Abstract
Conjugated linoleic acid (CLA) inhibits rat mammary carcinogenesis, in part by inducing apoptosis of preneoplastic and neoplastic mammary epithelial cells. The current study focused on the mechanism by which apoptosis is induced. In TM4t mammary tumor cells, trans-10,cis-12 (t10,c12)-CLA induced proapoptotic C/EBP-homologous protein (CHOP) concurrent with the cleavage of poly(ADP-ribose) polymerase. Knockdown of CHOP attenuated t10,c12-CLA-induced apoptosis. Furthermore, t10,c12-CLA induced the cleavage of endoplasmic reticulum (ER)-resident caspase-12, and a selective inhibitor of caspase-12 significantly alleviated t10,c12-CLA-induced apoptosis. Using electron microscopy, we observed that t10,c12-CLA treatment resulted in marked dilatation of the ER lumen. Together, these data suggest that t10,c12-CLA induces apoptosis through ER stress. To further explore the ER stress pathway, we examined the expression of the following upstream ER stress signature markers in response to CLA treatment: X-box binding protein 1 (XBP1) mRNA (unspliced and spliced), phospho-eukaryotic initiation factor (eIF) 2α, activating transcription factor 4 (ATF4), and BiP proteins. We found that t10,c12-CLA induced the expression and splicing of XBP1 mRNA as well as the phosphorylation of eIF2α. In contrast, ATF4 was induced modestly, but not significantly, and BiP was not altered. In summary, our data demonstrate that apoptosis induced by t10,c12-CLA is mediated, at least in part, through an atypical ER stress response that culminates in the induction of CHOP and the cleavage of caspase-12.
Keywords: mammary; trans-10,cis-12; CLA
The endoplasmic reticulum (ER) plays an important role in the biosynthesis of proteins and membrane lipids. Proteins are synthesized on the cytoplasmic surface of the ER, and those destined for secretion or for transmembrane compartments are translocated into the ER, where they undergo posttranslational modifications such as disulfide bond formation or glycosylation and are correctly folded and assembled to their final three-dimensional conformation. Any disruption to this closely regulated maturation event, such as may occur by changes in the ER microenvironment, can lead to the accumulation and/or aggregation of unfolded or misfolded proteins. As a result, a coordinated unfolded protein response or ER stress response is initiated within the cell (1), with the goal of rescuing the cell if the stress is not prolonged or severe.
The unfolded protein response is typically regulated by three ER transmembrane proteins, PERK (for double-stranded RNA-dependent protein kinase (PKR)-like endoplasmic reticulum-resident kinase), ATF6 (for activating transcription factor 6), and IRE1 (for inositol-requiring enzyme 1), which are bound to the ER-resident chaperone BiP/GRP78 (for 78 kDa glucose-regulated protein) in nonstressed cells (2). Upon sensing stress within the ER, BiP dissociates from these three transducers, facilitating their activation, and binds to and stabilizes the unfolded/misfolded proteins (3). Active PERK phosphorylates the eukaryotic initiation factor eIF2α, which inactivates it, resulting in translation attenuation (4). ATF6 is cleaved by the S1 and S2 proteases of the Golgi, and the cleaved ATF6 binds to endoplasmic reticulum stress response elements (ERSEs) in the promoters of the C/EBP-homologous protein (CHOP), X-box binding protein 1 (XBP1), and BiP genes, among others, resulting in their transcription (5–9). In addition, the endoRNase activity of active IRE1 cleaves a small intron from XBP1 mRNA, and translation of the resultant spliced XBP1 mRNA generates the active transcription factor XBP1, which can bind to ERSE and initiate the transcription of a distinct set of mRNAs, including the ER degradation-enhancing mannosidase-like protein and the proapoptotic protein CHOP (10, 11).
With prolonged or severe ER stress, cells undergo apoptosis. This ER response was recently recognized as a third major pathway implicated in apoptosis, in addition to the classical death receptor (“extrinsic”) and mitochondrial (“intrinsic”) pathways. The mechanism of ER stress-induced apoptosis is not fully understood; however, the proapoptotic protein CHOP and the ER-resident caspase-12 have been suggested to play key roles (11, 12).
Conjugated linoleic acid (CLA) is a family of C18 fatty acids with conjugated double bonds. Isomers of CLA, in particular c9,t11-CLA, are found naturally in dairy products and ruminant meats. In addition, synthetically produced CLA supplements, generally containing an equal mixture of trans-10,cis-12 (t10,c12)-CLA and c9,t11-CLA, are available in health food stores, and a mixture of these two isomers is being tested clinically for potential health benefits in obesity, including fat loss and improvement in insulin sensitivity (13, 14). Importantly, a number of studies have demonstrated that dietary CLA has preventive activity in models of mammary, gastric, and skin cancer (reviewed in Ref. 15), and both the c9,t11- and t10,c12-CLA isomers were found to be equally efficacious at inhibiting N-nitroso-N-methylurea-induced rat mammary carcinogenesis (16), metastasis from a transplantable mouse mammary tumor (17), and angiogenesis (18).
