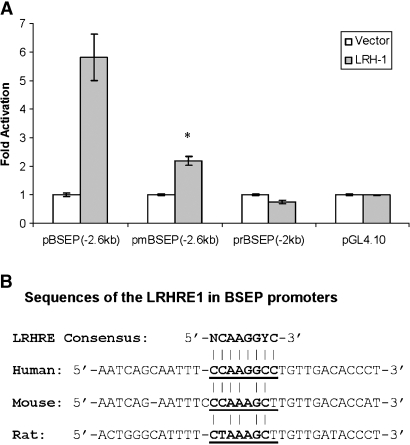
LRH-1 transactivates its target gene by binding to the LRHRE in the target gene promoters with the consensus sequence YCAAGGYCR (19–26). Bioinformatic analysis led to identification of four potential LRHREs in the BSEP promoter at nucleotides −175, −195, −1,195, and −1,510 bp (Fig. 4A). To determine whether any of those LRHREs are responsible for LRH-1-mediated activation of the BSEP promoter, each of the four sites was mutated by site-directed mutagenesis, resulting in mutants LRHRE1, -2, -3, and -4 Mut. The abilities of those mutants to respond to LRH-1 were evaluated in comparison with the wild type. As shown in Fig. 4B, mutation of the LRHRE3 and LRHRE4 sites had no significant effect on LRH-1-mediated transactivation of BSEP promoter, indicating that those two sites were not functionally involved in the transactivation. However, disruption of LRHRE1 resulted in a significant decrease in transactivation compared with the wild type (1.5- vs. 4.5-fold), suggesting that LRHRE1 is functionally involved in LRH-1-mediated transactivation of BSEP promoter. Surprisingly, disruption of LRHRE2 strongly enhanced the activation (∼13-fold induction), indicating a potential negative regulatory mechanism through LRHRE2. Together, these data showed that LRHRE1 was responsible for LRH-1 to mediate transactivation.
Fig. 4.
Mutational analyses of liver receptor homolog 1-responsive elements (LRHREs) in the BSEP promoter. A: The locations and sequences of the wild type and corresponding mutants of the four potential LRHREs in the BSEP promoter are presented. B: Mutagenesis was performed using the QuickChange site-directed mutagenesis kit. Three or four nucleotide substitutions in the LRHRE core sequence were introduced into pBSEP(−2.6kb), resulting in mutants LRHRE1 Mut, LRHRE2 Mut, LRHRE3 Mut, and LRHRE4 Mut. Huh 7 cells were cotransfected with the wild type, mutant reporters, or pGL4.10 vector (100 ng), with LRH-1 plasmid DNA (100 ng) or empty expression vector (100 ng) as a control. Luciferase activities were assayed at 48 h after transfection. Student's t-test was applied to analyze the differences between the wild type and mutants. The asterisks indicate significant differences (P < 0.05). C: Huh 7 cells were cotransfected with the wild type, mutants, or pGL4.10 vector (100 ng), the LRH-1 construct (100 ng), the FXR expression plasmid (50 ng), and the null-Renilla luciferase plasmid (10 ng). Sixteen hours after transfection, cells were treated with CDCA (10 μM) or vehicle DMSO (0.1%) for 30 h. Promoter activity was detected by the Dual-Luciferase Reporter Assay System. The asterisks indicate significant differences (P < 0.05). All data are presented as means ± SD of at least three separate experiments.
We had demonstrated previously that LRH-1 had a modulating effect on bile acid/FXR-mediated regulation of BSEP expression. We hypothesized that mutation of the functional LRHRE1 would have a repressive effect on bile acid/FXR-mediated activation of BSEP promoter. Indeed, as shown in Fig. 4C, cells cotransfected with LRHRE1 Mut, LRH-1, and FXR exhibited a significant decrease in both basal (DMSO) and CDCA-mediated activation, indicating that LRHRE1 is required for maximal activation of BSEP promoter by bile acid/FXR. Mutation in LRHRE2 markedly increased, whereas mutation in LRHRE3 and LRHRE4 had minimal effects on both basal (DMSO) and CDCA-mediated activation (Fig. 4C), consistent with the results from the previous experiment. Thus, we identified LRHRE1 as the functional cis element supporting LRH-1-mediated transactivation of BSEP.
