Abstract
Our understanding of sphingolipid metabolism and functions in the baker's yeast Saccharomyces cerevisiae has progressed substantially in the past 2 years. Yeast sphingolipids contain a C26-acyl moiety, all of the genes necessary to make these long-chain fatty acids have been identified, and a mechanism for how chain length is determined has been proposed. Advances in understanding how the de novo synthesis of ceramide and complex sphingolipids is regulated have been made, and they demonstrate that the Target Of Rapamycin Complex 2 (TORC2) controls ceramide synthase activity. Other work shows that TORC2 regulates the level of complex sphingolipids in a pathway using the Slm1 and Slm2 proteins to control the protein phosphatase calcineurin, which regulates the breakdown of complex sphingolipids. The activity of Slm1 and Slm2 has also been shown to be regulated during heat stress by phosphoinositides and TORC2, along with sphingoid long-chain bases and the Pkh1 and Pkh2 protein kinases, to control the actin cytoskeleton, the trafficking of nutrient transporters, and cell viability. Together, these results provide the first molecular insights into understanding previous genetic interaction data that indicated a connection between sphingolipids and the TORC2 and phosphoinositide signaling networks. This new knowledge provides a foundation for greatly advancing our understanding of sphingolipid biology in yeast.
Keywords: long-chain base, signal transduction, heat stress, actin, Target Of Rapamycin, phytosphingosine, very-long-chain fatty acids
Sphingolipids, sterols, and glycerophospholipids are the prime lipid components of eukaryotic membranes and are particularly abundant in the plasma membrane. Since the initial indication that sphingolipids might be second messengers for regulating signal transduction pathways (reviewed in Ref. 1), we have come to realize and appreciate their roles in regulating the signaling pathways that control essential cellular processes, including differentiation, migration, programmed cell death, and inflammation (2–5). Early on, it was recognized that sphingolipid signaling was important in human cancers, and their role has expanded to include roles in other human ailments, such as diabetes and heart disease, microbial infections, neurological disorders including Alzheimer's disease, and immune dysfunctions. Impressive as these advances are, there is much to be learned, and simpler model organisms will be important in elucidating mammalian sphingolipid signaling along with how and why sphingolipids and their metabolism evolved.
The common baker's yeast Saccharomyces cerevisiae has served in many ways to foster our understanding of sphingolipid metabolism and functions, beginning with the early work of Herbert Carter and his students in the 1950s and early 1960s, who played seminal roles in characterizing sphingolipid long-chain bases (LCBs) (6). This was followed by elucidation of the types of complex sphingolipids found in yeast cells and their route of synthesis by Lester and colleagues (reviewed in Refs. 7, 8). More recently, yeast has been used to identify nearly all of the genes that encode sphingolipid metabolic enzymes, and many of these were critical in identifying human homologs (9, 10). This review describes and integrates advances in understanding the sphingolipid metabolism and functions of S. cerevisiae during the past 2 years and focuses on the role of LCBs in signal transduction pathways that regulate growth, responses to stress, particularly heat stress, exocytosis of plasma membrane proteins, endocytosis, and actin cytoskeleton dynamics. Previous reviews should be consulted for topics that are not presented here and for more detailed coverage of areas only briefly touched upon in this review (8–14).
SPHINGOLIPID METABOLISM
Sphingolipids contain an LCB, a fatty acid, and a polar head group. The LCBs in yeast are dihydrosphingosine (DHS; sphinganine) and its 4-hydroxy derivative, phytosphingosine (PHS; 4-hydroxysphinganine). Yeast DHS can contain 16, 18, or 20 carbons and PHS can contain 18 or 20 carbons (15). The fatty acid in Saccharomyces sphingolipids is usually 26 carbons long and is saturated, but it can contain zero, one, or two hydroxyls (9). LCBs are normally required for cell viability; however, viable mutant strains lacking LCBs have been isolated and shown to be viable because they synthesize a compensatory set of novel C26-acyl glycerophospholipids that mimic some functions of sphingolipids (16, 17). These cells have impaired stress resistance that can be reversed by LCB addition, the first indication that LCBs or some other sphingolipids are important for stress protection (18).
As is the case with other lipids, sphingolipid synthesis begins in the endoplasmic reticulum (ER), where serine palmitoyltransferase (SPT) condenses serine with a fatty acyl-CoA to yield 3-ketodihydrosphingosine (ketosphinganine) and CO2 (Fig. 1). In yeast, SPT is a heterodimer made from the Lcb1 and Lcb2 proteins (reviewed in Ref. 19). These proteins are ubiquitous and have been found in all organisms that make sphingolipids: in a unique case, they have been found fused into a single gene in a viral genome (20). Yeast SPT has a third, small hydrophobic subunit, Tsc3, that is necessary for optimal enzyme activity and for growth at temperatures above ∼30°C, but its function remains unknown and it is not found in mammals (21). Recently, mammals were shown to make two types of Lcb2 subunits, SPTLC2 and SPTLC3, whose concentrations vary in a tissue-specific manner (22). Moreover, mammalian SPT appears to be a large molecular weight complex, possibly containing four SPTLC1 monomers associated with four total copies of SPTLC2 and SPTLC3, whose ratio in the complex varies in different cell types (23). It is not clear at this time whether the yeast SPT exists in such a large complex.
Fig. 1.
Outline of sphingolipid metabolism in Saccharomyces cerevisiae. Metabolic intermediates and complex sphingolipids are shown in boldface, genes are indicated by italics, and enzyme names are in regular lettering. Structures of the indicated compounds are presented in previous publications (9, 14). The cellular locations of these reactions are discussed in the text and in a recent discussion of intracellular sphingolipid trafficking (106). When grown aerobically, the fatty acid in complex sphingolipids is often hydroxylated at C2 and sometimes at C3, a reaction that requires Scs7 (not shown). Ceramides can be hydrolyzed by two ceramidases, Ydc1 and Ypc1, to yield a fatty acid and a long-chain base (LCB) (107, 108).
In the next reaction, 3-ketodihydrosphingosine is reduced to DHS by the Tsc10 protein (24). DHS is then amide-linked to a C26 fatty acid by either of two ceramide synthases (acyl-CoA:sphingosine N-acyltransferase), Lag1 and Lac1 (25, 26), to yield N-acylsphinganine (dihydroceramide), which is hydroxylated at C-4 by Sur2/Syr2 to give phytoceramide (Fig. 1). Phytoceramide can also be made by hydroxylating DHS to form PHS, which is then amide-linked to a C26 fatty acid (27, 28). Ceramide synthases in yeast require another protein, Lip1, whose function is unknown (29). This essential role of Lip1 is unique to yeast because mammals lack a sequence homolog and yeast cells expressing the mammalian Lass5 ceramide synthase enzyme do not require Lip1 (30). The C26 fatty acid in phytoceramide can be nonhydroxylated or can contain one hydroxyl at C-2 or two hydroxyls with the second at C-3 (31). The Scs7/Fah1 protein performs these hydroxylation reactions (27, 32, 33).
Impressive progress in understanding how fatty acids are elongated from C14–C18 to C26 in the ER by the very-long-chain fatty acid (VLCFA) synthetase complex has been made recently. Four reactions constitute the elongation cycle, catalyzed by a multienzyme complex that initiates with the condensation of malonyl-CoA with an acyl-CoA to form a 3-ketoacyl-CoA (Fig. 1, step 1). The 3-ketoacyl-CoA is reduced to 3-hydroxy acyl-CoA in the second step, followed by the dehydration of 3-hydroxy acyl-CoA to an enoyl intermediate in the third step. In the fourth and final step, the enoyl is reduced to yield an acyl-CoA that is two carbons longer than the starting compound. The Elo1 protein was shown early on to catalyze the elongation of C14 fatty acids to C16 (34). Two related proteins, Fen1 and Sur4, were subsequently discovered and shown to play a role in elongation up to C26 (35). Elo1, Fen1, and Sur4 belong to a family of elongase proteins (Elops) (36) that specify chain length and that were thought to catalyze the first reaction in the cycle. The second reaction was shown to be catalyzed by Ybr159w (37, 38), and the final reaction was thought to be catalyzed by Tsc13 (39).
The identity of the protein that catalyzed the third reaction was unknown, and it was unclear how fatty acid chain length was determined. This confusion has been clarified with the reconstitution of the yeast VLCFA synthetase complex in liposomes (40). These studies verified that one of two Elops (36), either Fen1 or Sur4 (35), catalyzes the first reaction and that the third, dehydration reaction is catalyzed by the newly discovered Phs1 protein (41). A seminal contribution of the reconstituted elongation system was to clarify the function of the Fen1 and Sur4 Elops and to suggest that each enzyme complex contains a single Elop. A VLCFA synthetase complex with Fen1 was shown to make C22 and C24 fatty acids, whereas a complex with Sur4 could make longer C26 fatty acids. Mutational analysis of Fen1 and Sur4 revealed that a transmembrane helix containing a lysine residue and facing the lumen of the ER determines fatty acid chain length. The lysine is hypothesized to act as part of a caliper mechanism to specify chain length (40).
Ceramides must be transported from the ER to the Golgi so that the polar head groups can be added. In yeast, there appears to be a vesicle and a nonvesicle transport system that seems to require direct contact between the ER and Golgi membranes (42). Mammals also use vesicle and nonvesicle ceramide transport. Nonvesicle transport is mediated by the CERT protein, which extracts ceramides from the lumenal surface of the ER and deposits them on the outer leaflet of the Golgi membrane (43, 44). No CERT-like protein has been found in yeast, but this could be attributable simply to a lack of amino acid sequence conservation.