At the cellular level, CLA exerts its effects by inducing apoptosis of mammary tumor cells in vitro and preneoplastic lesions in the rat mammary gland in vivo (19, 20). One of the pathways used by CLA to induce apoptosis is through the mitochondria, and we found that in p53 mutant mammary tumor cells, t10,c12-CLA induced a time- and concentration-dependent cleavage of caspases-9 and -3 as well as the release of cytochrome c from mitochondria to cytosol (20). Moreover, our data suggested that Bcl-2 played an important role in the apoptotic effect of CLA, because both mitochondrial and whole cell levels of Bcl-2 were decreased in cells treated with CLA and the overexpression of Bcl-2 significantly attenuated the apoptotic effect of CLA (20).
In addition to the mitochondrial membrane, Bcl-2 localizes to the ER membrane, where it has been shown to be functional in blocking apoptosis in some cell lines (21, 22). The protective effect of Bcl-2 in TM4t cells, as well as our current data that t10,c12-CLA induced a marked dilatation of the lumen of the ER, a phenomenon that has been linked to an ER stress response (23, 24), led us to investigate the role of ER stress in CLA-induced apoptosis.
MATERIALS AND METHODS
Cell culture and cell growth measurements
p53 mutant TM4t mouse mammary tumor cells were obtained from Dr. Dan Medina at Baylor College of Medicine (Houston, TX) and were cultured in DMEM-F12 supplemented with 2% adult bovine serum, 10 μg/ml insulin, 5 ng/ml epidermal growth factor, and 5 μg/ml gentamicin. Viable cell number was determined by counting the number of cells that excluded trypan blue (time dependence experiments) or by the thiazolyl blue tetrazolium bromide (MTT) assay [concentration dependence, small interfering RNA (siRNA), and caspase-12 inhibitor experiments]. For the latter, cells (104/well) were seeded in triplicate wells in 24-well plates, allowed to attach overnight, and then treated for 72 h without or with 10, 20, or 40 μM CLA. To evaluate viable cell number, 200 μl of MTT (5 mg/ml in PBS; Sigma-Aldrich, St. Louis, MO) was added to each well and the plates were incubated at 37°C for 4 h. After dissolving the purple formazan crystals in isopropanol, the centrifuged solutions were transferred to a 96-well microplate and read at a wavelength of 570 nm. The CLA isomers, ∼98% pure, were obtained from Larodan Fine Chemicals (Malmö, Sweden) and prepared as the sodium salt for use in culture (25). Thapsigargin (1 and 10 μM) and ionomycin (1 and 10 μg/ml) were obtained from Calbiochem (San Diego, CA) and Invitrogen (Carlsbad, CA), respectively.
Immunoblotting
For each experiment, 5 × 105 cells were plated in 100 mm dishes (0.5 × 105 cells/ml medium) then allowed to attach overnight before the addition of t10,c12-CLA. After CLA treatment for the indicated times and concentrations, attached and floating cells were collected and lysed with 300 μl of lysis buffer [1% (v/v) Triton X-100, 50 mM Tris, pH 8.0, 150 mM NaCl, 2 mM EDTA, 10 mM sodium phosphate, 10 mM sodium pyrophosphate, 5 mM sodium vanadate, 0.1% (w/v) SDS, 0.5% (w/v) sodium deoxycholate, 20 μg/ml leupeptin, 100 μg/ml soybean trypsin inhibitor, 120 μg/ml Pefabloc, 1 μM DTT, 1 μg/ml pepstatin A, 10 μg/ml aprotinin, and 1 μM NaF]. The protein concentration was measured using the Dc Protein Assay (Bio-Rad Laboratories). Lysates were separated by electrophoresis on a SDS/12% polyacrylamide gel and then transferred to polyvinylidene difluoride Western blotting membranes (Roche, Indianapolis, IN).
