Abstract
Sphingolipid biosynthesis and breakdown in yeast share many homologies in their pathways with higher eukaryotes (Dickson, R. C. 1998. Sphingolipid functions in Saccharomyces cerevisiae: comparison to mammals. Annu. Rev. Biochem. 67: 27–48). In mammals, ceramide can be generated through hydrolysis of sphingomyelin catalyzed by sphingomyelinase (SMase). To date, as many as five SMases have been identified molecularly, separated into three main groups: acid, alkaline, and neutral SMases (nSMases) (Marchesini, N., and Y. Hannun. 2004. Acid and neutral sphingomyelinases: roles and mechanisms of regulation. Biochem. Cell Biol. 82: 27–44). nSMase in mammals is represented by its homolog, inositol phosphosphingolipase C, codified by ISC1 in Saccharomyces cerevisiae (Sc) and Cryptococcus neoformans (Cn) and by CSS1 (Can't Stop Synthesizing cell wall) in Schizosaccharomyces pombe (Sp). Yeasts do not have sphingomyelin but instead have inositol phosphosphingolipids, which may function as orthologs of mammalian sphingomyelin. In this review, we will describe findings related to the function of ISC1, its localization, mechanisms, and its roles in cell response to different types of stresses. These studies serve as a foundation for the elucidation of the properties and functions of the extended family of nSMases.
Keywords: ceramide, sphingomyelinase, yeast
Ceramide has emerged as a highly studied bioactive and second-messenger lipid, with proposed roles in regulation of cell growth, cell death, senescence, and various stress responses (1, 2).
A large number of enzymes are involved in regulating the levels of ceramide. To identify crucial enzymes that participate in the biochemical pathways leading to ceramide accumulation, many laboratories have focused on dissecting the biochemical pathways, and these studies have shown that ceramide can be produced in at least two distinct ways: the first is through the de novo pathway, which commences by the condensation of l-serine and palmitoyl-CoA, and the second by hydrolysis of complex lipids, especially sphingomyelin (3). The budding yeast, Saccharomyces cerevisiae (Sc), a fast-growing and genetically modifiable eukaryote, has been used extensively in elucidation of biochemical pathways of sphingolipid metabolism and functions of bioactive sphingolipids. In this review, we summarize findings about the enzyme that catalyzes the hydrolysis of yeast complex lipids to produce ceramide, ISC1 in Sc. We will also review other homologs of ISC1 in Schizosaccharomyces pombe (Sp), and the pathogen Cryptococcus neoformans (Cn), and then discuss implications for mammalian studies.
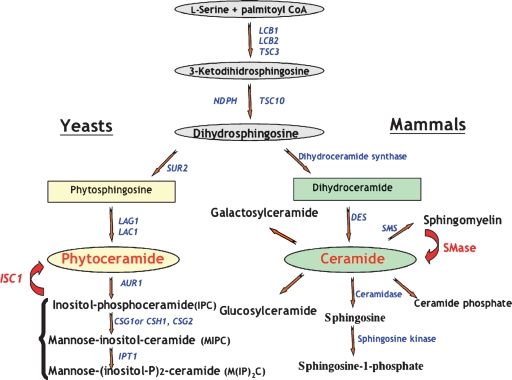
Yeast and mammals share many similarities in sphingolipid metabolism (4), and these include the first two steps in the de novo pathway: synthesis of 3-ketodihydrosphingosine catalyzed by Lcb1p and Lcb2p in yeast, and by Sptlc1p and Sptlc2p in mammals. Lcb1p and Sptlc1p share 33% homology in their sequence, whereas Lcb2p and Sptlc2p have 48% sequence homology. These enzymes were first identified in yeast and later in mouse and human. The second step, characterized by the formation of dihydrosphingosine (sphinganine), is catalyzed by the reductase Tsc10p in yeast and by its homolog (26% amino acid identity) Fvt1p in human. Following these very similar steps between yeast and