Upon reaching the Golgi membrane, ceramide incorporates into the outer leaflet and then either flips spontaneously or is flipped enzymatically (it is not clear which mechanism is important) to the inner leaflet so that it is accessible to the enzymes that attach polar head groups, which are in the lumen. The first head group to be added to the C1 OH of ceramides is inositol phosphate. This reaction is catalyzed by inositol phosphorylceramide synthase (IPC synthase) (45, 46) and yields the first complex sphingolipid, inositol phosphoceramide (IPC) (Fig. 1). The second complex sphingolipid, mannose inositol phosphoceramide (MIPC), is made by the transfer of mannose from GDP-mannose onto the inositol 2-OH moiety of IPC. The enzyme inositol phosphoceramide mannosyl transferase catalyzes this reaction and has two forms. One contains the Csg1 and Csg2 proteins, and the other contains the Csh1 and Csg2 proteins. The Csg1 and Csh1 subunits appear to be the catalytic subunits, whereas Csg2 performs a regulatory function (47). The Csg2 protein binds Ca2+, and transcription of the CSG2 gene is stimulated by high levels of Ca2+ in culture medium, as is the conversion of IPC to MIPC (48). These results may partly explain why csg2 mutants were isolated in a screen for mutations that give a calcium-sensitive growth phenotype (49). In the presence of high calcium, cells may have to convert IPC to MIPC at a faster rate to grow, either because IPC becomes toxic to an essential function in the Golgi apparatus or the plasma membrane or because MIPC or its product mannose-(inositol-P)2-ceramide [M(IP)2C] becomes rate-limiting for an essential process. This connection between calcium and complex sphingolipid synthesis is fascinating and needs to be examined more thoroughly. The calcium-sensitive phenotype of csg1 and csg2 mutants has been of great value in identifying genes involved in sphingolipid metabolism (24) and, as described below in this review, has provided new insights into how sphingolipid synthesis is regulated.
The terminal yeast complex sphingolipid made in the Golgi apparatus is M(IP)2C, made by the transfer of a second inositol phosphate from phosphatidylinositol to MIPC. Complex sphingolipids made in the Golgi apparatus move by vesicle transport primarily to the plasma membrane (50). Readers seeking more information about ceramide transport and sphingolipid metabolism in the Golgi apparatus should consult a previous review (14).
Although mammalian cells must turn over or break down complex sphingolipids to survive and prevent the accumulation of toxic molecules that cause debilitating human diseases termed sphingolipidoses (51), it has only fairly recently become apparent that S. cerevisiae cells even break down complex sphingolipids, let alone that the breakdown plays any observable physiological role. Complex sphingolipids in yeast constitute ∼30% of the phosphorylated membrane lipids and nearly 7% of the mass of the plasma membrane (52). Thus, it was experimentally challenging to biochemically detect the breakdown of a small fraction of complex sphingolipids and even more difficult to determine where the breakdown was occurring in cells. This technical challenge can be conquered by yeast genetics.
Because of amino acid sequence similarity to mammalian sphingomyelinases, the ISC1 gene was examined and found to be a phospholipase C-type enzyme that cleaved polar head groups from yeast sphingolipids (53, 54). Isc1 appears to be the only enzyme in yeast with such activity, and at this time it is the only known enzyme that breaks down complex sphingolipids in yeast. A short summary of the previously described roles of Isc1 is presented as an introduction to its recently discovered roles in the regulation of sphingolipid synthesis and stress protection.
Isc1 may have a role in tolerance to sodium and lithium ions (55), but the mechanism is unclear. Isc1 activity is required for growth on a nonfermentable carbon source such as during the diauxic shift, implying a role in respiration/mitochondria (56). A mitochondrial connection is supported by the observation that Isc1 moves from the ER to the mitochondria as cells progress from fermentative to respiratory growth during the diauxic shift (57). During this time, the specific activity of the enzyme increases by 3- to 5-fold and phytoceramide increases by 4-fold, but only if Isc1 is active. Lipids enriched in mitochondria, phosphatidylglycerol, and cardiolipin activate and appear to be physiologically important regulators of enzyme activity (56). Isc1 in cells grown to saturation (postdiauxic shift) was recently shown to reside in the outer leaflet of the mitochondrial membrane (58). Lipid analysis of purified mitochondria indicates a high content of α-hydroxy-fatty acyl-containing phytoceramides, suggesting that it is derived from complex sphingolipids by the action of Isc1. Cells with an intact ISC1 gene are less likely than isc1Δ cells to lose mitochondrial function and become incapable of using nonfermentable carbon sources. Likewise, cells with wild-type Isc1 activity are more resistant to oxidative stress as measured by hydrogen peroxide resistance, and they resist ethidium bromide-induced mitochondrial damage better than cells lacking Isc1 (58). Together, these studies demonstrate that Isc1 plays an important role in mitochondrial function during the diauxic shift. Future studies should reveal the functional role of phytoceramide in mitochondria. In log-phase cells, Isc1 is responsible for part of the increase in ceramide that occurs when yeast are heat shocked (54, 59, 60), and it also plays an uncharacterized role in sporulation (60). A new role for Isc1 in heat stress and the regulation of sphingolipid synthesis is presented in the next section.
A seminal advance in our understanding of how cells regulate de novo ceramide synthesis and, thus, the synthesis of complex sphingolipids was made recently by Powers and colleagues (61), who showed that the Target Of Rapamycin Complex 2 (TORC2) controls the activity of ceramide synthase. TOR is a conserved protein kinase that senses nutrients and stresses and coordinates metabolism both temporarily and spatially to control cell growth (62). All known eukaryotes contain two TOR protein complexes, TORC1, which is inhibited by rapamycin, a bacterial macrocyclic lactone, and TORC2, which is rapamycin-insensitive.
A connection between sphingolipids and the TORs was first suggested by the work of Beeler et al. (49), who were studying calcium homeostasis in yeast. They isolated a mutation in the CSG2 gene that conferred sensitivity to 100 mM Ca2+. A screen for temperature-sensitive mutations that bypassed the calcium sensitivity of csg2 cells identified several genes, including TOR2 and AVO3/TSC11, that encode components of TORC2 (24). TORC2 controls the organization of the actin cytoskeleton in yeast and mammals (62).
To elucidate the connection between TOR signaling and sphingolipids, Aronova et al. (61) isolated an allele of AVO3 (avo3-30) that diminished growth at 30°C. Temperature was a concern because previous alleles required higher temperatures to impair growth and high temperatures enhance DHS, PHS, and ceramide levels (12), which might mask the effect of an avo3 mutation on ceramide synthesis. At 25°C avo3-30 cells grew like wild-type cells, but growth slowed within 5–6 h after a shift to 30°C. Aronova et al. (61) found that 3 h after shifting avo3-30 cells to 30°C, the concentration of the major yeast ceramide species containing PHS and a C26 fatty acid was reduced by 5-fold compared with wild-type cells. The level of minor ceramide species having shorter fatty acid chains was reduced by ∼10-fold. Just as telling, the concentration of the major ceramides was reduced by 2-fold even in avo3-30 cells grown at 25°C. These results predict that avo3-30 cells have reduced ceramide synthase activity, a prediction that was confirmed by measuring enzyme activity in microsomes isolated from cells grown at 30°C.
Aronova et al. (61) next examined how TORC2s affect ceramide synthase activity. They focused on the Ypk2 protein kinase because it is activated by TORC2 (63) and because mutant ypk2 and avo3-30 cells show similar defects in cell wall integrity and actin polarization. Insightful data were obtained using a constitutively active allele of YPK2: all avo3-30 phenotypes including the ceramide deficiency were reversed. These results argue that Ypk2 acts downstream of TORC2 to activate ceramide synthase activity (Fig. 2). The actual molecular mechanism will require further work to determine whether Ypk2 directly phosphorylates a subunit of ceramide synthase to govern enzyme activity or whether some other Ypk2 substrate regulates the activity.
Fig. 2.
Target Of Rapamycin Complex 2 (TORC2) and calcineurin regulate de novo ceramide and sphingolipid synthesis in yeast. TORC2 is proposed to sense growth signals and phosphorylate the protein kinase Ypk2 (61), which, after being phosphorylated in its activation loop by Pkh2 (63) (and perhaps Pkh1), activates ceramide synthase. The Ca2+/calmodulin-regulated protein phosphatase calcineurin opposes the TORC2-Pkh1/2-Ypk2 pathway to downregulate ceramide synthase activity in response to heat and other stresses. Environmental factors such as heat stress that increase LCBs are proposed to act in a feed-forward manner to activate Pkh1/2 and possibly increase the rate of ceramide synthesis (not shown) (61).
Another interesting finding involved calcineurin, an evolutionarily conserved Ca2+/calmodulin-regulated protein phosphatase that downregulates cellular processes controlled by TORC2, including the synthesis of complex sphingolipids (see below). Aronova et al. (61) found that deleting the CNB1 gene, which encodes the calcineurin regulatory subunit B, restored ceramide levels in avo3-30 cells and promoted growth at 30°C. This finding suggests that calcineurin downregulates ceramide synthase activity (Fig. 2). Perhaps calcineurin dephosphorylates ceramide synthase. Surprisingly, deletion of CNB1 did not restore actin polarization to avo3-30 cells, implying that it is controlled by TORC2 in a distinctly different manner from ceramide synthesis activity.
Previous work had shown that blocking steps in sphingolipid synthesis downstream of DHS and PHS causes their accumulation (25, 26, 64), as does heat stress (59, 65). Aronova et al. (61) found that avo3-30 cells do indeed accumulate these LCBs, as expected for cells with reduced ceramide synthase activity. DHS and PHS are known to activate the Pkh1 and Pkh2 protein kinases, which, along with TORC2, activate Ypk2 (reviewed in Ref. 12). There is also an indication that DHS and PHS can act directly to partially activate Ypk2 (66). Thus, Aronova et al. (61) speculated that DHS and PHS act in a feed-forward manner to coregulate Ypk2 along with TORC2, thereby controlling ceramide synthase activity and the flux of LCBs that are incorporated into ceramides and complex sphingolipids (Fig. 2).