Antibodies
Primary antibodies against cleaved poly(ADP-ribose) polymerase (PARP) (9544), phospho(Ser51)-eIF2α (9721), eIF2α (9722), and caspase-12 (2202) were purchased from Cell Signaling Technology (Danvers, MA). BiP antibody (SPA-827) was purchased from Stressgen Biotechnologies (Victoria, British Columbia, Canada); this antibody also recognizes the chaperone GRP94. ATF4 (SC-200) and CHOP (SC-13968) antibodies were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Actin antibody (CP01) was purchased from Calbiochem (La Jolla, CA). The Bcl-2 antibody (D038-3) was purchased from Medical and Biological Laboratories Co. (Woburn, MA). These antibodies were used at the dilutions recommended by each supplier.
siRNA transfection
A Lipofectamine™ 2000 kit was used for the transfection. Negative control siRNA (4611) and CHOP siRNA (288792 and 288791) were purchased from Ambion (Austin, TX). The first CHOP siRNA targets exon 4 and the second targets exon 3. The sense and antisense siRNA sequences (5′→3′) were CGGAAACAGAGUGGUCAGUtt and ACUGACCACUCUGUUUCCGtt for the first siRNA (288792) and GCCUGGUAUGAGGAUCUGCtt and GCAGAUCCUCAUACCAGGCtt for the second siRNA (288791). For the MTT assay, TM4t cells were plated on 24-well plates (2 × 104 cells/well) and 20 pmol of siRNA was used for transfection. For Western blot, TM4t cells were plated in six-well plates (1 × 105 cells/well) and 75 pmol of siRNA was used for transfection. After transfection, cells were left in culture with the siRNA for 6 h, then the siRNA was removed by replacing with 0 or 40 μM CLA-containing medium. The assays were performed after 72 h of CLA treatment.
Inhibition of caspase-12
A synthetic peptide, Z-ATAD-FMK, that irreversibly inhibits caspase-12 activity was purchased from Biovision (Mountain View, CA; catalog No. 1079-100). TM4t cells (104/well for 24-well plates and 105/well for 6-well plates) were seeded and allowed to attach overnight. They were then pretreated with the caspase-12 inhibitor (1:1,000 or 1:500 in DMSO) for 30 min, followed by treatment with 0 or 40 μM CLA for 72 h. The inhibitor was left in the medium during CLA treatment. The viability of the cells was determined by MTT assay, and PARP cleavage was determined by Western blot analysis.
RT-PCR
Total RNA was isolated using TRIzol® Reagent (Life Technologies, Inc., Gaithersburg, MD; catalog No. 15596-010) according to the manufacturer's protocol. The cDNA reaction as well as the PCR were performed using the Titan One Tube RT-PCR kit (Roche; catalog No. 1939823).
The XBP1 primers (sense, 5′-AAACAGAGTAGCAGCGCAGACTGC-3′; antisense, 5′-TCCTTCTGGGTAGACCTCTGGGAG-3′) were synthesized by Integrated DNA Technologies, Inc. (Coralville, IA). A 2% agarose gel containing 0.5 μg/ml ethidium bromide was used to separate the PCR products, and the products were visualized with a MultiImager system (Alpha Innotech Corp., San Leandro, CA).
Electron microscopy
One million cells were plated on 100 mm dishes at a concentration of 1 × 105 cells/ml, then allowed to attach overnight before the addition of 0 or 40 μM t10,c12- or c9,t11-CLA. After 72 h of treatment, cells were trypsinized and washed twice in PBS, then resuspended and fixed with 3% glutaraldehyde for 1 h on ice. The samples then were sent to the electron microscopy laboratories at Roswell Park Cancer Institute and the University at Buffalo. Sections were viewed and photographs were taken with a JEOL 100 CXII electron microscope. To quantify the dilatation of the ER lumen after CLA treatment, the width of the largest ER lumen in each cell was measured in 10 cells per group.
Statistical analysis
With one exception, ANOVA followed by Bonferroni's procedure was used to determine statistical differences between control and experimental groups. For the determination of statistical differences between the sizes of the ER lumen, Kruskal-Wallis ANOVA on ranks was used, followed by the Student-Newman-Keuls test for all pairwise multiple comparisons. P < 0.05 was considered statistically significant.
RESULTS
t10,c12-CLA induces apoptosis of TM4t mouse mammary tumor cells
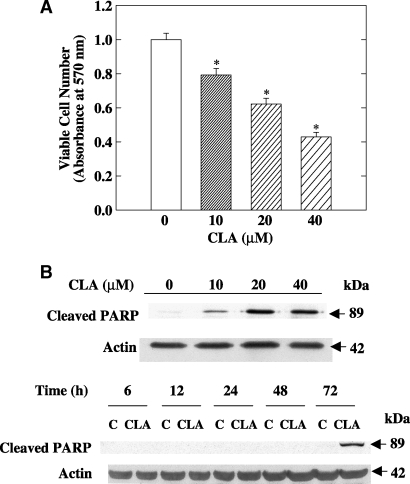
TM4t cells were cultured with different concentrations of t10,c12-CLA for 72 h, and viable cell number was assessed using the MTT assay. Compared with the control, t10,c12-CLA decreased viable cell number in a concentration-dependent manner (Fig. 1A). Concomitantly, cleavage of PARP, a marker of the apoptotic response (26), was observed, an effect that was maximal after 72 h of treatment with 10, 20, or 40 μM t10,c12-CLA (Fig. 1B and data not shown). A decrease in the number of viable cells was detected as early as 48 h (30% decrease with 40 μM t10,c12-CLA), although PARP cleavage was minimal at this time (Fig. 1B). Microscopic observation and data from annexin V-PI double staining confirmed that t10,c12-CLA induces the apoptosis of TM4t cells (20).