LRH-1 specifically bound to LRHRE1
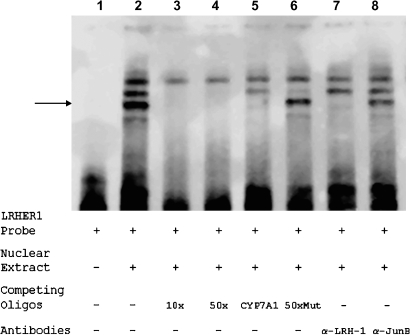
To determine whether LRH-1 specifically binds to LRHRE1, a series of EMSAs were performed with a biotin-labeled LRHRE1 oligonucleotide as a probe and nuclear extracts prepared from LRH-1-transfected Huh 7 cells. As shown in Fig. 5, lane 2, three major bands were shifted after mixing the probe with nuclear extracts, indicating that multiple DNA/protein complexes were formed. To determine which band represents the specific binding of LRH-1 to the LRHRE1 oligonucleotide, a series of competition assays were carried out. As shown in lanes 3 and 4, the middle and bottom bands were readily competed out by 10× or 50× unlabeled LRHRE1 oligonucleotide, indicating that those two bands represent two specific complexes formed by nuclear proteins with the LRHRE1 oligonucleotide. Competition with an oligonucleotide-containing CYP7A1 LRHRE resulted in disappearance of the bottom band (Fig. 4A, lane 5), whereas such band was not competed out by a mutant LRHRE1 oligonucleotide in which the core sequence of LRHRE1 was mutated (Fig. 5, lane 6), suggesting that the bottom band represents the specific complex formed by LRH-1 and the LRHRE1 oligonucleotide. To further confirm this finding, supershift assays with antibodies against LRH-1 or Jun-B as a negative control were performed. As shown in Fig. 5, lane 7, preincubation of LRH-1 antibodies with nuclear extracts resulted in disappearance of the bottom band, suggesting that the binding of LRH-1 antibodies to LRH-1 interferes with its binding to the LRHRE1 oligonucleotide, preventing the formation of the DNA/LRH-1 complexes. As expected, no effect was detected with preincubation with Jun-B antibodies (Fig. 5, lane 8), indicating the specificity of the effect. Thus, the data confirmed that the bottom band resulted from the specific binding of LRH-1 to the LRHRE1 oligonucleotide.
Fig. 5.
LRH-1 specifically bound to LRHRE1 in vitro. Electrophoretic mobility shift assay (EMSA) was performed using the LightShift Chemiluminescent EMSA kit (Pierce). Nuclear extracts were prepared from Huh 7 cells transfected with LRH-1 and subjected to EMSA analysis with biotin-labeled LRHRE1-specific probe. The specificity of the binding was established with competition assays using various competing oligonucleotides, including unlabeled LRHRE1 (10× and 50×) (lanes 3, 4), mutated LRHRE1 (50×Mut) (lane 6), and the LRHRE from human cholesterol 7α-hydroxylase (CYP7A1; 50×) (lane 5). Identification of LRH-1 in the shifted complex was carried out by a supershift assay using polyclonal antibodies against human LRH-1 (lane 7). As a negative control, antibodies against JunB were included in the experiments (lane 8). The shifted band representing the specific binding of LRH-1 to the LRHRE1 oligonucleotide is indicated by the arrow.
It should be mentioned that the top band shifted in Fig. 5, lanes 2–8, represented a nonspecific complex, whereas the middle band represented a specific complex formed by other nuclear proteins (other than LRH-1) with the sequences flanking the LRHRE1 site in the LRHRE1 oligonucleotide, which contained 12 and 7 additional nucleotides on the 5′ and 3′ sides of the LRHRE1 site, respectively. Two lines of evidence support this conclusion. First, this band was competed out by unlabeled wild-type as well as mutant LRHRE1 oligonucleotides (Fig. 5, lanes 3, 4, 6) but not by the CYP7A1 LRHRE (Fig. 5, lane 5). Second, preincubation of nuclear extracts with antibodies against LRH-1 had no effect on the formation of this complex. Thus, the data indicate that other nuclear factors were potentially involved in the transactivation of BSEP, and it will be important to identify those nuclear factors in future studies.
LRH-1 was recruited to LRHRE1
After demonstrating the specific binding of LRH-1 to LRHRE1 in EMSAs, we next performed ChIP experiments to determine whether LRH-1 is specifically recruited to the element in the BSEP promoter. Chromatins were prepared from Huh 7 cells transfected with LRH-1 and immunoprecipitated with anti-LRH-1 antibodies. A primer set, pBSEP(−260b) and pBSEP(+35b), flanking the LRHRE1 site, was used to detect the presence of LRHRE1-containing chromatin DNA. As shown in Fig. 6A, an LRHRE1-containing PCR fragment was readily detected using precipitated chromatins with antibodies against LRH-1, whereas no PCR products were amplified in a sample precipitated with IgG antibodies, indicating the specificity of the assay. To further validate this finding, a second set of primers, pBSEP(−805b) and pBSEP(−995b), located 630 bp upstream of the LRHRE1 site, was used as a negative control to amplify a 190 bp fragment. As shown in Fig. 6A, a PCR product with the expected size was readily detected with input chromatin DNA. However, no PCR products were detected with chromatin DNA immunoprecipitated with IgG or LRH-1 antibodies, confirming the specificity of the ChIP assay. We used two additional sets of primers to determine whether LRH-1 is recruited to LRHRE3 and LRHRE4. As expected, minimal or no recruitment of LRH-1 to the LRHRE3 and LRHRE4 sites was detected (Fig. 6A). Together, these data clearly demonstrate that LRH-1 was specifically recruited to the LRHRE1 element. It should be mentioned that the recruitment of LRH-1 to the LRHRE1 site was also detected using nuclear extract prepared from Huh 7 cells untransfected with LRH-1 expression plasmid, although at a lower level (data not shown), consistent with the low levels of LRH-1 expression in Huh 7 cells.