These studies in yeast begin to reveal how cells promote ceramide and sphingolipid synthesis when conditions favor growth and how they reduce synthesis when stresses threaten cells and impede growth. Because the early steps in de novo sphingolipid synthesis are reasonably conserved, mammalian TORC2 may play a role in regulating de novo ceramide synthesis. However, mammals have at least six ceramide synthases that make ceramides with fatty acids of a particular chain length (67). These enzymes also display unique temporal and cell-specific expression patterns. Given this diversity, it is unlikely that mTORC2 regulates all of these enzymes, but it could govern some in certain cell types. The mammalian homolog of Ypk2 is the serum- and glucocorticoid-inducible protein kinase, which may transmit the signal from mTORC2 to some ceramide synthases. But it would not be surprising if other members of the AGC kinase family, including Akts/PKBs, transmitted the mTORC2 signal to ceramide synthases, because Akt/PKB functions downstream of mTORC2 (62). Mammalian ceramide synthases are already receiving much attention, but the yeast results should stimulate a more focused interest and may pave the way to understanding how de novo ceramide synthesis is regulated in mammals. Such understanding could lead to better chemotherapeutic drugs, some of which promote killing by enhancing de novo ceramide synthesis in ways that are not understood (68, 69).
It has been more than 10 years since heat stress was observed to induce an increase in LCBs (59, 65), but the molecular mechanism(s) underlying the increase has remained unclear. The increases are substantial, but transient. For example, C18-DHS and C18-PHS increase by 2- to 3-fold and C20-DHS and C20-PHS increase by >100-fold, with the peaks appearing at 5–10 min after the temperature shift. Thereafter, LCBs return to basal values, even though the cells remain at an increased temperature. Cowart and Hannun (70) have presented evidence that the transient increase in LCBs is at least partly driven by an increased uptake of serine from the culture medium and is not attributable to changes in the specific activity of SPT (Fig. 1). However, heat-induced increases in C20-DHS and C20-PHS were reduced by ∼75% in cells lacking the Tsc3 subunit of SPT, indicating that this protein, which is only essential for growth at increased temperatures (21), plays a role in the synthesis of C20-LCBs. Previous studies with mammalian cells indicated that substrate availability played a role in determining the rate of LCB production (71, 72). The availability of the two SPT substrates, fatty acyl-CoAs and serine, was examined for effects on LCB synthesis, and serine was found to be important (70). Using a mutant strain defective in serine synthesis (ser3Δser33Δ), serine taken up from the culture medium was shown to be responsible for the heat-induced increase in LCBs. In addition, the rate of serine uptake was shown to be stimulated by heat. Two other factors besides heat that are known to increase amino acid uptake, the acidification of the culture medium and the addition of glucose to glucose-starved cells, also increased LCB production. Thus, the rate of de novo LCB synthesis in yeast is at least partially controlled by the rate of serine uptake.
CELLULAR PROCESSES REGULATED BY LCBS
LCBs were first suspected of being intracellular signaling molecules or second messengers when their concentration was observed to rapidly but transiently increase after heat stress caused by shifting cells from 25°C to 37°C or 39°C (59, 65). Such a transient increase in concentration is a classic trait of second messengers.
Insight into the signal transduction pathways regulated by LCBs was found first while attempting to identify genes whose overexpression bypassed the growth inhibition caused by myriocin (73). Myriocin inhibits SPT and blocks cell growth by curtailing sphingolipid synthesis (74). Sun et al. (73) found that the YPK1 gene, when present on a multicopy vector, bypassed the myriocin block. Ypk1 is a protein kinase that plays a role in cell wall maintenance and actin cytoskeleton dynamics (75, 76), endocytosis (77), and translation during nitrogen starvation and nutrient sensing (78). Ypk1 and its paralog Ypk2 are structural and functional homologs of mammalian serum- and glucocorticoid-inducible kinase (79).
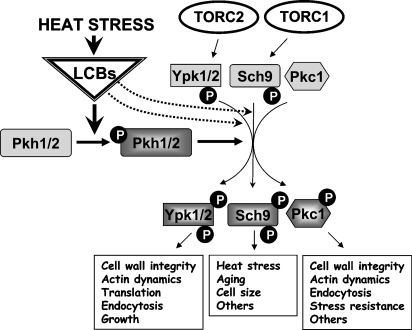
Because the protein kinase Pkh1 was known to phosphorylate and activate Ypk1 (79), multiple copies of PKH1 were examined and found to also bypass the growth inhibition caused by myriocin (73). These results suggested that some sphingolipid activated the Pkh1/Ypk1-Ypk2 signaling pathway. Pkh1 is a homolog of mammalian phosphoinositide-dependent protein kinase 1 (PDK1), which is well known for being activated by binding to 3-phosphoinositides via a pleckstrin homology domain. However, Pkh1 has no pleckstrin homology domains and was not activated by phosphoinositides in vitro (79). Sun et al. (73) showed that myriocin-treated cells lacked a phosphorylated and presumably active form of Ypk1 and that this form reappeared in vivo when PHS was added to the culture medium, even with myriocin still present. These experiments supported, but did not prove, the hypothesis that PHS activated Pkh1 or its homolog Pkh2, which then phosphorylated and activated Ypk1. After these initial experiments, many laboratories contributed data supporting the hypothesis that LCBs act to activate Pkh1/2, which then phosphorylate and contribute to the activation of kinases, including Ypk1/2, Pkc1, and Sch9 (Fig. 3). Contributions made by individual laboratories to our understanding of the LCB→Pkh1/2 pathway have been described in detail in previous reviews (11, 12).
Fig. 3.
Summary of the signaling pathways regulated by LCBs in S. cerevisiae. Heat stress is known to produce a transient increase in LCBs, which then activate Pkh1 and Pkh2. In vitro, Pkh2 is more strongly stimulated by LCBs than is Pkh1 (66), but the situation in vivo has not been analyzed directly by enzyme activity measurements. Studies in vitro suggest that LCBs can directly produce small increases in the activity of Ypk1, Ypk2, and Sch9 (indicated by dotted lines), whereas the stimulation of Pkc1 has not been examined (66). Pkh1/2 phosphorylate Ypk1, Ypk2, Sch9, and Pkc1 in their activation loop (PDK1 site), but the proteins are not enzymatically active. To become active, they also need to be phosphorylated in a hydrophobic region (PDK2 site) and in a turn motif in their C termini. Phosphorylation of these sites in Ypk2 is mediated by TORC2 (63), and for Sch9 phosphorylation is mediated by TORC1 (109). Ypk1/2 and Pkc1 are shown working in parallel pathways to control cell wall integrity, but data also support an alternative pathway in which Ypk1/2 work upstream of Pkc1 (75, 76).
One aspect of the LCB-regulated signaling pathways and cellular processes that is not elucidated and, therefore, is not shown in Fig. 3, is where the individual components of the pathway reside in a yeast cell. For example, recent observations suggest that there are specialized pools of PHS, one of which controls some fraction of Ypk1 activity that protects cells from oxidative stress (80). These experiments began with the Survival Factor 1 gene, SVF1, that is required for the diauxic shift (the transition from respiration to fermentation) and whose function in this process can be complemented by the mammalian Bcl-Xl protein, which is involved in antiapoptotic or prosurvival functions (81). Results from genetic interaction analyses and measurements of LCBs suggest a role for Svf1 in regulating the generation of a pool of C18-PHS and C18-PHS phosphate that modulate Ypk1 functions. Gene interaction results argue that these pools are formed by the concerted action of the LCB kinase Lcb4 and the LCB phosphate phosphatase Lcb3 but not by Sur2. Svf1 is proposed to regulate the activity of Lcb3 and Lcb4, but the mechanism remains to be determined. These data are the first to suggest that the growth and survival of yeast cells depends on specialized intracellular pools of LCBs that regulate signal transduction pathways.
Previous results (24) implied a connection or connections between the LCB→Pkh1/2 and TOR signaling pathways, but the mechanism was unclear. This void in our knowledge is beginning to be filled with the realization that these two pathways, along with the calcineurin signaling pathway, modulate the phosphorylation/dephosphorylation cycle of the Slm1 and Slm2 proteins during heat stress to control actin polarization, endocytosis, and sphingolipid metabolism (82–84).
SLM1 and SLM2 have overlapping functions, and at least one of them is required for viability (85). A distinguishing feature of the Slm proteins is a pleckstrin homology domain that enables them to bind phosphatidylinositol 4,5-bisphosphate (PIP2). PIP2 is made transiently, for example during heat shock, on the inner leaflet of the plasma membrane, where it serves to initially anchor the Slm proteins (85, 86). The strength of Slm binding to the membrane is strengthened by interactions with the Avo2 and Bit61 subunits of TORC2 (85, 87), and this interaction also facilitates the phosphorylation of Slm1 and Slm2 by TORC2 (85). Dual binding enables the Slm proteins to mediate effects downstream of both the PIP2 and TORC2 signaling pathways that control roles of the actin cytoskeleton essential for growth, cell wall integrity, and receptor-mediated endocytosis.