Fig.1.
Trans-10,cis-12-conjugated linoleic acid (t10,c12-CLA) decreases viable cell number and induces apoptosis of TM4t mouse mammary tumor cells. A: The effect of CLA on viable cell number at 72 h was examined using the thiazolyl blue tetrazolium bromide (MTT) assay. Each point represents the mean ± SEM of triplicate wells. * P < 0.05 compared with control. B: CLA induces the cleavage of poly(ADP-ribose) polymerase (PARP) in a concentration- and time-dependent manner. TM4t mammary tumor cells were treated with different concentrations of t10,c12-CLA for 72 h (upper panel) or with 0 or 40 μM t10,c12-CLA for the indicated times (lower panel) and then cell lysates were evaluated by Western blot analysis. Each result is representative of two or three independent experiments.
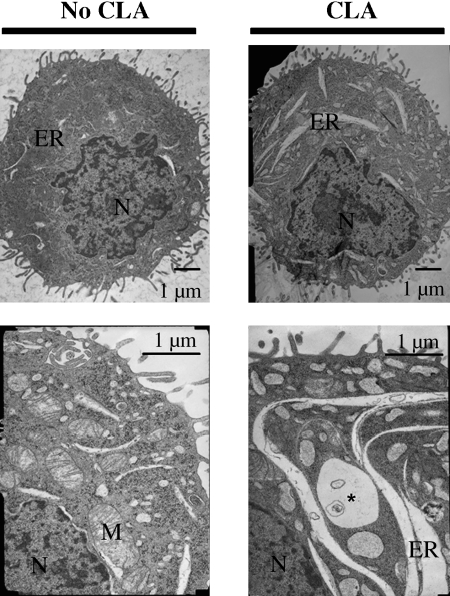
t10,c12-CLA induces a marked dilatation of the ER
Electron microscopy was used as a tool to obtain leads on the mechanism by which t10,c12-CLA induced apoptosis. As shown in the representative cells in Fig. 2, treatment of TM4t cells with t10,c12-CLA for 72 h induced a marked dilatation of the ER, which was seen in 8 of 10 cells examined. To quantify the extent of dilatation, the width of the largest lumen in each cell was determined. This measurement indicated a statistically significant 2.5-fold increase in lumen width, from 0.18 ± 0.07 μm in the control group to 0.44 ± 0.21 μm in the t10,c12-CLA group (n = 10, P < 0.05). ER dilatation was detectable within 48 h of treatment with 40 μM t10,c12-CLA but not at 12 or 24 h (data not shown). These data suggested that t10,c12-CLA might induce an ER stress response (23, 24), a notion that was investigated as described below.
Fig. 2.
t10,c12-CLA induces a marked dilatation of the endoplasmic reticulum (ER). TM4t cells were treated for 72 h with 40 μM t10,c12-CLA and examined by electron microscopy. Compared with the control, CLA induced an extensive increase in the size of the ER. M, mitochondria; N, nucleus. The asterisk indicates a multivesicular body.