Fig. 6.
LRH-1 was specifically recruited to LRHRE1 in the BSEP promoter. A: Huh 7 cells seeded on six-well plates were transfected with LRH-1 expression plasmid (2 μg), and chromatins with optimized size ranging from 400 to 800 bp were prepared as described in Materials and Methods. After immunoprecipitation with antibodies against LRH-1 or IgG as a negative control, three sets of PCR amplification were performed to specifically detect the presence of LRHRE1-, LRHRE3-, or LRHRE4-containing chromatin DNA. A negative control primer set was included to amplify a 190 bp fragment 630 bp upstream of the LRHRE1 site. PCR amplification was carried out in a final volume of 25 μl with 2 μl of eluted chromatin DNA, input DNA, or water (mock) as template with 40 cycles. PCR products were resolved on a 2% agarose gel. B: Chromatins were prepared from normal human liver tissue and immunoprecipitated with antibodies against LRH-1 or IgG. The presence of LRHRE1-containing chromatin DNA was detected by PCR amplification using a set of primers flanking LRHRE1. PCR amplification using the negative control primer set was also performed as a control. C: Chromatins were prepared from Huh 7 cells transfected with LRH-1 plasmid and subsequently treated with CDCA (10 μM) or vehicle DMSO (0.1%). The presence of LRHRE1-containing chromatin DNA was detected by PCR amplification using a set of primers flanking LRHRE1.
To confirm the finding from Huh 7 cells, we next performed ChIP assays using chromatins prepared from human liver tissue. Consistent with the data from Huh 7 cells, an LRHRE1-containing PCR fragment was robustly detected, which was actually stronger than the signal detected with input chromatins (Fig. 6B). Again, minimal signals were detected using the negative control primer set or chromatins immunoprecipitated with IgG antibodies. Thus, we demonstrated that LRH-1 was specifically recruited to the LRHRE1 site in the BSEP promoter in both Huh 7 cells and human primary hepatocytes. To determine whether such recruitment is affected by the activation of the bile acid/FXR signaling pathway, ChIP assays were carried out using chromatins prepared from Huh 7 cells transfected with LRH-1, followed by treatment with CDCA or vehicle DMSO. As shown in Fig. 6C, CDCA treatment had no effect on LRH-1 recruitment to LRHRE1, suggesting that the recruitment of LRH-1 to the BSEP promoter is independent of bile acid/FXR-mediated activation.
Regulatory role of SHP in BSEP expression
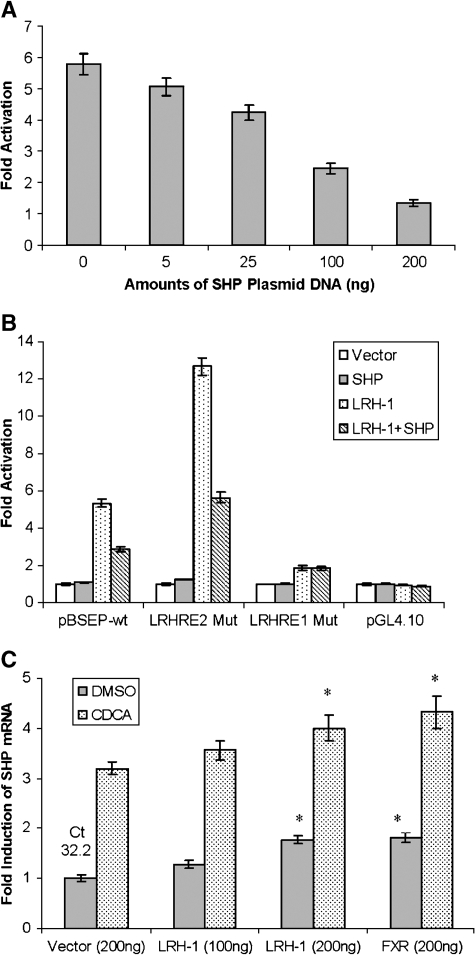
It is well established that SHP negatively regulates a set of LRH-1 target genes through heterodimerization with LRH-1 (10–14, 21, 22). The FXR/SHP/LRH-1 pathway represents a major negative feedback mechanism for bile acids to regulate those LRH-1 target genes. To explore the possibility that SHP has a regulatory role in BSEP expression by interacting with LRH-1, the effect of SHP on the LRH-1-mediated transactivation of the BSEP promoter was investigated. Huh 7 cells were cotransfected with the BSEP promoter reporter pBSEP(−2.6kb) and LRH-1 in the absence or presence of increasing amounts of SHP expression plasmids (0, 10, 25, 100, and 200 ng). As shown in Fig. 7A, SHP dose-dependently repressed the LRH-1-mediated transactivation of the BSEP promoter. These data are consistent with the notion that SHP is a negative regulator for the LRH-1 target gene by heterodimerizing with LRH-1.