Heat stress was recently shown to initiate the dephosphorylation and rephosphorylation of the Slm proteins (83–85). Dephosphorylation was shown to be mediated by the protein phosphatase calcineurin, which binds to the Slm proteins (82–84) and plays roles in a variety of stress responses. Because heat also generates a transient increase in PHS and DHS, which then activate the Pkh1/2 signaling pathway (Fig. 3), it was hypothesized that PHS played a role in the phosphorylation of the Slm proteins. Several types of experiments support this hypothesis, but the most physiologically significant used myriocin to reduce the concentration of PHS and DHS and demonstrate a reduction in the heat-induced phosphorylation of Slm1 and Slm2 (83, 84). The idea that the Slm proteins depend upon sphingolipid signaling or metabolism or both to carry out their essential role in cell growth was supported also by the observation that deletion of either slm1 or slm2 made cells more sensitive to myriocin (83, 84). Furthermore, combining a slm1Δ mutation with a nonessential mutation in a sphingolipid metabolic gene, including fen1Δ, lcb4Δ, or csg2Δ, generated cells that were more sensitive to myriocin than the single mutants. Likewise, the actin cytoskeleton was more disrupted in the double mutant than in the single mutants, supporting the idea that Slm function depends upon sphingolipids (84). Finally, phosphorylation of Slm1 has been shown to depend upon the Pkh kinases. In cells carrying a temperature-sensitive allele of pkh1ts and having pkh2 deleted, the basal and heat-induced level of phosphorylated Slm was reduced (84). This phosphorylation seems to be independent of TORC2.
Attempts to determine whether Pkh1 directly phosphorylates Slm1 and Slm2 suggest that they do not, because His6-Pkh1 produced in and purified from Escherichia coli failed to phosphorylate purified GST-Slm1 or GST-Slm2 in vitro. However, these experiments are inconclusive, because Pkh1 made in E. coli may not be properly activated or the in vitro reaction conditions may not be suitable for phosphorylation of the Slm proteins. Whether or not the Slm proteins are phosphorylated by one of the kinases that acts downstream of Pkh1/2 is also unresolved. Serine 659 appears to be the site phosphorylated by the Pkh1/2 pathway, and this phosphorylated residue is essential for Slm1 to support growth at higher but not at lower temperatures (84). Although the biochemical function or activity of Slm1 and Slm2 is unknown, it seems likely that they function downstream of the LCB→Pkh1/2 pathway to control the polarization of the actin cytoskeleton. This conclusion is based upon a mutant strain having a negatively charged aspartic acid residue in place of serine 659. The negative charge mimics phosphorylation and produces constitutive Slm1 activity, which, when overexpressed, is able to restore actin polarization at increased temperatures in the pkh1ts pkh2Δ mutant strain (84). However, the SLM1S659D allele does not restore the growth of pkh1ts pkh2Δ cells at increased temperatures, indicating that Slm1 does not mediate all essential Pkh1/2 functions. Understanding the biochemical action of the Slm proteins would provide an important advance in understanding how the LCB→Pkh1/2 pathway regulates the actin cytoskeleton.
The PIP2 pathway was recently found to regulate the synthesis of IPC via the Slm1 and Slm2 proteins, which downregulate the activity of Isc1 (Fig. 1), the enzyme that cleaves polar head groups from complex sphingolipids (82). Previous studies had implicated the PIP2 pathway in regulating IPC levels, but the mechanism was unclear. PIP2 is synthesized from phosphatidylinositol by the sequential action of Stt4, a phosphatidylinositol 4-kinase that makes phosphatidylinositol 4-phosphate (PI4P), which is then converted to PIP2 by the Mss4 enzyme. Previous data had implied a connection between the PIP2 pathway and sphingolipids. A screen for mutations that could bypass the calcium sensitivity of csg2Δ cells identified mss4 (24). In addition, the screen for genes that bypassed growth inhibition by myriocin also uncovered MSS4 as a multicopy suppressor gene in addition to identifying YPK1 (88). In recently reported studies, synthetic genetic array analysis was used to search for genes that regulate or work downstream of PI4P generated by Stt4. Strains in the nonessential yeast deletion collection were combined with a temperature-sensitive stt4ts strain to identify double mutants with slow or impaired growth (82). This screen identified FEN1 and SUR4, required for the synthesis of C26 fatty acids that are a necessary component of sphingolipids (Fig. 1), and thereby implicated sphingolipids as targets of the PIP2 pathway.
The synthetic genetic array results were followed up by several types of experiments, and the key results will be discussed here. Analyses of mutants that bypassed the calcium sensitivity of csg2Δ suggested that they restored growth by reducing the level of a species of IPC termed IPC-C. Therefore, the levels of this and other complex sphingolipids plus ceramide were analyzed by radiolabeling cells grown at 26°C and 38°C with [3H]serine and measuring radioactive sphingolipids by thin-layer chromatography and autoradiography (82). At 26°C, the levels of IPCs, MIPC, and ceramide were reduced in stt4ts cells compared with wild-type cells, and the difference was even greater in cells grown at 38°C. Similar results were observed with mss4ts cells. These and other data confirm the hypothesis that Stt4 and Mss4 regulate IPC levels, and they do so independently of their known role in the activation of the Rho1/Pkc1 cell wall integrity pathway (reviewed in Ref. 89).
Because Slm1 and Slm2 operate downstream of the PIP2 and TORC2 pathways, they were examined for effects on IPC synthesis. IPC-C and other complex sphingolipids were less abundant in slm1ts slm2Δ cells grown at 26°C or 38°C, similar to what was found in stt4ts and mss4ts cells. The connection between the Slm proteins and sphingolipids was further strengthened by analyzing the effect of slm1 and slm2 mutations on csg2Δ cells. Interestingly, slm1ts slm2 csg2Δ cells did not grow, which implies a functional relation between the Slm protein and Csg2. By deleting one or the other SLM gene, it was observed that slm1Δcsg2Δ cells grew at 26°C but not 38°C and slm2Δcsg2Δ cells grew at both temperatures. The difference between these two types of double mutants is probably a reflection of Slm1 being more abundant than Slm2. These data suggest a functional interaction between Slm1 and Csg2. This idea is strengthened by the finding that the actin cytoskeleton is completely depolarized in slm1Δcsg2Δ cells grown at 38°C, whereas the single mutants show no defect.
The relationship between Slm1 and Csg2 was explored further by assessing the phosphorylation of Slm1 during heat stress. Although Slm1 phosphorylation increased during a 60 min heat shock in wild-type cells, there was almost no increase in csg2Δ cells. Because phosphorylation is mediated by TORC2, one interpretation of these data is that TORC2 function is disrupted by heat stress in csg2Δ cells. However, another interpretation is that a protein phosphatase is activated when csg2 is deleted. Because calcineurin subunits had been found to interact with Slm2 and both Slm2 and Slm1 have putative calcineurin binding motifs, the phosphorylation of Slm1 was examined in csg2Δ cells treated with the calcineurin inhibitor FK506. Phosphorylation of Slm1 was partially restored in FK506-treated cells, consistent with calcineurin being responsible for dephosphorylation in csg2Δ cells. Measurement of calcineurin activity showed that it was increased by ∼3-fold in csg2Δ cells, increased slightly in slm1Δ cells, and increased by 10-fold in slm1Δcsg2Δ double mutant cells. Likewise, it was increased in mss4ts and tor2ts cells. Together, these data indicate that Slm1, Slm2, and Csg2 cooperate to downregulate calcineurin activity (Fig. 4). Furthermore, they imply the regulation of IPC metabolism by calcineurin, and analysis of sphingolipids showed that activating calcineurin by adding 100 mM Ca2+ to cells accelerated the conversion of IPC to MIPC.
Fig. 4.
Phosphatidylinositol 4,5-bisphosphate (PI4,5P2) and the Slm1 and Slm2 proteins regulate the synthesis and turnover of complex sphingolipids and the dynamics of the actin cytoskeleton. The enzymes Stt4 and Mss4, which synthesize PI4,5P2 along with the TORC2 protein kinase, are proposed to activate Slm1 and Slm2, which then downregulate the turnover of complex sphingolipids, particularly inositol phosphoceramide (IPC), by the Isc1 enzyme. The Slm proteins also regulate the calcium/calmodulin-regulated protein phosphatase calcineurin, which dephosphorylates and inactivates the Slm proteins and also interacts with Csg1/2 in an unknown manner to regulate the conversion of IPC to mannose inositol phosphoceramide by the Csg1/2 enzymes. Slm regulation of the Rho1/Pkc1 pathway is independent from the regulation of sphingolipid metabolism. Genetic interactions suggest that IPC plays a role in actin organization, but the mechanism is unknown. PI, phosphatidylinositol; PI4P, phosphatidylinositol 4-phosphate.
How might calcineurin regulate the level of IPCs in cells? Tabuchi et al. (82) focused on Isc1, because it degrades IPCs. They found a 2-fold increase in IPCs in isc1Δ cells. Also, deletion of ISC1, similar to deletion of calcineurin, restored IPC levels in slm1ts slm2Δ cells to wild-type levels and reversed the ts phenotype. Moreover, deletion of ISC1 rescued the nonviable phenotype of slm1Δcsg2Δ cells. These data imply that misregulation of the hyperactivation of Isc1 and calcineurin most likely explains the phenotypes associated with the loss of Slm1 and Slm2 activity. In fact, inactivation of both ISC1 and calcineurin rescued the actin and viability defects seen in slm1ts slm2Δ cells at the restrictive temperature.
The data of Tabuchi et al. (82) are summarized in the model diagrammed in Fig. 4. They propose that the Slm proteins function downstream of the PIP2 and TORC2 signaling pathways to enable cells to respond to stresses. Part of the response involves recruitment of the Slms to the plasma membrane by binding to PIP2, followed by phosphorylation and activation mediated by TORC2. Activated Slms then downregulate Isc1 activity and control the breakdown of complex sphingolipids, especially IPC-C. The Slms also downregulate calcineurin phosphatase activity, which interacts with Csg2 in an unknown manner to regulate the conversion of IPC-C to MIPC. Dephosphorylation of the Slm proteins by calcineurin acts as a negative feedback loop to modulate Slm function. In this context, the Slm proteins serve to modulate changes in membrane composition and/or architecture in response to stresses. Other results from Tabuchi et al. (82) demonstrate the PIP2 control of the Rho1/Pkc1 pathway independent of its control of sphingolipid metabolism. Finally, it is likely that IPC, possibly a specific pool of IPC-C, regulates actin organization and viability. Identifying this pool of IPC-C could provide important clues for understanding how such regulation occurs.