mammals, differences in the pathways between the two species begin to be distinguished. Dihydrosphingosine is hydroxylated by Sur2p in yeast to produce phytosphingosine, which is then acylated by the action of the ‘ceramide’ synthases, Lag1 and Lac1. In mammals, dihydrosphingosine is directly acylated to dihydroceramide formed by the addition of C16–C24 alkyl chains fatty acid through the action of enzymes with high homology to Lag1p/Lac1p, the six Lass's/CerSs (4). In mammals, dihydroceramide can be converted to ceramide by the introduction of a double bond between C4 and C5 on the sphingoid base; this reaction is catalyzed by dihydroceramide desaturase. From phytoceramide, yeast synthesizes the following complex sphingolipids: inositol phosphorylceramide (IPC) by adding inositol phosphate, and then mannosylinositol phosphorylceramide (MIPC) and mannosyldiinositol phosphorylceramide (M(IP)2C) by further additions. Mammals synthesize many complex sphingolipids, such us sphingomyelin, by transferring phosphocholine from phosphatidylcholine to ceramide, galactosylceramide, by adding galactose to ceramide, and glucosylceramide, by adding glucose. Sphingosine is formed by the deacylation of ceramide through the action of several ceramidases, and ceramide phosphate is formed by phosphorylation of ceramide by ceramide kinase (Fig. 1).
Fig. 1.
Sphingolipid metabolic pathways in yeast and mammals. Enzymes are in blue, sphingomyelinase (SMase) and ISC1 are in red. The big red arrows represent the phospholipase-C reactions catalyzed by ISC1 in yeast and by SMase in mammals.
Breakdown of the complex sphingolipids of yeast, IPC, MIPC, and M(IP)2C, occurs by the action of a phospholipase C-type reaction to produce ceramide. In mammals, breakdown of sphingomyelin is catalyzed by one of several sphingomyelinases (SMases) distinguished by their pH optima (5). This review will focus on the yeast ISC1 as a representative member of the recently appreciated family of neutral SMases (nSMases).
Sc NEUTRAL SPHINGOMYELINASE: ISC1
An early study discovered that yeasts contain enzymatic activity capable of hydrolyzing mammalian sphingomyelin at neutral pH, although Sc does not contain this lipid (6). Subsequently, the enzymatic hydrolysis of inositol phosphosphingolipids (IPSs) in budding yeast was described in a study using [3H] inositol-labeled sphingolipids, which led to the detection of a membrane-associated phospholipase C-type activity that produced ceramide from IPC, MIPC, and (M(IP)2C). These studies suggested the existence of an enzyme that acts on yeast complex sphingolipids, as well as on sphingomyelin (7). Then, the ISC1(YER019w) gene was discovered as codifying for inositol phosphosphingolipase C-type based on distant homology to bacterial-secreted SMases (8). Overexpression of the enzyme resulted in increased levels of yeast phytoceramides, whereas knockout resulted in accumulation of yeast complex sphingolipids. Activity of Isc1p was reconstituted in lipid-detergent mixed micelles and was found to depend on the presence of Mg2+ with an optimum pH of 7.5; activity was very low at acidic or alkaline pH (8). Moreover, the enzyme was found to act on exogenously supplied mammalian sphingomyelin. Since then, an important body of work has been produced by several laboratories to elucidate the biological roles and functions of Isc1p.