Fig. 7.
Regulatory role of small heterodimer partner (SHP) in BSEP expression. A: Huh 7 cells were transfected with the BSEP promoter reporter pBSEP(−2.6kb) (100 ng), the LRH-1 expression plasmid (100 ng), and the null-Renilla luciferase plasmid (10 ng) in the absence or presence of increasing amounts of SHP constructs (0, 5, 25, 100, and 200 ng). Luciferase activities were detected at 36 h after transfection with the Dual-Luciferase Reporter Assay System. B: Huh 7 cells were cotransfected with 100 ng of the wild-type BSEP promoter reporter pBSEP(−2.6kb) (pBSEP-wt), LRHRE1 Mut, LRHRE2 Mut, or pGL4.10 vector, with expression empty vector (100 ng), LRH-1 (100 ng), SHP (50 ng), or a combination of LRH-1 and SHP. Luciferase activities were detected at 36 h after transfection. C: Huh 7 cells seeded on 12-well plates were transfected with LRH-1, FXR, or vector as a control, followed by treatment of transfected cells with CDCA (10 μM) or vehicle DMSO (0.1%) for 30 h. Total RNA was isolated and subjected to cDNA synthesis and real-time PCR using the TaqMan Gene Expression Assay with a SHP-specific probe. The mRNA levels for GAPDH were detected as internal standards. The PCR cycle time (Ct) in cells transfected with vector control is given at the top of the column. The asterisks indicate significant differences (P < 0.05) between vector control and treatment groups by Student's t-test. All data are presented as means ± SD of at least three separate experiments.
We next performed experiments to determine whether such repression by SHP is dependent on the presence of the functional LRHRE1 identified in previous studies. The effects of SHP on the LRH-1-mediated activation of pBSEP(−2.6kb) (wild type), LRHRE1 Mut, and LRHRE2 Mut were evaluated. As shown in Fig. 7B, SHP repressed the LRH-1-mediated activation of wild-type pBSEP(−2.6kb) and LRHRE2 Mut by ∼50%, but no effect was detected in cells transfected with LRHRE1 Mut. Although LRHRE1 Mut exhibited a low level of activation (1.5-fold) by LRH-1, such activation was not repressed further by SHP. These data suggested that the SHP-mediated repression was dependent on the presence of the functional LRHRE1 in the BSEP promoter.
The finding that SHP repressed the LRH-1-mediated transactivation of the BSEP promoter raised the intriguing question of why BSEP expression is induced by bile acids through FXR activation but not repressed through the FXR/SHP/LRH-1 pathway. We hypothesized that activation mediated by LRH-1 and FXR was dominant over the SHP-mediated repression in BSEP regulation. To support this hypothesis, we performed experiments to establish the expression profile of SHP in responding to LRH-1, FXR, and bile acid, the same treatments used to establish the BSEP expression profile in Fig. 3A, followed by comparison of the expression profiles of BSEP and SHP. As shown in Fig. 7C, overexpression of LRH-1 and FXR increased both basal and CDCA-induced SHP mRNA expression, a pattern strikingly similar to that observed for BSEP expression (Fig. 3A). Such paralleling but not inverse expression patterns between BSEP and SHP appeared not to be in agreement with the finding that SHP repressed BSEP promoter transactivation (Fig. 7A, B) but were consistent with our hypothesis that activation mediated by LRH-1 and FXR is dominant over the SHP repression pathway in BSEP regulation. A similar phenomenon has been reported for the regulation of SHP (10) and Ostα and Ostβ (23).