Overall, the realization that the PIP2 and TORC2 pathways use the Slm proteins and calcineurin to regulate sphingolipid metabolism provides a framework for a detailed mechanistic understanding of how sphingolipid synthesis and breakdown are integrated with the need for cells to grow and to be able to interrupt growth to respond to life-threatening stresses.
SECRETORY PATHWAY, ENDOCYTOSIS, AND THE PLASMA MEMBRANE
The very-long-chain C26 fatty acid in yeast sphingolipids must perform unique and essential functions, because mutant cells with shorter chain fatty acids are very feeble (reviewed in Ref. 7). Recent studies on the proton-pumping H+-ATPase, Pma1, reveal some of these functions (90). Pma1 has been a valuable model for determining how proteins are transported from the ER to the plasma membrane, and readers are directed to published reviews for more detailed information (91, 92). Like all integral membrane proteins, Pma1 becomes part of the hydrophobic membrane in the ER, where lipids must contribute to the microenvironment necessary for the correct folding of Pma1 into a functional conformation. Pma1 progresses next to the Golgi apparatus via vesicle transport and then onward to the plasma membrane, where it acts to maintain intracellular pH and establish a proton gradient, which is essential for transporting nutrients into yeast cells.
These new studies used strains carrying the semidominant SLC1-1 mutation, which carries a Q44L substitution in the Slc1 protein, a 1-acyl-sn-glycerol-3-phosphate acyltransferase (17). This mutation enables strains carrying lcb1Δ, which is normally lethal because cells cannot make LCBs, to survive by catalyzing the incorporation of C26 fatty acids into the sn-2 position of glycerolipids, which then mimic yeast ceramides and serve as substrates for the enzymes that add the polar head groups found in yeast sphingolipids (Fig. 1). Thus, because of the lcb1Δ mutation, SLC1-1 cells lack LCBs and ceramides but they contain a set of novel glycerol-based lipids with the polar head groups and C26 fatty acid found in yeast sphingolipids (16). When SLC1-1 cells are fed LCBs, they make sphingolipids and behave like wild-type cells.
Sphingolipids and sterols associate with each other in membranes to form microdomains or raft-like structures (93, 94) with high affinity for specific membrane proteins, including Pma1 (95). The new work by Gaigg, Toulmay, and Schneiter (90) shows that the C26-containing inositol glycerophospholipids in SLC1-1 cells can functionally substitute for sphingolipids in forming detergent-resistant membrane domains containing Pma1 and another raft-associated protein, Gas1. Gaigg, Toulmay, and Schneiter (90) also demonstrated that the C26-containing inositol glycerophospholipids in SLC1-1 cells substitute for the functions of sphingolipids in the secretory pathway and facilitate the normal delivery of Pma1 to the plasma membrane. The essential function(s) of the C26 acyl chain in the suppressor lipids was demonstrated by introducing a sur4Δ (elo3Δ) mutation into the SLC1-1 strain. Sur4 is the Elop necessary for elongating fatty acids up to 26 carbon atoms in length (Fig. 1). sur4Δ cells make C22-containing sphingolipids and are viable (35), but SLC1-1 lcb1Δsur4Δ cells are not viable unless they are fed an LCB so that they can make sphingolipids. These experiments establish that the C26 acyl group in suppressor lipids is essential and that two of its essential roles are to direct Pma1 to the plasma membrane and to stabilize it in a detergent-resistant membrane environment (90).
Gaigg, Toulmay, and Schneiter (90) also examined endocytosis in SLC1-1 lcb1Δ cells that lack sphingolipids, because the stability of Pma1 in the plasma membrane of these cells could simply be attributable to a block in endocytosis. However, this seems not to be the case, because uptake of the fluorescent dye Lucifer Yellow, a marker for fluid-phase endocytosis, was similar in SLC1-1 lcb1Δ cells that contained or lacked sphingolipids and in wild-type cells. These results create a conundrum, because previous data showed that LCBs are essential for endocytosis (96, 97). Gaigg, Toulmay, and Schneiter (90) suggest that the protein phosphatase PP2A, a negative regulator of endocytosis (98), may be less active in SLC1-1 lcb1Δ cells because they lack ceramide, which is thought to be an activator of PP2A (99, 100). This explanation probably represents only part of a very complex picture, because the absence of sphingolipids in SLC1-1 lcb1Δ cells likely increases some and decreases other regulatory pathways to distort cellular processes.
Results from another recent study (30) exemplify how yeast cells can adapt to changes in their sphingolipids and still remain viable. In that study, a strain lacking the yeast ceramide synthases Lag1 and Lac1 (Fig. 1) and expressing the mammalian ceramide synthase Lass5 made sphingolipids containing primarily DHS, rather than the usual PHS, and C16 and C18 fatty acids rather than the usual C24 and C26 fatty acids. Although 97% of the sphingolipids in these cells lacked C24 or C26 fatty acids, these VLCFAs were still essential, because a block in the fatty acid elongation cycle produced by deleting TSC13 (Fig. 1) was lethal. The authors suggest that the VLCFAs may be necessary for the synthesis of glycerolipids with a VLCFA in the sn-1 position (101).
Another plasma membrane protein, the general amino acid permease Gap1, depends in various ways upon sphingolipids (102). Gap1 synthesized in lcb1-100 cells at a restrictive temperature is transported to the plasma membrane but is not incorporated into detergent-resistant lipid rafts, lacks amino acid transport activity, and is rapidly endocytosed in a ubiquitin-mediated manner. The authors conclude that these phenotypes result from Gap1 synthesis in the ER in the absence of sphingolipids and that sphingolipids provide a microenvironment for producing Gap1 with a conformation necessary for transport activity, the association with lipid rafts, and normal stability.
The Slm proteins mentioned above have also been shown to play roles in endocytosis. Sphingolipids are normally involved in regulating the turnover and endocytosis of the uracil permease Fur4 during heat stress (97, 103–105). However, in cells lacking Slm protein activity, Fur4 was quite stable during heat shock and accumulated in a ubiquitinated form that would normally be a substrate for endocytosis but that fails to be endocytosed in slm1ts slm2Δ cells (83). Thus, the Slm proteins are required for a step in the endocytosis of Fur4 that follows ubiquitylation. The Slm proteins, however, are not required generally for endocytosis, because slm mutant cells do endocytose Lucifer Yellow at high temperatures (83).
SUMMARY AND FUTURE DIRECTIONS
We are now beginning to understand how sphingolipid synthesis and turnover are regulated, but current knowledge is far from complete and serves as a foundation upon which to build a more mechanistic understanding of regulatory mechanisms and how they are connected to and integrated with other cellular metabolism that promotes growth and survival. The central role of LCBs in regulating cellular processes is expanding, and it is becoming clearer that advancement in some areas is limited by our experimental inability to determine where changes in LCBs occur in cells. For example, there appear to be pools of PHS and IPCs with specialized roles, but their location is unknown. Likewise, it is not clear whether LCBs regulate Ypk1, Ypk2, and Sch9 directly (Fig. 3) and, if they do respond directly, where this occurs in cells. We assume that LCBs are the only upstream activators of Pkh1 and Pkh2, but this is based upon a very limited amount of experimental evidence, none of which involves direct enzymatic assay of the kinase activity of these enzymes and their dependence on LCBs. There may be other upstream activators.
Finally, nearly all of our current understanding of sphingolipid metabolism and functions comes from studies done with rapidly dividing cells growing in log phase and with glucose as the carbon source. Sphingolipids are likely to play novel roles under different growth conditions and in different parts of their life cycle, including stationary phase, spore formation, and germination, and in mature spores.
Acknowledgments
The advice, encouragement, knowledge, and friendship of the author's longtime colleague and collaborator, Robert Lester, is gratefully acknowledged.
Published, JLR Papers in Press, February 23, 2008.
Footnotes
Work in the author's laboratory was supported by Grant AG-024377 from the National Institutes of Health and by core facilities supported by Grant P20 RR-020171 from the National Center for Research Resources, a component of the National Institutes of Health.
References
- 1.Hannun Y. A., and R. M. Bell. 1989. Functions of sphingolipids and sphingolipid breakdown products in cellular regulation. Science. 243 500–507. [DOI] [PubMed] [Google Scholar]
- 2.Ogretmen B., and Y. A. Hannun. 2004. Biologically active sphingolipids in cancer pathogenesis and treatment. Nat. Rev. Cancer. 4 604–616. [DOI] [PubMed] [Google Scholar]
- 3.Oskeritzian C. A., S. Milstien, and S. Spiegel. 2007. Sphingosine-1-phosphate in allergic responses, asthma and anaphylaxis. Pharmacol. Ther. 115 390–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Savtchouk I. A., F. J. Mattie, and A. A. Ollis. 2007. Ceramide: from embryos to tumors. Sci. STKE. 2007 jc1–jc2. [DOI] [PubMed] [Google Scholar]
- 5.El Alwani M., B. X. Wu, L. M. Obeid, and Y. A. Hannun. 2006. Bioactive sphingolipids in the modulation of the inflammatory response. Pharmacol. Ther. 112 171–183. [DOI] [PubMed] [Google Scholar]
- 6.Carter H. E., and H. S. Hendrickson. 1963. Biochemistry of the sphingolipids. XV. Structure of phytosphingosine and dehydrophytosphingosine. Biochemistry. 2 389–393. [DOI] [PubMed] [Google Scholar]
- 7.Dickson R. C., and R. L. Lester. 1999. Metabolism and selected functions of sphingolipids in the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta. 1438 305–321. [DOI] [PubMed] [Google Scholar]
- 8.Dickson R. C., and R. L. Lester. 1999. Yeast sphingolipids. Biochim. Biophys. Acta. 1426 347–357. [DOI] [PubMed] [Google Scholar]
- 9.Dickson R. C., and R. L. Lester. 2002. Sphingolipid functions in Saccharomyces cerevisiae. Biochim. Biophys. Acta. 1583 13–25. [DOI] [PubMed] [Google Scholar]
- 10.Sims K. J., S. D. Spassieva, E. O. Voit, and L. M. Obeid. 2004. Yeast sphingolipid metabolism: clues and connections. Biochem. Cell Biol. 82 45–61. [DOI] [PubMed] [Google Scholar]
- 11.Cowart L. A., and L. M. Obeid. 2007. Yeast sphingolipids: recent developments in understanding biosynthesis, regulation, and function. Biochim. Biophys. Acta. 1771 421–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dickson R. C., C. Sumanasekera, and R. L. Lester. 2006. Functions and metabolism of sphingolipids in Saccharomyces cerevisiae. Prog. Lipid Res. 45 447–465. [DOI] [PubMed] [Google Scholar]
- 13.Cowart, L. A., and Y. A. Hannun. 2004. Baker's yeast: a rising foundation for eukaryotic sphingolipid-mediated cell signalling. In Topics in Current Genetics. Vol. 6. G. Daum, editor. Springer-Verlag, Berlin. 383–401.