Sequence, structure, and mechanism of Isc1p
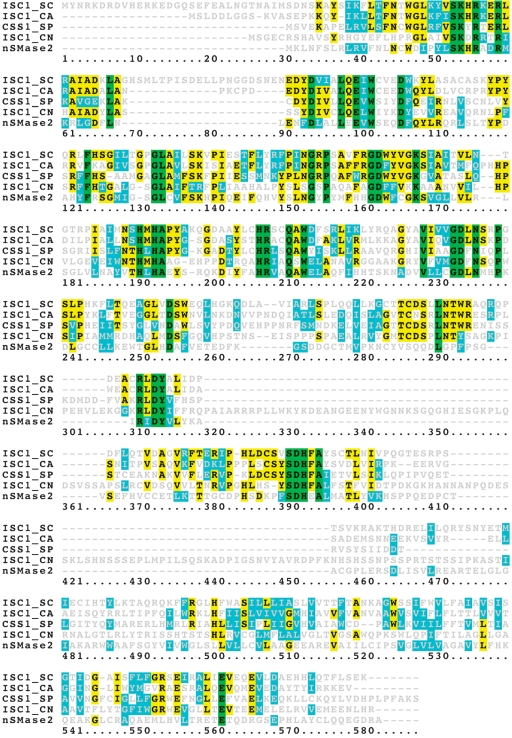
nSMase sequence analysis from Bacillus ceureus and Staphylococcus aureus, helped the identification of Isc1p as a homolog in yeast (9) and nSMase1 and nSMase2 as homologs in mammals (10). These enzymes are considered members of a single enzymatic family (11). Comparing the protein sequences between Isc1p in Sc, nSMase2 in human, Css1p in Sp, and Isc1p in Cn, 44 residues are conserved, and these residues include D163 and K168, and E100, D234, and H390, which have been identified as essential for Isc1p activity (Fig. 2).
Fig. 2.
Alignment of the entire protein sequences from Saccharomyces cerevisiae (Sc), Candida albicans, Schizosaccharomyces pombe (Sp), and Cryptococcus neoformans (Cn), and nSMase2. Identical conserved residues are shown in green, identical residues (conserved only in 2, 3, or 4 species) are in yellow, and similar residues are in blue. Sequences for Isc1p, Sc; Css1p, Sp; and neutral SMase2 (nSMase2), human are from https://www.proteome.com. Sequences for Isc1p in Cn and Candida albicans are from NCBI ABF47139 (27) and XP 711695 (28), respectively. Aligned with biology WorkBench version 3.2, at http://workbench.sdsc.edu.
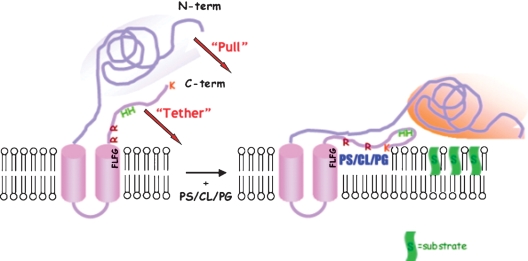
Early studies disclosed that Isc1p activity required anionic phospholipids as cofactors. To elucidate the structural requirements for anionic phospholipid-selective binding of Isc1p, site-directed mutagenesis and deletions of a portion of the gene were performed (12). Phosphatidylserine (PS), cardiolipin (CL), and phosphatidylglycerol (PG) were found to activate Isc1p (8) and to bind to it, whereas other zwitterionic phospholipids were without effect. This study showed that the C terminus of Isc1p interacted with the anionic phospholipids PS, CL, and PG, and this domain of the enzyme was found to share sequence similarity to other proteins, such as protein kinase C and PS decarboxylase, especially in regions thought to interact with PS. These studies led to the proposal of a “tether-and-pull” model, whereby the C terminus of Isc1p associates with the membrane in the presence of PS, CL, and PG (the tether), and as a result, the N-terminal catalytic domain is “pulled” to the membrane to interact with the lipid substrates (Fig. 3). To identify Isc1p-specific domains and amino acid residues required for catalysis, individual mutations of conserved amino acid residues were made to identify those that played a crucial role in activity (13). The mutagenesis of three highly conserved amino acids, E100, D232, and H390, in the catalytic domain completely abrogated ISC1 activity. These residues are also present in bacterial SMases and human neutral SMases (nSMases) 1 and 2, as well as in the distally related DNaseI, suggesting similar mechanisms of hydrolysis (14).
Fig. 3.