Species differences in the LRH-1-mediated transactivation of BSEP promoters
We had demonstrated that LRH-1 activated the human BSEP promoter. To determine whether such activation is conserved among mouse and rat BSEP, the effects of LRH-1 on mouse and rat BSEP promoter transactivation were investigated. Huh 7 cells were cotransfected with the mouse BSEP promoter reporter pmBSEP(−2.6kb) or the rat BSEP promoter reporter prBSEP(−2kb), with LRH-1 expression plasmid or vector as a control. The promoter activities were detected by the dual luciferase assay. As shown in Fig. 8A, similar to the finding with the human BSEP promoter, LRH-1 significantly activated the mouse BSEP promoter, although at a relatively lower level. However, such transactivation was not observed with the rat BSEP promoter. In contrast, slightly decreased transactivation was detected with the rat BSEP promoter, although the decrease was not statistically significant. These data suggested that the LRH-1-mediated transactivation of the BSEP promoter was conserved between human and mouse but not with rat. It should be mentioned that BSEP promoters derived from mouse and rat had relatively much higher levels of basal activity than the human BSEP promoter (23.5- and 23.1-fold increase, respectively; data not shown). Sequence analysis revealed that the LRHRE1 site was not completely conserved between human and mouse or rat. Two and three nucleotides were substituted in the corresponding mouse and rat sequence, respectively, compared with the human LRHRE1 (Fig. 8B). Therefore, it is possible that the LRHRE1-like site in mouse is still functional in responding to LRH-1, whereas the three substitutions in the rat sequence result in total loss of the site. It should be mentioned that a similar species difference has been reported in the LRH-1-mediated transactivation of ASBT (20).
Fig. 8.
Species differences in the LRH-1-mediated transactivation of the BSEP promoter. A: Huh 7 cells seeded on 24-well plates were transfected with 100 ng of the human, mouse, or rat BSEP promoter reporter [pBSEP(−2.6kb), pmBSEP(−2.6kb), or prBSEP(−2kb)], 10 ng of the null-Renilla luciferase plasmid, and 100 ng of LRH-1 expression plasmid or vector as a control. Promoter activities were detected with the Dual-Luciferase Reporter Assay System at 36 h after transfection. The asterisk indicates a significant difference between vector- and LRH-1-treated cells by Student's t-test (P < 0.05). Data are presented as means ± SD of at least three separate experiments. B: Alignment of the corresponding LRHRE1 sequences in human, mouse, and rat BSEP promoters. The sequence of the consensus LRHRE is given at the top of the alignment.
DISCUSSION
As the canalicular effluxer of bile acids, the expression of BSEP is regulated by its substrates bile acids. Bile acids markedly induce BSEP expression by activating the nuclear receptor FXR (15, 16). Such feed-forward regulation of BSEP expression by bile acids represents a mechanism for eliminating hepatic bile acids and preventing hepatic injury as a result of the excessive accumulation of toxic bile acids. In this study, we showed that BSEP expression was also transcriptionally regulated by LRH-1, which functioned as a modulator in the bile acid/FXR-mediated regulation of BSEP expression, most likely by modulating the basal expression of BSEP. Our finding is consistent with the results obtained with LRH-1 knockout mice, in which BSEP expression was decreased compared with that in wild-type mice (30). Thus, BSEP represented another such target gene that is regulated directly by both FXR and LRH-1 activation pathways as SHP (10), Ostα and Ostβ (23).
It is well established that CYP7A1 is the rate-limiting enzyme for bile acid synthesis (1, 2). LRH-1 is required for the maximal hepatic expression of CYP7A1 (19, 20). With our new finding that LRH-1 also transcriptionally regulates BSEP expression, it can be concluded that LRH-1 plays a supporting role to FXR in maintaining homeostatic bile acid levels in liver by coordinately regulating CYP7A1 and BSEP for bile acid synthesis and elimination, respectively. Such coordinated regulation of CYP7A1 and BSEP through the LRH-1 signaling pathway favors cholesterol conversion into bile acids and subsequent elimination. Therefore, activation of the LRH-1 signaling pathway may represent a new strategy for the removal of excessive cholesterol from the body, and LRH-1 agonists potentially have therapeutic effects on hypercholesterolemia and related cardiovascular diseases.