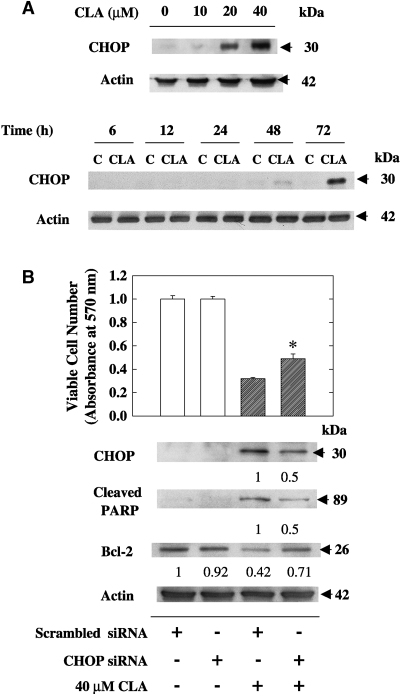
t10,c12-CLA induces the proapoptotic protein CHOP, and knockdown of CHOP attenuates CLA-induced apoptosis
CHOP, also known as GADD153, is a proapoptotic transcription factor that is activated as a result of ER stress (11). In TM4t cells, we found that CHOP was present at undetectable to very low levels in unstressed cells but was induced slightly after 48 h of treatment with 40 μM t10,c12-CLA and markedly at 72 h (Fig. 3A). At the latter time point, 20 μM, but not 10 μM, t10,c12-CLA was also effective at inducing CHOP. To investigate its role in t10,c12-CLA-induced apoptosis, we used the siRNA technique to knock down CHOP expression in t10,c12-CLA-treated cells. As seen in Fig. 3B, 40 μM t10,c12-CLA reduced viable cell number, accompanied by an increase in both CHOP protein and apoptosis, the latter indicated by the cleavage of PARP. Knockdown of CHOP with an siRNA that targeted exon 4 reduced CHOP levels and PARP cleavage by 50% in t10,c12-CLA-treated cells and attenuated the t10,c12-CLA reduction in viable cell number (Fig. 3B). Similarly, CHOP knockdown diminished the PARP cleavage induced by 20 μM t10,c12-CLA (data not shown). Furthermore, based on our previous demonstration that t10,c12-CLA decreased Bcl-2 levels (20) and studies suggesting an inverse relationship between CHOP and Bcl-2 (27, 28), we also asked whether knockdown of CHOP would restore Bcl-2 levels in t10,c12-CLA-treated cells. This indeed was the case, with Fig. 3B demonstrating that knockdown of CHOP partially blocked the t10,c12-CLA-induced loss of Bcl-2. Similar observations were made using an siRNA that targeted exon 3 (data not shown).
Fig. 3.
t10,c12-CLA induces the proapoptotic protein C/EBP-homologous protein (CHOP), and knockdown of CHOP by small interfering RNA (siRNA) attenuates CLA-induced apoptosis. A: Induction of CHOP by CLA. TM4t mammary tumor cells were treated with various concentrations of t10,c12-CLA for 72 h or with 0 or 40 μM t10,c12-CLA for various times, and the expression of CHOP was determined by Western blot. A concentration-dependent response after 72 h of treatment and a time-dependent response with 40 μM CLA, as evaluated by Western blot analysis, are shown. B: Effect of CHOP knockdown on the efficacy of t10,c12-CLA at reducing cell number and Bcl-2 expression and inducing PARP cleavage. TM4t cells were transfected with scrambled or CHOP (exon 4) siRNA for 6 h, then 0 or 40 μM t10,c12-CLA was added for 72 h. Each point represents the mean ± SEM of triplicate wells. * Significantly different from the scrambled siRNA control plus CLA group (lane 3). Numbers under the blots represent the fold change, which was calculated relative to the actin loading control. Each result is representative of two or three individual experiments.
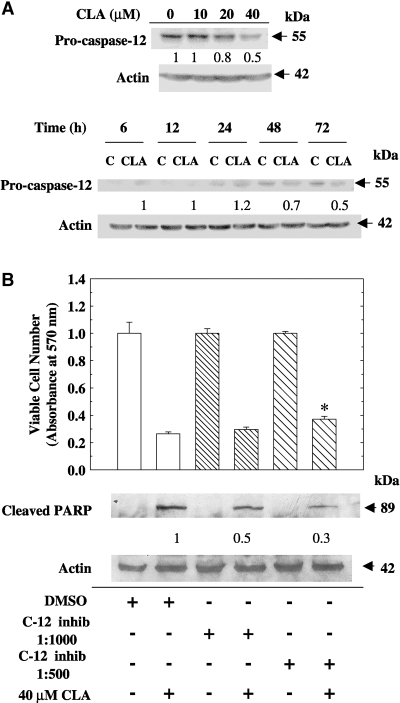
t10,c12-CLA induces caspase-12 cleavage, and a specific caspase-12 inhibitor attenuates CLA-induced apoptosis
In addition to CHOP, ER stress-induced apoptosis can also be mediated through the activation of caspase-12. This enzyme is localized to the outer membrane of the ER and is specifically cleaved as a result of ER stress (12). When TM4t cells were treated with t10,c12-CLA, 20% and 50% decreases in full-length caspase-12 were observed at 20 and 40 μM concentrations, respectively (Fig. 4A). Decrease of full-length caspase-12, suggestive of its cleavage, was first detected at 48 h (30%) but was more extensive (50%) at 72 h (Fig. 4A). To evaluate the significance of caspase-12 activation in CLA-induced apoptosis, we used a specific caspase-12 inhibitor, Z-ATAD-FMK. This inhibitor modestly increased viable cell number in the cells that had been cultured with CLA (P < 0.05). More dramatic was the 70% reduction in PARP cleavage in CLA-treated cells at the higher caspase-12 inhibitor concentration (Fig. 4B), suggesting an important role for this enzyme in CLA-induced apoptosis.