- 14.Funato K., B. Vallee, and H. Riezman. 2002. Biosynthesis and trafficking of sphingolipids in the yeast Saccharomyces cerevisiae. Biochemistry. 41 15105–15114. [DOI] [PubMed] [Google Scholar]
- 15.Lester R. L., and R. C. Dickson. 2001. High-performance liquid chromatography analysis of molecular species of sphingolipid-related long chain bases and long chain base phosphates in Saccharomyces cerevisiae after derivatization with 6-aminoquinolyl-N-hydroxysuccinimidyl carbamate. Anal. Biochem. 298 283–292. [DOI] [PubMed] [Google Scholar]
- 16.Lester R. L., G. B. Wells, G. Oxford, and R. C. Dickson. 1993. Mutant strains of Saccharomyces cerevisiae lacking sphingolipids synthesize novel inositol glycerophospholipids that mimic sphingolipid structures. J. Biol. Chem. 268 845–856. [PubMed] [Google Scholar]
- 17.Nagiec M. M., G. B. Wells, R. L. Lester, and R. C. Dickson. 1993. A suppressor gene that enables Saccharomyces cerevisiae to grow without making sphingolipids encodes a protein that resembles an Escherichia coli fatty acyltransferase. J. Biol. Chem. 268 22156–22163. [PubMed] [Google Scholar]
- 18.Patton J. L., B. Srinivasan, R. C. Dickson, and R. L. Lester. 1992. Phenotypes of sphingolipid-dependent strains of Saccharomyces cerevisiae. J. Bacteriol. 174 7180–7184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hanada K. 2003. Serine palmitoyltransferase, a key enzyme of sphingolipid metabolism. Biochim. Biophys. Acta. 1632 16–30. [DOI] [PubMed] [Google Scholar]
- 20.Han G., K. Gable, L. Yan, M. J. Allen, W. H. Wilson, P. Moitra, J. M. Harmon, and T. M. Dunn. 2006. Expression of a novel marine viral single-chain serine palmitoyltransferase and construction of yeast and mammalian single-chain chimera. J. Biol. Chem. 281 39935–39942. [DOI] [PubMed] [Google Scholar]
- 21.Gable K., H. Slife, D. Bacikova, E. Monaghan, and T. M. Dunn. 2000. Tsc3p is an 80-amino acid protein associated with serine palmitoyltransferase and required for optimal enzyme activity. J. Biol. Chem. 275 7597–7603. [DOI] [PubMed] [Google Scholar]
- 22.Hornemann T., S. Richard, M. F. Rutti, Y. Wei, and A. von Eckardstein. 2006. Cloning and initial characterization of a new subunit for mammalian serine-palmitoyltransferase. J. Biol. Chem. 281 37275–37281. [DOI] [PubMed] [Google Scholar]
- 23.Hornemann T., Y. Wei, and A. von Eckardstein. 2007. Is the mammalian serine palmitoyltransferase a high-molecular-mass complex? Biochem. J. 405 157–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beeler T., D. Bacikova, K. Gable, L. Hopkins, C. Johnson, H. Slife, and T. Dunn. 1998. The Saccharomyces cerevisiae TSC10/YBR265w gene encoding 3-ketosphinganine reductase is identified in a screen for temperature-sensitive suppressors of the Ca2+-sensitive csg2 Delta mutant. J. Biol. Chem. 273 30688–30694. [DOI] [PubMed] [Google Scholar]
- 25.Guillas I., P. A. Kirchman, R. Chuard, M. Pfefferli, J. C. Jiang, S. M. Jazwinski, and A. Conzelmann. 2001. C26-CoA-dependent ceramide synthesis of Saccharomyces cerevisiae is operated by Lag1p and Lac1p. EMBO J. 20 2655–2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schorling S., B. Vallee, W. P. Barz, H. Riezman, and D. Oesterhelt. 2001. Lag1p and Lac1p are essential for the acyl-CoA-dependent ceramide synthase reaction in Saccharomyces cerevisiae. Mol. Biol. Cell. 12 3417–3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haak D., K. Gable, T. Beeler, and T. Dunn. 1997. Hydroxylation of Saccharomyces cerevisiae ceramides requires Sur2p and Scs7p. J. Biol. Chem. 272 29704–29710. [DOI] [PubMed] [Google Scholar]
- 28.Grilley M. M., S. D. Stock, R. C. Dickson, R. L. Lester, and J. Y. Takemoto. 1998. Syringomycin action gene SYR2 is essential for sphingolipid 4-hydroxylation in Saccharomyces cerevisiae. J. Biol. Chem. 273 11062–11068. [DOI] [PubMed] [Google Scholar]
- 29.Vallee B., and H. Riezman. 2005. Lip1p: a novel subunit of acyl-CoA ceramide synthase. EMBO J. 24 730–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cerantola V., C. Vionnet, O. F. Aebischer, T. Jenny, J. Knudsen, and A. Conzelmann. 2007. Yeast sphingolipids do not need to contain very long chain fatty acids. Biochem. J. 401 205–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith S. W., and R. L. Lester. 1974. Inositol phosphorylceramide, a novel substance and a chief member of a major group of yeast sphingolipids containing single inositol phosphate. J. Biol. Chem. 249 3395–3405. [PubMed] [Google Scholar]
- 32.Mitchell A. G., and C. E. Martin. 1997. Fah1p, a Saccharomyces cerevisiae cytochrome b5 fusion protein, and its Arabidopsis thaliana homolog that lacks the cytochrome b5 domain both function in the alpha-hydroxylation of sphingolipid-associated very long chain fatty acids. J. Biol. Chem. 272 28281–28288. [DOI] [PubMed] [Google Scholar]
- 33.Dunn T. M., D. Haak, E. Monaghan, and T. J. Beeler. 1998. Synthesis of monohydroxylated inositolphosphorylceramide (IPC-C) in Saccharomyces cerevisiae requires Scs7p, a protein with both a cytochrome b5-like domain and a hydroxylase/desaturase domain. Yeast. 14 311–321. [DOI] [PubMed] [Google Scholar]
- 34.Toke D. A., and C. E. Martin. 1996. Isolation and characterization of a gene affecting fatty acid elongation in Saccharomyces cerevisiae. J. Biol. Chem. 271 18413–18422. [DOI] [PubMed] [Google Scholar]
- 35.Oh C. S., D. A. Toke, S. Mandala, and C. E. Martin. 1997. ELO2 and ELO3, homologues of the Saccharomyces cerevisiae ELO1 gene, function in fatty acid elongation and are required for sphingolipid formation. J. Biol. Chem. 272 17376–17384. [DOI] [PubMed] [Google Scholar]
- 36.Jakobsson A., R. Westerberg, and A. Jacobsson. 2006. Fatty acid elongases in mammals: their regulation and roles in metabolism. Prog. Lipid Res. 45 237–249. [DOI] [PubMed] [Google Scholar]
- 37.Beaudoin F., K. Gable, O. Sayanova, T. Dunn, and J. A. Napier. 2002. A Saccharomyces cerevisiae gene required for heterologous fatty acid elongase activity encodes a microsomal beta-keto-reductase. J. Biol. Chem. 277 11481–11488. [DOI] [PubMed] [Google Scholar]
- 38.Han G., K. Gable, S. D. Kohlwein, F. Beaudoin, J. A. Napier, and T. M. Dunn. 2002. The Saccharomyces cerevisiae YBR159w gene encodes the 3-ketoreductase of the microsomal fatty acid elongase. J. Biol. Chem. 277 35440–35449. [DOI] [PubMed] [Google Scholar]
- 39.Kohlwein S. D., S. Eder, C. S. Oh, C. E. Martin, K. Gable, D. Bacikova, and T. Dunn. 2001. Tsc13p is required for fatty acid elongation and localizes to a novel structure at the nuclear-vacuolar interface in Saccharomyces cerevisiae. Mol. Cell. Biol. 21 109–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Denic V., and J. S. Weissman. 2007. A molecular caliper mechanism for determining very long-chain fatty acid length. Cell. 130 663–677. [DOI] [PubMed] [Google Scholar]
- 41.Schuldiner M., S. R. Collins, N. J. Thompson, V. Denic, A. Bhamidipati, T. Punna, J. Ihmels, B. Andrews, C. Boone, J. F. Greenblatt, et al. 2005. Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell. 123 507–519. [DOI] [PubMed] [Google Scholar]
- 42.Funato K., and H. Riezman. 2001. Vesicular and nonvesicular transport of ceramide from ER to the Golgi apparatus in yeast. J. Cell Biol. 155 949–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hanada K., K. Kumagai, S. Yasuda, Y. Miura, M. Kawano, M. Fukasawa, and M. Nishijima. 2003. Molecular machinery for non-vesicular trafficking of ceramide. Nature. 426 803–809. [DOI] [PubMed] [Google Scholar]
- 44.Kumagai K., S. Yasuda, K. Okemoto, M. Nishijima, S. Kobayashi, and K. Hanada. 2005. CERT mediates intermembrane transfer of various molecular species of ceramides. J. Biol. Chem. 280 6488–6495. [DOI] [PubMed] [Google Scholar]
- 45.Nagiec M. M., E. E. Nagiec, J. A. Baltisberger, G. B. Wells, R. L. Lester, and R. C. Dickson. 1997. Sphingolipid synthesis as a target for antifungal drugs—complementation of the inositol phosphorylceramide synthase defect in a strain of Saccharomyces cerevisiae by the AUR1 gene. J. Biol. Chem. 272 9809–9817. [DOI] [PubMed] [Google Scholar]