In this study, we explored the potential regulatory role of SHP in BSEP expression and found that SHP dose-dependently repressed LRH-1-mediated transactivation of BSEP promoter, consistent with its heterodimerization with LRH-1 to exert a negative regulatory effect on LRH-1 target genes (10–14, 21, 22). Supporting such a repressive effect of SHP on BSEP expression is the finding that BSEP is upregulated in SHP knockout mice (31). However, in this study, we also demonstrated that BSEP and SHP exhibited similar expression profiles in responding to the overexpression of LRH-1 and FXR and to CDCA treatment (Figs. 3A, 7C), which appears inconsistent with SHP's repressive role in BSEP expression. Therefore, it was suggested that BSEP expression was repressed by SHP but that activation mediated by LRH-1 and FXR was dominant over SHP-mediated repression. Such a phenomenon has been reported for SHP (10), Ostα and Ostβ (23). One characteristic feature of transcriptional regulation for those genes as well as BSEP is that they are direct targets for both LRH-1 and FXR. It appears that those targets are positively and negatively regulated through the FXR activation pathway and the FXR/SHP/LRH-1 repression cascade, respectively. Although the physiological significance has not been clearly defined for such dual regulatory mechanisms, it can be concluded that the mechanism provides another layer of regulation to more precisely titrate the expression of those target genes in responding to different cellular situations, such as increases of bile acid level.
Species differences in LRH-1-mediated transactivation have been reported (21). In this study, we showed that, similar to the finding with the human BSEP promoter, LRH-1 activated the mouse BSEP promoter, whereas such transactivation was not observed with the rat BSEP promoter. Failure of the rat BSEP promoter to respond to LRH-1 was consistent with the absence of the LRHRE site in the rat BSEP promoter. Although the LRHRE in the mouse BSEP promoter was not perfectly conserved, such a LRHRE-like site may be still functional. However, additional studies are required to definitively establish the reason for such species differences. Compared with other LRH-1 target genes, including CYP7A1 and ABCG5/8, the human BSEP promoter was much more robustly activated by LRH-1 (data not shown) (13, 25), indicating gene-specific differences in LRH-1-mediated activation. Such gene-specific differences in activation may result from variations in the sequences of the LRHREs in the respective promoters. Indeed, LRHRE1 in the BSEP promoter differs from the LRHREs in human CYP7A1 and ABCG5/8 by 2 and 3 bases, respectively. Therefore, the LRH-1-mediated transactivation exhibits clear species- and gene-specific variations.
We determined that LRHRE1 was responsible for mediating the transactivation of the BSEP promoter by LRH-1. Interestingly, disruption of the nearby LRHRE2 site resulted in strongly enhanced transactivation by LRH-1 and FXR (Fig. 4B, C). One possible explanation for such an enhancement is that LRH-1 also binds to the LRHRE2 site and that such binding results in competitive inhibition of the transactivation via the nearby LRHRE1 site. Supporting this possibility is the finding that LRH-1 also specifically bound to the LRHRE2 site detected by EMSA (data not shown). Another possibility is that the binding of transcriptional factors other than LRH-1 to the LRHRE2 site interferes in the transactivation signaling pathway mediated through the LRHRE1 site. We are currently investigating those possible mechanisms.
In summary, this study demonstrates that LRH-1 transcriptionally regulates BSEP expression through the functional LRHRE1 in the BSEP promoter and functions as a modulator in the bile acid/FXR-mediated regulation of BSEP expression. The finding indicates that LRH-1 plays a supporting role to FXR in maintaining hepatic bile acid levels by coordinately regulating CYP7A1 and BSEP for bile acid synthesis and elimination, respectively.
Acknowledgments
The authors thank Dr. Wang Li for providing LRH-1 and SHP expression constructs and Dr. David Mangelsdorf for providing FXR expression plasmid.
Abbreviations
ASBT, apical sodium-dependent bile acid transporter
BSEP, bile salt export pump
CDCA, chenodeoxycholic acid
ChIP, chromatin immunoprecipitation
CYP7A1, cholesterol 7α-hydroxylase
eGFP, enhanced green fluorescent protein
EMSA, electrophoretic mobility shift assay
FXR, farnesoid X receptor
LRH-1, liver receptor homolog 1
LRHRE, liver receptor homolog 1-responsive element
Ostα and Ostβ, organic solute transporters α and β
SHP, small heterodimer partner
siRNA, small interfering RNA
Published, JLR Papers in Press, February 12, 2008.
Footnotes
This work was partially supported by funding from the New Investigator Program for Pharmacy Faculty, American Association of Colleges of Pharmacy; by a Medical Research Grant from the Rhode Island Foundation; by Rhode Island-IDeA Network of Biomedical Research Excellence Grant P20 RR-016457 from the National Center for Research Resources/National Institutes of Health; and by a Faculty Research Development Grant, University of Rhode Island. B.Y. was supported by National Institutes of Health Grants R01 GM-61988 and R01 ES-07965.