Fig. 4.
t10,c12-CLA induces the cleavage of caspase-12, and a caspase-12-specific inhibitor attenuates CLA-induced apoptosis. A: Induction of caspase-12 cleavage by CLA. TM4t cells were treated with various concentrations of t10,c12-CLA for 72 h or with 0 or 40 μM t10,c12-CLA for various times, and the expression of caspase-12 was determined by Western blot. B: Effect of a specific caspase-12 inhibitor, Z-ATAD-FMK, on viable cell number and PARP cleavage. TM4t cells were treated in the absence or presence of 40 μM t10,c12-CLA, with or without the caspase-12 inhibitor (c-12 inhib) Z-ATAD-FMK at a 1:1,000 or 1:500 dilution, for 72 h. Upper panel: Viable cell number was determined using the MTT assay; each point represents the mean ± SEM of triplicate wells. * Significantly different from the DMSO vehicle/CLA group (lane 2). Lower panel: Western blot shows that the caspase-12 inhibitor attenuated CLA-induced PARP cleavage. Numbers under the blots represent the fold change, which was calculated relative to the actin loading control. Each result is representative of two or three individual experiments.
t10,c12-CLA induces an ER stress response
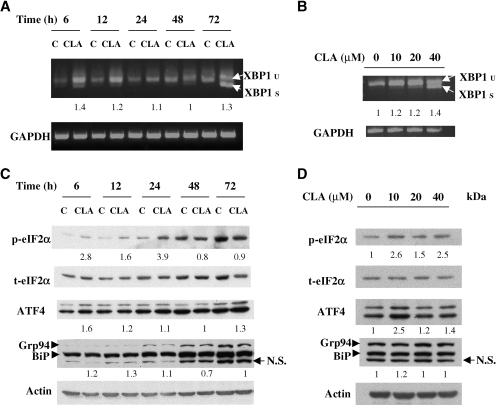
The CLA-induced morphological changes within the ER, together with the induction of CHOP and the cleavage of caspase-12, provide strong support for the hypothesis that CLA induces apoptosis through ER stress. To further evaluate this concept, we examined upstream components of the ER stress response. We first examined XBP1 mRNA, which is spliced by the endoRNase activity of active IRE1 (6, 29), one of the transducers of the ER stress signal, and whose transcription can be increased by cleaved ATF6 (6), the transcriptionally active form of another transducer. Treatment of cells with t10,c12-CLA induced the splicing of XBP1 mRNA, with the most dramatic effect observed with 40 μM t10,c12-CLA and at 72 h (Fig. 5A, B). Additionally, total XBP1 mRNA levels were rapidly (6 h) increased by t10,c12-CLA. Although we could not examine the expression of IRE1 and ATF6 because of the commercial unavailability of suitable mouse-reactive antibodies, these data suggest the activation of these two pathways.
Fig. 5.
t10,c12-CLA induces an ER stress response. A, B: t10,c12-CLA induces the expression and splicing of the mRNA encoding the X-box binding protein 1 (XBP1) transcription factor. TM4t cells were treated with 0 or 40 μM t10,c12-CLA for various times (A) or with various concentrations of t10,c12-CLA for 72 h (B), and the expression of unspliced (u) and spliced (s) XBP1 mRNA was determined by RT-PCR. Each panel is representative of two or three individual experiments. C, D: Effect of t10,c12-CLA on the expression of phospho-eukaryotic initiation factor (eIF) 2α, total eIF2α, activating transcription factor 4 (ATF4), BiP, and 94 kDa glucose-regulated protein (GRP94). TM4t mammary tumor cells were treated with 0 or 40 μM t10,c12-CLA for different times (C) or with various concentrations of t10,c12-CLA for 24 h (D), and protein expression was evaluated by Western blot. The t10,c12-CLA-induced expression of phospho-eIF2α was statistically significant at 6, 12, and 24 h (P < 0.05). The modest induction of ATF4 was not statistically significant. The expression of BiP and GRP94 was not altered. Fold induction of phospho-eIF2α was calculated relative to total eIF2α, and that of other proteins was calculated relative to the actin loading control; the numbers under GRP94/BiP refer to the quantification of BiP. N.S., nonspecific.