Model for Isc1p access to substrates (S) on the membrane. The C terminus of Isc1p tethers the protein to the membrane, and the N terminus is pulled closer to the membrane that contains the substrate. Reproduced from (12) with permission from JBC. PS, phosphatidylserine; CL, cardiolipin; PG, phosphatidylglycerol.
Inspection of the sequence of ISC1 revealed a short segment that is conserved in the entire family, the P Loop-Like domain. The substitution of D163 and K168 to alanine in this domain resulted in a complete abrogation of Isc1p activity. Additionally, the study showed that G162A, G167A, and S169A are required for Isc1p catalytic efficiency. Most of the amino acid residues, E100, G162, D163, K168, D234, and H390 involved in catalytic activity were found conserved between Sc, Cn, Candida albicans (Ca), and Sp and mammalian homolog nSMase2, demonstrating an evolutionary conservation of Isc1p sequence (Fig. 2).
Functions of ISC1 as revealed from deletion studies
Deletion of the ISC1 gene caused complete loss of phospholipase C (PLC) activity for all IPSs (IPC, MIPC, and M(IP)2C), indicating that ISC1 is probably the only gene encoding IPS-PLC activities. In addition to the loss of PLC activity, the ISC1 gene deletion was characterized by several phenotypes. One study reported that lacking the ISC1 gene rendered cells less tolerant to high concentrations of salt (NaCl and LiCl) (15). Although the biochemical role of Isc1p in ion tolerance remains unknown, the same report described a depressed expression of ENA1, an ATPase involved in ion transport.
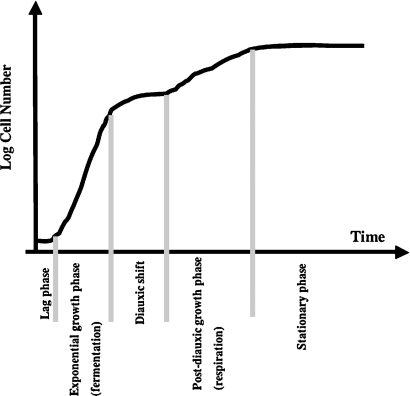
Further studies showed that the ISC1-deleted (isc1Δ) strain had a slow growth phenotype compared with wild-type under normal growth conditions (16). During logarithmic growth, yeast cells utilize dextrose to produce ethanol, by fermentation; this phase is also known as the prediauxic phase. When the cells have used all the dextrose present in the media to make ethanol, they switch carbon source and start to utilize the ethanol available in the media to produce CO2. This respiration phase is also known as the postdiauxic phase (Fig. 4). A major finding of this study was that Isc1p localized in the endoplasmic reticulum (ER) during the prediauxic phase and in the mitochondria during the postdiauxic phase (8). A substantial growth defect of isc1Δ strains when grown on a nonfermentable carbon source (NFCS) such us glycerol was described in a subsequent study (17).
Fig. 4.
Growth phases in the budding yeast, Sc. Yeast growth on fermentable carbon sources (such as dextrose) is distinguished by four phases. When cells are inoculated into a fresh medium, they initially undergo a lag in growth, known as the lag phase. After the lag phase, yeast cells start a logarithmic growth phase in which the cells use dextrose as a carbon source to produce ethanol. This is the phase of fermentation, also known as the prediauxic phase. When dextrose becomes rare in the media, the cells start utilizing the ethanol available in the media (produced during the fermentation growth) to produce carbon dioxide; this is respiration. This second phase of growth is commonly known as the postdiauxic phase. A stationary phase follows the postdiauxic phase; during this phase, yeast cells do not divide, and thus the growth reaches a plateau.