References
- 1.Russell D. W. 2003. The enzymes, regulation, and genetics of bile acid synthesis. Annu. Rev. Biochem. 72 137–174. [DOI] [PubMed] [Google Scholar]
- 2.Fuchs M. 2003. Bile acid regulation of hepatic physiology. III. Regulation of bile acid synthesis: past progress and future challenges. Am. J. Physiol. Gastrointest. Liver Physiol. 284 G551–G557. [DOI] [PubMed] [Google Scholar]
- 3.Noe J., B. Stieger, and P. J. Meier. 2002. Functional expression of the canalicular bile salt export pump of human liver. Gastroenterology. 123 1659–1666. [DOI] [PubMed] [Google Scholar]
- 4.Byrne J. A., S. S. Strautnieks, G. Mieli-Vergani, C. F. Higgins, K. J. Linton, and R. J. Thompson. 2002. The human bile salt export pump: characterization of substrate specificity and identification of inhibitors. Gastroenterology. 123 1649–1658. [DOI] [PubMed] [Google Scholar]
- 5.Meier P. J., and B. Stieger. 2002. Bile salt transporters. Annu. Rev. Physiol. 64 635–661. [DOI] [PubMed] [Google Scholar]
- 6.Kullak-Ublick G. A., B. Stieger, and P. J. Meier. 2004. Enterohepatic bile salt transporters in normal physiology and liver disease. Gastroenterology. 126 322–342. [DOI] [PubMed] [Google Scholar]
- 7.Shneider B. L., P. A. Dawson, D. M. Christie, W. Hardikar, W. H. Wong, and F. J. Sucky. 1995. Cloning and molecular characterization of the ontogeny of a rat ileal sodium-dependent bile acid transporter. J. Clin. Invest. 95 745–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shneider B. L. 2001. Intestinal bile acid transport: biology, physiology, and pathophysiology. J. Pediatr. Gastroenterol. Nutr. 32 407–417. [DOI] [PubMed] [Google Scholar]
- 9.Dawson P. A., M. Hubbert, J. Haywood, A. L. Craddock, N. Zerangue, W. V. Christian, and N. Ballatori. 2005. The heteromeric organic solute transporter alpha-beta, Ostalpha-Ostbeta, is an ileal basolateral bile acid transporter. J. Biol. Chem. 280 6960–6968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu T. T., M. Makishima, J. J. Repa, K. Schoonjans, T. A. Kerr, J. Auwerx, and D. J. Mangelsdorf. 2000. Molecular basis for feedback regulation of bile acid synthesis by nuclear receptors. Mol. Cell. 6 507–515. [DOI] [PubMed] [Google Scholar]
- 11.Goodwin B., S. A. Jones, R. R. Price, M. A. Watson, D. D. McKee, L. B. Moore, C. Galardi, J. G. Wilson, M. C. Lewis, M. E. Roth, et al. 2000. A regulatory cascade of the nuclear receptors FXR, SHP-1, and LRH-1 represses bile acid biosynthesis. Mol. Cell. 6 517–526. [DOI] [PubMed] [Google Scholar]
- 12.Chiang J. Y., R. Kimmel, C. Weinberger, and D. Stroup. 2000. Farnesoid X receptor responds to bile acids and represses cholesterol 7alpha-hydroxylase gene (CYP7A1) transcription. J. Biol. Chem. 275 10918–10924. [DOI] [PubMed] [Google Scholar]
- 13.Chen W., E. Owsley, Y. Yang, D. Stroup, and J. Y. Chiang. 2001. Nuclear receptor-mediated repression of human cholesterol 7alpha-hydroxylase gene transcription by bile acids. J. Lipid Res. 42 1402–1412. [PubMed] [Google Scholar]
- 14.Davis R. A., J. H. Miyake, T. Y. Hui, and N. J. Spann. 2002. Regulation of cholesterol-7alpha-hydroxylase: BAREly missing a SHP. J. Lipid Res. 43 533–543. [PubMed] [Google Scholar]
- 15.Ananthanarayanan M., N. Balasubramanian, M. Makishima, D. J. Mangelsdorf, and F. J. Suchy. 2001. Human bile salt export pump promoter is transactivated by the farnesoid X receptor/bile acid receptor. J. Biol. Chem. 276 28857–28865. [DOI] [PubMed] [Google Scholar]
- 16.Plass J. R., O. Mol, J. Heegsma, M. Geuken, K. N. Faber, P. L. Jansen, and M. Muller. 2002. Farnesoid X receptor and bile salts are involved in transcriptional regulation of the gene encoding the human bile salt export pump. Hepatology. 35 589–596. [DOI] [PubMed] [Google Scholar]