The third transducer of the ER stress signal is PERK. Although we were unable to measure active, phosphorylated PERK, we found that phosphorylation of eIF2α, a downstream target of phospho-PERK (4), was rapidly, but transiently, increased in t10,c12-CLA-treated cells (Fig. 5C). This increase was most notable after 6 h of treatment (Fig. 5C) and at a lower concentration of CLA (Fig. 5D). Although phosphorylated eIF2α is a general translation initiation inhibitor, translation of selected mRNAs, including the transcription factor ATF4, is increased upon the phosphorylation of eIF2α (30). ATF4 was induced modestly by t10,c12-CLA, but this induction was not statistically significant. Additionally, expression of BiP protein, the chaperone sensor of the ER stress response, was not altered by t10,c12-CLA, nor was the chaperone GRP94, which is recognized by the BiP antibody (Fig. 5C, D). In contrast to treatment with t10,c12-CLA, BiP levels were increased after 24 h of treatment with the classic ER stress inducers thapsigargin and ionomycin (data not shown), demonstrating that the TM4t cells are capable of mounting a typical ER stress response.
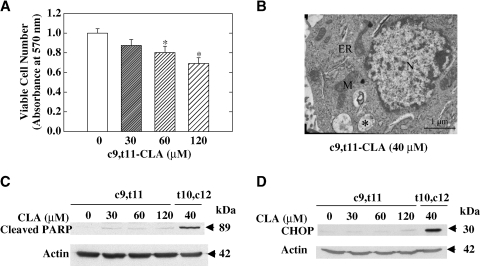
The c9,t11 isomer of CLA does not disrupt ER homeostasis or induce ER stress
To evaluate the specificity of the ER stress-inducing effect of t10,c12-CLA, we examined the effect of another CLA isomer, c9,t11-CLA, on specific parameters of the ER stress response. Both CLA isomers have been shown to exhibit anticancer effects in many model systems (15), although their mechanisms may differ. c9,t11-CLA modestly decreased viable cell number in the TM4t mammary tumor model (31% decrease at 120 μM; Fig. 6A), although it was significantly less potent in this regard than t10,c12-CLA (58% decrease at 40 μM; Fig. 1A). Electron microscopy demonstrated that, in contrast to t10,c12-CLA, c9,t11-CLA at the same 40 μM concentration did not induce ER dilatation (Fig. 6B); the width of the ER lumen in c9,t11-CLA-treated cells was 0.20 ± 0.10 μm, not significantly different from that of control cells. Furthermore, PARP cleavage was minimally increased by c9,t11-CLA, an effect that was not concentration-dependent and that was markedly less than that induced by a lower concentration of t10,c12-CLA in the same experiment (Fig. 6C). This suggests that c9,t11-CLA may not induce apoptosis, at least not to any significant extent. Finally, the proapoptotic protein CHOP was not induced by c9,t11-CLA after 72 h of treatment, even at a relatively high concentration (120 μM) (Fig. 6D).
Fig. 6.
The c9,t11 isomer of CLA did not disrupt ER homeostasis or induce ER stress-mediated apoptosis in TM4t mammary tumor cells. A: The effect of c9,t11-CLA on viable cell number after 72 h of treatment was examined using the MTT assay. Each point represents the mean ± SEM of triplicate wells. * Significantly different from control. B: The effect of c9,t11-CLA on ER morphology. No ER dilatation was observed by electron microscopy after 72 h of treatment with 40 μM c9,t11-CLA. M, mitochondria; N, nucleus. The asterisk indicates a multivesicular body. C: The effect of c9,t11-CLA on apoptosis. Cleavage of PARP after 72 h of treatment with the indicated concentration of c9,t11- or t10,c12-CLA. D: CHOP was not induced by c9,t11-CLA, even at a high concentration. CHOP was evaluated by Western blot after 72 h of treatment with various concentrations of c9,t11-CLA or with 40 μM t10,c12-CLA. Each panel is representative of two or three independent experiments.
DISCUSSION
t10,c12-CLA induces apoptosis through ER stress response pathways
The current study presents morphological and biochemical evidence that the t10,c12 isomer of CLA, but not c9,t11-CLA, induces apoptosis and suggests that specific ER stress response pathways play an important role in the t10,c12-CLA apoptotic response and, potentially, in the mechanism by which t10,c12-CLA exerts its chemopreventive activity. Notably, the data demonstrate that the IRE1/XBP1 and caspase-12 arms of the ER stress pathway are activated by t10,c12-CLA, resulting in apoptosis through the induction of the proapoptotic protein CHOP and the activation of caspase-12. Moreover, contributing roles for both CHOP and caspase-12 in t10,c12-CLA-induced apoptosis were demonstrated by CHOP knockdown and caspase-12 inhibitor experiments.