- 46.Levine T. P., C. A. Wiggins, and S. Munro. 2000. Inositol phosphorylceramide synthase is located in the Golgi apparatus of Saccharomyces cerevisiae. Mol. Biol. Cell. 11 2267–2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Uemura S., A. Kihara, J. Inokuchi, and Y. Igarashi. 2003. Csg1p and newly identified Csh1p function in mannosylinositol phosphorylceramide synthesis by interacting with Csg2p. J. Biol. Chem. 278 45049–45055. [DOI] [PubMed] [Google Scholar]
- 48.Uemura S., A. Kihara, S. Iwaki, J. Inokuchi, and Y. Igarashi. 2007. Regulation of the transport and protein levels of the inositol phosphorylceramide mannosyltransferases Csg1 and Csh1 by the Ca2+-binding protein Csg2. J. Biol. Chem. 282 8613–8621. [DOI] [PubMed] [Google Scholar]
- 49.Beeler T., K. Gable, C. Zhao, and T. Dunn. 1994. A novel protein, CSG2p, is required for Ca2+ regulation in Saccharomyces cerevisiae. J. Biol. Chem. 269 7279–7284. [PubMed] [Google Scholar]
- 50.Hechtberger P., and G. Daum. 1995. Intracellular transport of inositol-containing sphingolipids in the yeast, Saccharomyces cerevisiae. FEBS Lett. 367 201–204. [DOI] [PubMed] [Google Scholar]
- 51.Raas-Rothschild A., I. Pankova-Kholmyansky, Y. Kacher, and A. H. Futerman. 2004. Glycosphingolipidoses: beyond the enzymatic defect. Glycoconj. J. 21 295–304. [DOI] [PubMed] [Google Scholar]
- 52.Patton J. L., and R. L. Lester. 1991. The phosphoinositol sphingolipids of Saccharomyces cerevisiae are highly localized in the plasma membrane. J. Bacteriol. 173 3101–3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sawai H., Y. Okamoto, C. Luberto, C. Mao, A. Bielawska, N. Domae, and Y. A. Hannun. 2000. Identification of ISC1 (YER019w) as inositol phosphosphingolipid phospholipase C in Saccharomyces cerevisiae. J. Biol. Chem. 275 39793–39798. [DOI] [PubMed] [Google Scholar]
- 54.Wells G. B., R. C. Dickson, and R. L. Lester. 1998. Heat-induced elevation of ceramide in Saccharomyces cerevisiae via de novo synthesis. J. Biol. Chem. 273 7235–7243. [DOI] [PubMed] [Google Scholar]
- 55.Betz C., D. Zajonc, M. Moll, and E. Schweizer. 2002. ISC1-encoded inositol phosphosphingolipid phospholipase C is involved in Na+/Li+ halotolerance of Saccharomyces cerevisiae. Eur. J. Biochem. 269 4033–4039. [DOI] [PubMed] [Google Scholar]
- 56.Vaena de Avalos S., X. Su, M. Zhang, Y. Okamoto, W. Dowhan, and Y. A. Hannun. 2005. The phosphatidylglycerol/cardiolipin biosynthetic pathway is required for the activation of inositol phosphosphingolipid phospholipase C, Isc1p, during growth of Saccharomyces cerevisiae. J. Biol. Chem. 280 7170–7177. [DOI] [PubMed] [Google Scholar]
- 57.de Avalos S. V., Y. Okamoto, and Y. A. Hannun. 2004. Activation and localization of inositol phosphosphingolipid phospholipase C, Isc1p, to the mitochondria during growth of Saccharomyces cerevisiae. J. Biol. Chem. 279 11537–11545. [DOI] [PubMed] [Google Scholar]
- 58.Kitagaki H., L. A. Cowart, N. Matmati, S. Vaena de Avalos, S. A. Novgorodov, Y. H. Zeidan, J. Bielawski, L. M. Obeid, and Y. A. Hannun. 2007. Isc1 regulates sphingolipid metabolism in yeast mitochondria. Biochim. Biophys. Acta. 1768 2849–2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jenkins G. M., A. Richards, T. Wahl, C. Mao, L. Obeid, and Y. Hannun. 1997. Involvement of yeast sphingolipids in the heat stress response of Saccharomyces cerevisiae. J. Biol. Chem. 272 32566–32572. [DOI] [PubMed] [Google Scholar]
- 60.Cowart L. A., Y. Okamoto, X. Lu, and Y. A. Hannun. 2006. Distinct roles for de novo versus hydrolytic pathways of sphingolipid biosynthesis in Saccharomyces cerevisiae. Biochem. J. 393 733–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Aronova S., K. Wedaman, P. A. Aronov, K. Fontes, K. Ramos, B. D. Hammock, and T. Powers. 2008. TOR complex 2 regulates sphingolipid biosynthesis at the conserved step of de novo ceramide formation via the AGC kinase Ypk2. Cell Metab. 7 148–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wullschleger S., R. Loewith, and M. N. Hall. 2006. TOR signaling in growth and metabolism. Cell. 124 471–484. [DOI] [PubMed] [Google Scholar]
- 63.Kamada Y., Y. Fujioka, N. N. Suzuki, F. Inagaki, S. Wullschleger, R. Loewith, M. N. Hall, and Y. Ohsumi. 2005. Tor2 directly phosphorylates the AGC kinase Ypk2 to regulate actin polarization. Mol. Cell. Biol. 25 7239–7248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wu W. I., V. M. McDonough, J. T. Nickels, Jr., J. Ko, A. S. Fischl, T. R. Vales, A. H. Merrill, Jr., and G. M. Carman. 1995. Regulation of lipid biosynthesis in Saccharomyces cerevisiae by fumonisin B1. J. Biol. Chem. 270 13171–13178. [DOI] [PubMed] [Google Scholar]
- 65.Dickson R. C., E. E. Nagiec, M. Skrzypek, P. Tillman, G. B. Wells, and R. L. Lester. 1997. Sphingolipids are potential heat stress signals in Saccharomyces. J. Biol. Chem. 272 30196–30200. [DOI] [PubMed] [Google Scholar]
- 66.Liu K., X. Zhang, R. L. Lester, and R. C. Dickson. 2005. The sphingoid long chain base phytosphingosine activates AGC-type protein kinases in Saccharomyces cerevisiae including Ypk1, Ypk2, and Sch9. J. Biol. Chem. 280 22679–22687. [DOI] [PubMed] [Google Scholar]
- 67.Pewzner-Jung Y., S. Ben-Dor, and A. H. Futerman. 2006. When do Lasses (longevity assurance genes) become CerS (ceramide synthases)? Insights into the regulation of ceramide synthesis. J. Biol. Chem. 281 25001–25005. [DOI] [PubMed] [Google Scholar]
- 68.Bose R., M. Verheij, A. Haimovitz-Friedman, K. Scotto, Z. Fuks, and R. Kolesnick. 1995. Ceramide synthase mediates daunorubicin-induced apoptosis: an alternative mechanism for generating death signals. Cell. 82 405–414. [DOI] [PubMed] [Google Scholar]
- 69.Senkal C. E., S. Ponnusamy, M. J. Rossi, J. Bialewski, D. Sinha, J. C. Jiang, S. M. Jazwinski, Y. A. Hannun, and B. Ogretmen. 2007. Role of human longevity assurance gene 1 and C18-ceramide in chemotherapy-induced cell death in human head and neck squamous cell carcinomas. Mol. Cancer Ther. 6 712–722. [DOI] [PubMed] [Google Scholar]
- 70.Cowart L. A., and Y. A. Hannun. 2007. Selective substrate supply in the regulation of yeast de novo sphingolipid synthesis. J. Biol. Chem. 282 12330–12340. [DOI] [PubMed] [Google Scholar]
- 71.Merrill A. H., Jr., E. Wang, and R. E. Mullins. 1988. Kinetics of long-chain (sphingoid) base biosynthesis in intact LM cells: effects of varying the extracellular concentrations of serine and fatty acid precursors of this pathway. Biochemistry. 27 340–345. [DOI] [PubMed] [Google Scholar]
- 72.Smith E. R., and A. H. Merrill, Jr. 1995. Differential roles of de novo sphingolipid biosynthesis and turnover in the “burst” of free sphingosine and sphinganine, and their 1-phosphates and N-acyl-derivatives, that occurs upon changing the medium of cells in culture. J. Biol. Chem. 270 18749–18758. [DOI] [PubMed] [Google Scholar]
- 73.Sun Y., R. Taniguchi, D. Tanoue, T. Yamaji, H. Takematsu, K. Mori, T. Fujita, T. Kawasaki, and Y. Kozutsumi. 2000. Sli2 (Ypk1), a homologue of mammalian protein kinase SGK, is a downstream kinase in the sphingolipid-mediated signaling pathway of yeast. Mol. Cell. Biol. 20 4411–4419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Miyake Y., Y. Kozutsumi, S. Nakamura, T. Fujita, and T. Kawasaki. 1995. Serine palmitoyltransferase is the primary target of a sphingosine-like immunosuppressant, ISP-1/myriocin. Biochem. Biophys. Res. Commun. 211 396–403. [DOI] [PubMed] [Google Scholar]