Both sets of results, Isc1p localization in the ER prediauxic and mitochondria postdiauxic and the growth defect on an NFCS, suggested a critical role of Isc1p in mitochondrial metabolism and/or in the regulation of pre/post-diauxic shift. Moreover, the same study described in vitro experiments that defined a role for the mitochondrial lipids PG and CL, formed as downstream products of Pgs1p, the phosphatidylglycerolphosphate synthase in the activation of Isc1p. The data suggested that PG and CL function as upstream regulators of Isc1p (17). Subsequent work linked Isc1p to sphingolipid metabolism in mitochondria in budding yeast (18). An important finding was the determination of Isc1p localization in mitochondria to the outer mitochondrial membrane as an integral membrane protein. In this study, yeast mitochondria were found to be highly enriched in very long chain α-hydroxylated phyto-ceramides. Mutants missing Isc1p had an altered mitochondrial lipid profile with lower content of α-hydroxylated phytoceramide compared with wild-type. Nevertheless, it remains unclear where or how Isc1p interacts with complex sphingolipids in this organelle. In addition to the sublocalization of Isc1p and the lipid profile changes due to Isc1p presence or loss, mitochondrial function was studied in the isc1Δ mutant. It was found that the mutant was more sensitive to hydrogen peroxide and ethidium bromide. This suggested a defect in mitochondria, implicating sphingolipids as possible regulators of this organelle's performance, especially in the postdiauxic phase.
Other studies investigated involvement of Isc1p in stress responses, especially heat stress. It was found that following heat stress, Isc1p mediated production of long- and very long chain dihydroceramide species distinct from the de novo pathway (19). Microarray data in this investigation revealed the role of Isc1p in transcriptional regulation during heat stress (20). Several genes implicated in mating, meiosis, sporulation, osmotic stress, and fermentation were found to be misregulated in the isc1Δ strain. This tied in to the phenotypes found previously in the isc1Δ strain, such as slow growth, high salt sensitivity, and growth defect on NFCSs. Interestingly, a recent report described Isc1p as a downstream target of Slm1p and Slm2p, two proteins involved in binding to phosphatidylinositol 4,5 bisphosphate and Torc2p (21). It was found that loss of Slm1p and Slm2p function resulted in defects of sphingolipid biosynthesis, specifically decreased IPC levels in the Slm1ts/Slm2Δ mutant. Deletion of the ISC1 gene from this mutant to obtain Slm1ts/Slm2Δ/isc1Δ restored the levels of IPC to those in the wild type. These data indicated that hyperactivation of Isc1p is implicated in the loss of function of Slm1 and Slm2.
A genome screen for methyl methanesulfonate (MMS)-sensitive mutants showed that the isc1Δ mutant was sensitive to this genotoxic agent (22). Cross sensitivity of MMS-sensitive mutants to hydroxyurea, a ribonucleotide reductase inhibitor, was evaluated, and isc1Δ was found to be sensitive to this drug as well.
A very recent work found that Isc1p played a key role in oxidative stress resistance and chronological lifespan (23). The study showed that Isc1p regulated cellular redox homeostasis through modulation of iron levels. Loss of ISC1 rendered cells hypersensitive to H2O2 compared with the wild-type cells. An investigation of the transcriptome was also undertaken, and the results showed that the lack of ISC1 led to an increase of mRNA levels of 72 genes, and a decrease of 142 genes. Several of these genes were related to iron metabolism, and further study revealed an increase in iron levels of 50% in the exponential phase. The increase was higher during the postdiauxic phase. Another interesting finding of this work was the observation that isc1Δ mutants had a premature aging phenotype: viability in aged isc1Δ was 100 times less than in the wild type.
Taken together, these results show that Isc1p plays crucial roles in very important cell processes, including growth, cell division, sporulation, ion protection, heat shock response, genotoxic protection, oxidative stress response, and aging.
ISC1 IN OTHER YEASTS
Cn nSMase:ISC1
ISC1 was cloned from the fungal pathogen Cn and was shown to metabolize fungal inositol sphingolipids (24). Loss of IPC and PLC activity in the Cn isc1Δ strain increased the survival of immunocompetent mice infected with a strain lacking ISC1 compared with those infected with the wild-type strain. This result suggested that Isc1p enhances Cn survival in macrophages and that it is critical for controlling the dissemination of the pathogen to the brain. Another characteristic found during the investigation was the hyperencapsulation of the isc1Δ mutant strain. The capsule of Cn prevents a host pathogen antigenic response and limits inflammation by inhibiting leukocyte migration. Because the microbes in the phagolysosome are exposed to low pH and reactive oxygen species (ROS), the investigators examined the sensitivity of isc1Δ to low pH and ROS. The mutant was found to be sensitive to both acidic pH and exposure to ROS, and the combination of the two was lethal.