- 17.Deng R., D. Yang, A. Radke, J. Yang, and B. Yan. 2007. Hypolipidemic agent guggulsterone regulates the expression of human bile salt export pump: dominance of transactivation over FXR-mediated antagonism. J. Pharmacol. Exp. Ther. 320 1153–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deng R. 2007. Therapeutic effects of guggul and its constituent guggulsterone: cardiovascular benefits. Cardiovasc. Drug Rev. 25 375–390. [DOI] [PubMed] [Google Scholar]
- 19.Nitta M., S. Ku, C. Brown, A. Y. Okamoto, and B. Shan. 1999. CPF: an orphan nuclear receptor that regulates liver-specific expression of the human cholesterol 7α-hydroxylase gene. Proc. Natl. Acad. Sci. USA. 96 6660–6665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.del Castillo-Olivares A., and G. Gil. 2000. Role of FXR and FTF in bile acid-mediated suppression of cholesterol 7alpha-hydroxylase transcription. Nucleic Acids Res. 28 3587–3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen F., L. Ma, P. A. Dawson, C. J. Sinal, E. Sehayek, F. J. Gonzalez, J. Breslow, M. Ananthanarayanan, and B. L. Shneider. 2003. Liver receptor homologue-1 mediates species- and cell line-specific bile acid-dependent negative feedback regulation of the apical sodium-dependent bile acid transporter. J. Biol. Chem. 278 19909–19916. [DOI] [PubMed] [Google Scholar]
- 22.Li H., F. Chen, Q. Shang, L. Pan, B. L. Shneider, J. Y. Chiang, B. M. Forman, M. Ananthanarayanan, G. S. Tint, G. Salen, et al. 2005. FXR-activating ligands inhibit rabbit ASBT expression via FXR-SHP-FTF cascade. Am. J. Physiol. Gastrointest. Liver Physiol. 288 G60–G66. [DOI] [PubMed] [Google Scholar]
- 23.Frankenberg T., A. Rao, F. Chen, J. Haywood, B. L. Shneider, and P. A. Dawson. 2006. Regulation of the mouse organic solute transporter alpha-beta, Ostalpha-Ostbeta, by bile acids. Am. J. Physiol. Gastrointest. Liver Physiol. 290 G912–G922. [DOI] [PubMed] [Google Scholar]
- 24.Malerod L., M. Sporstol, L. K. Juvet, S. A. Mousavi, T. Gjoen, T. Berg, N. Roos, and W. Eskild. 2005. Bile acids reduce SR-BI expression in hepatocytes by a pathway involving FXR/RXR, SHP, and LRH-1. Biochem. Biophys. Res. Commun. 336 1096–1105. [DOI] [PubMed] [Google Scholar]
- 25.Freeman L. A., A. Kennedy, J. Wu, S. Bark, A. T. Remaley, S. Santamarina-Fojo, and H. B. Brewer, Jr. 2004. The orphan nuclear receptor LRH-1 activates the ABCG5/ABCG8 intergenic promoter. J. Lipid Res. 45 1197–1206. [DOI] [PubMed] [Google Scholar]
- 26.Luo Y., C. P. Liang, and A. R. Tall. 2001. The orphan nuclear receptor LRH-1 potentiates the sterol-mediated induction of the human CETP gene by liver X receptor. J. Biol. Chem. 276 24767–24773. [DOI] [PubMed] [Google Scholar]
- 27.Delerive P., C. M. Galardi, J. E. Bisi, E. Nicodeme, and B. Goodwin. 2004. Identification of liver receptor homolog-1 as a novel regulator of apolipoprotein AI gene transcription. Mol. Endocrinol. 18 2378–2387. [DOI] [PubMed] [Google Scholar]
- 28.Deng R., D. Yang, J. Yang, and B. Yan. 2006. Oxysterol 22(R)-hydroxycholesterol induces the expression of the bile salt export pump through nuclear receptor farsenoid X receptor but not liver X receptor. J. Pharmacol. Exp. Ther. 317 317–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu F. J., X. Song, D. Yang, R. Deng, and B. Yan. 2008. The far and distal enhancers in the CYP3A4 gene coordinate the proximal promoter in responding similarly to the pregnane X receptor but differentially to hepatocyte nuclear factor-4alpha. Biochem. J. 409 243–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mataki C., B. C. Magnier, S. M. Houten, J. S. Annicotte, C. Argmann, C. Thomas, H. Overmars, W. Kulik, D. Metzger, J. Auwerx, et al. 2007. Compromised intestinal lipid absorption in mice with a liver-specific deficiency of liver receptor homolog 1. Mol. Cell. Biol. 27 8330–8339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kerr T. A., S. Saeki, M. Schneider, K. Schaefer, S. Berdy, T. Redder, B. Shan, D. W. Russell, and M. Schwarz. 2002. Loss of nuclear receptor SHP impairs but does not eliminate negative feedback regulation of bile acid synthesis. Dev. Cell. 2 713–720. [DOI] [PMC free article] [PubMed] [Google Scholar]