- 75.Schmelzle T., S. B. Helliwell, and M. N. Hall. 2002. Yeast protein kinases and the RHO1 exchange factor TUS1 are novel components of the cell integrity pathway in yeast. Mol. Cell. Biol. 22 1329–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Roelants F. M., P. D. Torrance, N. Bezman, and J. Thorner. 2002. Pkh1 and Pkh2 differentially phosphorylate and activate Ypk1 and Ykr2 and define protein kinase modules required for maintenance of cell wall integrity. Mol. Biol. Cell. 13 3005–3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.deHart A. K., J. D. Schnell, D. A. Allen, and L. Hicke. 2002. The conserved Pkh-Ypk kinase cascade is required for endocytosis in yeast. J. Cell Biol. 156 241–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gelperin D., L. Horton, A. DeChant, J. Hensold, and S. K. Lemmon. 2002. Loss of ypk1 function causes rapamycin sensitivity, inhibition of translation initiation and synthetic lethality in 14-3-3-deficient yeast. Genetics. 161 1453–1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Casamayor A., P. D. Torrance, T. Kobayashi, J. Thorner, and D. R. Alessi. 1999. Functional counterparts of mammalian protein kinases PDK1 and SGK in budding yeast. Curr. Biol. 9 186–197. [DOI] [PubMed] [Google Scholar]
- 80.Brace J. L., R. L. Lester, R. C. Dickson, and C. M. Rudin. 2007. SVF1 regulates cell survival by affecting sphingolipid metabolism in Saccharomyces cerevisiae. Genetics. 175 65–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Vander Heiden M. G., J. S. Choy, D. J. VanderWeele, J. L. Brace, M. H. Harris, D. E. Bauer, B. Prange, S. J. Kron, C. B. Thompson, and C. M. Rudin. 2002. Bcl-x(L) complements Saccharomyces cerevisiae genes that facilitate the switch from glycolytic to oxidative metabolism. J. Biol. Chem. 277 44870–44876. [DOI] [PubMed] [Google Scholar]
- 82.Tabuchi M., A. Audhya, A. B. Parsons, C. Boone, and S. D. Emr. 2006. The phosphatidylinositol 4,5-biphosphate and TORC2 binding proteins Slm1 and Slm2 function in sphingolipid regulation. Mol. Cell. Biol. 26 5861–5875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bultynck G., V. L. Heath, A. P. Majeed, J. M. Galan, R. Haguenauer-Tsapis, and M. S. Cyert. 2006. Slm1 and Slm2 are novel substrates of the calcineurin phosphatase required for heat stress-induced endocytosis of the yeast uracil permease. Mol. Cell. Biol. 26 4729–4745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Daquinag A., M. Fadri, S. Y. Jung, J. Qin, and J. Kunz. 2007. The yeast PH domain proteins Slm1 and Slm2 are targets of sphingolipid signaling during the response to heat stress. Mol. Cell. Biol. 27 633–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Audhya A., R. Loewith, A. B. Parsons, L. Gao, M. Tabuchi, H. Zhou, C. Boone, M. N. Hall, and S. D. Emr. 2004. Genome-wide lethality screen identifies new PI4,5P2 effectors that regulate the actin cytoskeleton. EMBO J. 23 3747–3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yu J. W., J. M. Mendrola, A. Audhya, S. Singh, D. Keleti, D. B. DeWald, D. Murray, S. D. Emr, and M. A. Lemmon. 2004. Genome-wide analysis of membrane targeting by S. cerevisiae pleckstrin homology domains. Mol. Cell. 13 677–688. [DOI] [PubMed] [Google Scholar]
- 87.Fadri M., A. Daquinag, S. Wang, T. Xue, and J. Kunz. 2005. The pleckstrin homology domain proteins Slm1 and Slm2 are required for actin cytoskeleton organization in yeast and bind phosphatidylinositol-4,5-bisphosphate and TORC2. Mol. Biol. Cell. 16 1883–1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kobayashi T., H. Takematsu, T. Yamaji, S. Hiramoto, and Y. Kozutsumi. 2005. Disturbance of sphingolipid biosynthesis abrogates the signaling of Mss4, phosphatidylinositol-4-phosphate 5-kinase, in yeast. J. Biol. Chem. 280 18087–18094. [DOI] [PubMed] [Google Scholar]
- 89.Levin D. E. 2005. Cell wall integrity signaling in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 69 262–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gaigg B., A. Toulmay, and R. Schneiter. 2006. Very long-chain fatty acid-containing lipids rather than sphingolipids per se are required for raft association and stable surface transport of newly synthesized plasma membrane ATPase in yeast. J. Biol. Chem. 281 34135–34145. [DOI] [PubMed] [Google Scholar]
- 91.Schneiter R., and A. Toulmay. 2007. The role of lipids in the biogenesis of integral membrane proteins. Appl. Microbiol. Biotechnol. 73 1224–1232. [DOI] [PubMed] [Google Scholar]
- 92.Toulmay A., and R. Schneiter. 2007. Lipid-dependent surface transport of the proton pumping ATPase: a model to study plasma membrane biogenesis in yeast. Biochimie. 89 249–254. [DOI] [PubMed] [Google Scholar]
- 93.Simons K., and W. L. Vaz. 2004. Model systems, lipid rafts, and cell membranes. Annu. Rev. Biophys. Biomol. Struct. 33 269–295. [DOI] [PubMed] [Google Scholar]
- 94.Ramstedt B., and J. P. Slotte. 2002. Membrane properties of sphingomyelins. FEBS Lett. 531 33–37. [DOI] [PubMed] [Google Scholar]
- 95.Bagnat M., S. Keranen, A. Shevchenko, and K. Simons. 2000. Lipid rafts function in biosynthetic delivery of proteins to the cell surface in yeast. Proc. Natl. Acad. Sci. USA. 97 3254–3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zanolari B., S. Friant, K. Funato, C. Sutterlin, B. J. Stevenson, and H. Riezman. 2000. Sphingoid base synthesis requirement for endocytosis in Saccharomyces cerevisiae. EMBO J. 19 2824–2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chung N., G. Jenkins, Y. A. Hannun, J. Heitman, and L. M. Obeid. 2000. Sphingolipids signal heat stress-induced ubiquitin-dependent proteolysis. J. Biol. Chem. 275 17229–17232. [DOI] [PubMed] [Google Scholar]
- 98.Friant S., B. Zanolari, and H. Riezman. 2000. Increased protein kinase or decreased PP2A activity bypasses sphingoid base requirement in endocytosis. EMBO J. 19 2834–2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Nickels J. T., and J. R. Broach. 1996. A ceramide-activated protein phosphatase mediates ceramide-induced G1 arrest of Saccharomyces cerevisiae. Genes Dev. 10 382–394. [DOI] [PubMed] [Google Scholar]
- 100.Gurunathan S., M. Marash, A. Weinberger, and J. E. Gerst. 2002. t-SNARE phosphorylation regulates endocytosis in yeast. Mol. Biol. Cell. 13 1594–1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Schneiter R., B. Brugger, C. M. Amann, G. D. Prestwich, R. F. Epand, G. Zellnig, F. T. Wieland, and R. M. Epand. 2004. Identification and biophysical characterization of a very-long-chain-fatty-acid-substituted phosphatidylinositol in yeast subcellular membranes. Biochem. J. 381 941–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lauwers E., G. Grossmann, and B. Andre. 2007. Evidence for coupled biogenesis of yeast Gap1 permease and sphingolipids: essential role in transport activity and normal control by ubiquitination. Mol. Biol. Cell. 18 3068–3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Volland C., D. Urban-Grimal, G. Geraud, and R. Haguenauer-Tsapis. 1994. Endocytosis and degradation of the yeast uracil permease under adverse conditions. J. Biol. Chem. 269 9833–9841. [PubMed] [Google Scholar]
- 104.Hearn J. D., R. L. Lester, and R. C. Dickson. 2003. The uracil transporter Fur4p associates with lipid rafts. J. Biol. Chem. 278 3679–3686. [DOI] [PubMed] [Google Scholar]
- 105.Dupre S., and R. Haguenauer-Tsapis. 2003. Raft partitioning of the yeast uracil permease during trafficking along the endocytic pathway. Traffic. 4 83–96. [DOI] [PubMed] [Google Scholar]
- 106.Futerman A. H., and H. Riezman. 2005. The ins and outs of sphingolipid synthesis. Trends Cell Biol. 15 312–318. [DOI] [PubMed] [Google Scholar]
- 107.Mao C., R. Xu, A. Bielawska, Z. M. Szulc, and L. M. Obeid. 2000. Cloning and characterization of a Saccharomyces cerevisiae alkaline ceramidase with specificity for dihydroceramide. J. Biol. Chem. 275 31369–31378. [DOI] [PubMed] [Google Scholar]
- 108.Mao C., R. Xu, A. Bielawska, and L. M. Obeid. 2000. Cloning of an alkaline ceramidase from Saccharomyces cerevisiae. An enzyme with reverse (CoA-independent) ceramide synthase activity. J. Biol. Chem. 275 6876–6884. [DOI] [PubMed] [Google Scholar]
- 109.Urban J., A. Soulard, A. Huber, S. Lippman, D. Mukhopadhyay, O. Deloche, V. Wanke, D. Anrather, G. Ammerer, H. Riezman, et al. 2007. Sch9 is a major target of TORC1 in Saccharomyces cerevisiae. Mol. Cell. 26 663–674. [DOI] [PubMed] [Google Scholar]