Localization of Isc1p in Cn remains unknown, and no information is available on regulators of Isc1p. More studies need to be done in this organism to reveal the function, regulation, and localization of ISC1 and its potential role in gene regulation in Cn, as well as investigating the intriguing role of Isc1p in the virulence of Cn. The Cn data suggest investigating the role of Isc1p in the virulence of another pathogenic yeast, Ca. The amino acid sequence of Ca Isc1p shares the same conserved sequences as the Cn. Finding an inhibitor of Isc1p may be useful in combating pathogenic yeasts.
Sp nSMase: CSS1
Preliminary studies identified four temperature-sensitive lethal mutants in a visual screen, where a nonvital dye, phloxin B, distributed unevenly within the mutant cells (25). Further studies by the same group focused on one of the mutants, named css1-2. Results showed that css1-2 lost the capability to coordinate glycan polymer synthesis with cell growth at nonpermissive temperatures; thus, cells continued to produce cell wall material even when it was not needed (26). CSS1 was cloned and found to share sequence homology (28%) (Fig. 2) to ISC1 and to nSMases. CSS1 was deleted, and it was found that in this yeast, css1Δ was not viable. Css1p was tagged with 13-myc at its C terminus and was found to localize at the plasma membrane. The ability of Css1p to hydrolyze IPC was assayed by overexpressing the CSS1 gene in budding yeast that lacked the ISC1 gene, isc1Δ, and it was shown that CSS1 hydrolyzed IPC to the same level as the wild-type cells containing ISC1. Thus, Css1p is a homolog of Isc1p, and the two genes have similar biochemical patterns but appear to be distinct in their localization and biological roles.
FUTURE DIRECTIONS
Investigations reviewed in this paper demonstrate a key function of Isc1p and related enzymes in the hydrolysis of complex sphingolipids to produce the bioactive ceramide (dihydro-and phyto-ceramides) with a tremendous impact on a large array of cellular functions. These findings make Isc1p a target for many experimental studies to answer important questions about this enzyme, its localization, mechanisms of activation, and function of its substrates and products. Detailed structural studies to define the domains involved in both activity and in localization would be very useful. Biochemical studies should determine whether Isc1p has one preferred substrate among IPC, MIPC, or M(IP)2C. It would also be interesting to find out how is this enzyme is involved in drug resistance. The role in the diauxic shift remains a very intriguing and fascinating question, specifically, what is the role of Isc1p in the signaling pathways that drive gene expression and metabolic adaptation in the postdiauxic phase? Revealing the molecular and biochemical roles of Isc1p in yeast sphingolipid metabolism would certainly help investigators in the field understand how these molecules mechanistically govern important functions of the cell. The homology of Isc1p to nSMase1 and nSMase2, the mammalian members of the nSMase family, gives a great advantage in elucidating new roles of these enzymes in cell regulatory mechanisms. Studies and discoveries in yeast will be in the vanguard in providing insightful knowledge in the far more complex sphingolipid metabolism in mammalian cells.
Acknowledgments
The authors would like to thank H. Kitagaki, L. A. Cowart, and D. Montefusco for their comments and suggestions.
Published, JLR Papers in Press, February 27, 2008.
Footnotes
This work was supported by National Institutes of Health Grants GM-63265 (Y.A.H.) and P20 RR-016461 (N.M.) (PACD program).
References
- 1.Hannun Y. A. 1994. The sphingomyelin cycle and the second messenger function of ceramide. J. Biol. Chem. 269 3125–3128. [PubMed] [Google Scholar]
- 2.Spiegel S., and A. H. Merrill, Jr. 1996. Sphingolipid metabolism and cell growth regulation. FASEB J. 10 1388–1397. [DOI] [PubMed] [Google Scholar]
- 3.Hirabayashi, Y., Y. Igarashi, and A. H. Merrill, Jr. 2006. Sphingolipids synthesis, transport and cellular signaling. In Sphingolipid Biology. Y. Hirabayashi, Y. Igarashi, and A. H. Merrill, Jr., editors, Tokyo.
- 4.Dickson R. C. 1998. Sphingolipid functions in Saccharomyces cerevisiae: comparison to mammals. Annu. Rev. Biochem. 67 27–48. [DOI] [PubMed] [Google Scholar]
- 5.Marchesini N., and Y. Hannun. 2004. Acid and neutral sphingomyelinases: roles and mechanisms of regulation. Biochem. Cell Biol. 82 27–44. [DOI] [PubMed] [Google Scholar]
- 6.Ella K. M., C. Qi, J. W. Dolan, R. P. Thompson, and K. E. Meier. 1997. Characterization of a sphingomyelinase activity in Saccharomyces cerevisiae. Arch. Biochem. Biophys. 340 101–110. [DOI] [PubMed] [Google Scholar]
- 7.Wells G. B., R. C. Dickson, and R. L. Lester. 1998. Heat-induced elevation of ceramide in Saccharomyces cerevisiae via de novo synthesis. J. Biol. Chem. 273 7235–7243. [DOI] [PubMed] [Google Scholar]
- 8.Sawai H., Y. Okamoto, C. Luberto, C. Mao, A. Bielawska, N. Domae, and Y. A. Hannun. 2000. Identification of ISC1 (YER019w) as inositol phosphosphingolipid phospholipase C in Saccharomyces cerevisiae. J. Biol. Chem. 275 39793–39798. [DOI] [PubMed] [Google Scholar]
- 9.Sawai H., N. Domae, N. Nagan, and Y. A. Hannun. 1999. Function of the cloned putative neutral sphingomyelinase as lyso-platelet activating factor-phospholipase C. J. Biol. Chem. 274 38131–38139. [DOI] [PubMed] [Google Scholar]
- 10.Tomiuk S., K. Hofmann, M. Nix, M. Zumbansen, and W. Stoffel. 1998. Cloned mammalian neutral sphingomyelinase: functions in sphingolipid signaling? Proc. Natl. Acad. Sci. USA. 95 3638–3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clarke C. J., C. F. Snook, M. Tani, N. Matmati, N. Marchesini, and Y. A. Hannun. 2006. The extended family of neutral sphingomyelinases. Biochemistry. 45 11247–11256. [DOI] [PubMed] [Google Scholar]
- 12.Okamoto Y., S. Vaena De Avalos, and Y. A. Hannun. 2002. Structural requirements for selective binding of ISC1 to anionic phospholipids. J. Biol. Chem. 277 46470–46477. [DOI] [PubMed] [Google Scholar]
- 13.Okamoto Y., S. Vaena de Avalos, and Y. A. Hannun. 2003. Functional analysis of ISC1 by site-directed mutagenesis. Biochemistry. 42 7855–7862. [DOI] [PubMed] [Google Scholar]
- 14.Matsuo Y., A. Yamada, K. Tsukamoto, H. Tamura, H. Ikezawa, H. Nakamura, and K. Nishikawa. 1996. A distant evolutionary relationship between bacterial sphingomyelinase and mammalian DNase I. Protein Sci. 5 2459–2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Betz C., D. Zajonc, M. Moll, and E. Schweizer. 2002. ISC1-encoded inositol phosphosphingolipid phospholipase C is involved in Na+/Li+ halotolerance of Saccharomyces cerevisiae. Eur. J. Biochem. 269 4033–4039. [DOI] [PubMed] [Google Scholar]
- 16.Vaena de Avalos S., Y. Okamoto, and Y. A. Hannun. 2004. Activation and localization of inositol phosphosphingolipid phospholipase C, Isc1p, to the mitochondria during growth of Saccharomyces cerevisiae. J. Biol. Chem. 279 11537–11545. [DOI] [PubMed] [Google Scholar]
- 17.Vaena de Avalos S., X. Su, M. Zhang, Y. Okamoto, W. Dowhan, and Y. A. Hannun. 2005. The phosphatidylglycerol/cardiolipin biosynthetic pathway is required for the activation of inositol phosphosphingolipid phospholipase C, Isc1p, during growth of Saccharomyces cerevisiae. J. Biol. Chem. 280 7170–7177. [DOI] [PubMed] [Google Scholar]
- 18.Kitagaki H., L. A. Cowart, N. Matmati, S. Vaena de Avalos, S. A. Novgorodov, Y. H. Zeidan, J. Bielawski, L. M. Obeid, and Y. A. Hannun. 2007. Isc1 regulates sphingolipid metabolism in yeast mitochondria. Biochim. Biophys. Acta. 1768 2849–2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cowart L. A., Y. Okamoto, X. Lu, and Y. A. Hannun. 2006. Distinct roles for de novo versus hydrolytic pathways of sphingolipid biosynthesis in Saccharomyces cerevisiae. Biochem. J. 393 733–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cowart L. A., and Y. A. Hannun. 2005. Using genomic and lipidomic strategies to investigate sphingolipid function in the yeast heat-stress response. Biochem. Soc. Trans. 33 1166–1169. [DOI] [PubMed] [Google Scholar]
- 21.Tabuchi M., A. Audhya, A. B. Parsons, C. Boone, and S. D. Emr. 2006. The phosphatidylinositol 4,5-biphosphate and TORC2 binding proteins Slm1 and Slm2 function in sphingolipid regulation. Mol. Cell. Biol. 26 5861–5875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chang M., M. Bellaoui, C. Boone, and G. W. Brown. 2002. A genome-wide screen for methyl methanesulfonate-sensitive mutants reveals genes required for S phase progression in the presence of DNA damage. Proc. Natl. Acad. Sci. USA. 99 16934–16939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Almeida T., M. Marques, D. Mojzita, M. A. Amorim, R. D. Silva, B. Almeida, P. Rodrigues, P. Ludovico, S. Hohmann, P. Moradas-Ferreira, et al. 2007. Isc1p plays a key role in hydrogen peroxide resistance and chronological lifespan through modulation of iron levels and apoptosis. Mol. Biol. Cell. 19 865–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shea J. M., T. B. Kechichian, C. Luberto, and M. Del Poeta. 2006. The cryptococcal enzyme inositol phosphosphingolipid-phospholipase C confers resistance to the antifungal effects of macrophages and promotes fungal dissemination to the central nervous system. Infect. Immun. 74 5977–5988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Balasubramanian M. K., D. McCollum, L. Chang, K. C. Wong, N. I. Naqvi, X. He, S. Sazer, and K. L. Gould. 1998. Isolation and characterization of new fission yeast cytokinesis mutants. Genetics. 149 1265–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feoktistova A., P. Magnelli, C. Abeijon, P. Perez, R. L. Lester, R. C. Dickson, and K. L. Gould. 2001. Coordination between fission yeast glucan formation and growth requires a sphingolipase activity. Genetics. 158 1397–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shea J. M., T. B. Kechichian, C. Luberto, and M. Del Poeta. 2006. The cryptococcal enzyme inositol phosphosphingolipid-phospholipase C confers resistance to the antifungal effects of macrophages and promotes fungal dissemination to the central nervous system. Infect. Immun. 74 5977–5988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jones T., N. A. Federspiel, H. Chibana, J. Dungan, S. Kalman, B. B. Magee, G. Newport, Y. R. Thorstenson, N. Agabian, P. T. Magee, et al. 2004. The diploid genome sequence of Candida albicans. Proc. Natl. Acad. Sci. 101 7329–7334. [DOI] [PMC free article] [PubMed] [Google Scholar]