Apoptosis of TM4t cells in response to t10,c12-CLA was delayed, with significant PARP cleavage not detected until 72 h after the start of CLA treatment. This followed the induction of CHOP and the loss of full-length caspase-12 at 48 h, further supporting the role of these two proteins in the apoptotic effect of CLA. Previous studies have demonstrated the proapoptotic activity of CHOP (11, 31, 32) as well as the ability of other chemopreventive agents to induce it, including retinoids (33), phenethylisothiocyanate (34), curcumin (35), selenium (36, 37), and celecoxib (38). Our study not only adds another chemopreventive agent to this list but, more importantly, extends the studies with the other agents to demonstrate that the potent induction of CHOP by CLA most likely involves the induction of XBP1, which directly regulates its transcription (11). CHOP transcription is also regulated by cleaved ATF6, which binds with NF-Y to ERSE (8), and by ATF4, which binds to the CHOP promoter at a composite C/EBP-ATF site (39). ATF6 represents a rapid transcriptional response mechanism in that its activation requires only cleavage and nuclear translocation (40).
Because of the delayed induction of CHOP, our data do not suggest a direct role for ATF6 in its CLA-induced increase, although this still remains a possibility. Rather, the rapid (6 h) induction of XBP1 mRNA in CLA-treated cells suggests that the ATF6 pathway contributes indirectly to CHOP-induced apoptosis by transcriptional activation of the XBP1 gene. XBP1 mRNA is then spliced by CLA-activated IRE1, and after translation, XBP1 protein binds to ERSE in the CHOP promoter (11). Previous studies showed that overexpression of “cleaved” ATF6 activated the XBP1 promoter, whereas a dominant negative ATF6 was inhibitory (8), suggesting an important role for this transcription factor in XBP1 transcription. More recent studies using ATF6-null mouse embryonic fibroblasts demonstrated that ATF6 is not required for XBP1 transcription, although it is contributory (induction was less robust in the absence of ATF6) (41). We interpret our data as suggesting that the early increase in XBP1 mRNA in response to t10,c12-CLA results from the cleavage of ATF6 and the subsequent binding of ATF6 to the XBP1 promoter. Later, XBP1 protein may activate the transcription of both CHOP and its own gene (6). Finally, although ATF4 can also bind to the CHOP and BiP promoters (42, 43), the lack of early induction of either protein suggests that this branch of the ER stress response is not activated significantly, because the translation of ATF4 in response to the unfolded protein response is generally rapid (30).
Two mechanisms can be advanced for how CLA-induced CHOP may mediate apoptosis. First, based on a study demonstrating that overexpression of CHOP led to a marked reduction in transcription of the Bcl-2 gene (28) and on our observation that t10,c12-CLA decreased Bcl-2 levels in TM4t cells (20), we asked whether CHOP knockdown would increase Bcl-2 levels in CLA-treated cells. As shown in Fig. 3, the answer to this question was yes, suggesting a role for Bcl-2 loss in CHOP-mediated apoptosis. A second potential mechanism by which CLA may induce apoptosis is through the activation of death receptor 5, which has a CHOP binding site in its promoter and is upregulated as a result of ER stress (44, 45). Future studies will address this possibility.
In addition to CHOP, caspase-12 plays a contributing role in CLA-induced apoptosis, because we found that the specific caspase-12 inhibitor Z-ATAD-FMK partially attenuated the apoptotic effect. A role for caspase-12 in ER stress has been somewhat controversial. First, mouse embryonic fibroblasts from one line of caspase-12-deficient mice exhibited partial apoptotic resistance to ER stress-inducing agents (12); however, those from another line did not differ from wild-type mouse embryonic fibroblasts in terms of apoptotic sensitivity (46). This discrepancy was attributed to a different targeting strategy, although the initial data do suggest at least a partial requirement for caspase-12. Second, although mice and most other species tested express a functional caspase-12, the human caspase-12 has polymorphisms that limit the expression of a full-length protein to a small subset of individuals (47); moreover, in that population, caspase-12 did not appear to be involved in ER stress-induced apoptosis. In humans, it has been suggested that ER-associated caspase-4 or -7 may play a role in ER stress-induced apoptosis (48, 49).
The ER stress response induced by t10,c12-CLA is atypical
One unexpected observation in this study was that the expression of the ER chaperone BiP was not altered by CLA treatment. This was not attributable to the cell line used in these studies, because BiP could be induced by the classical ER stress inducers thapsigargin and ionomycin (data not shown). BiP appeared to function in response to CLA, however, because IRE1 and possibly ATF6, to which BiP is bound in healthy cells, became activated, suggesting that BiP had dissociated from them. A similar lack of induction of BiP protein concurrent with the induction of ER stress and apoptosis was also seen with another fatty acid, palmitate, in pancreatic β-cells (50). Because BiP plays a protective role during ER stress (51), its lack of induction would enhance the apoptotic effect of CLA. In addition to the lack of effect on BiP, our data do not provide strong support for the CLA-induced activation of the PERK arm of the ER stress pathway. Thus, ATF4 was not activated significantly, and although phosphorylation of the translation initiation factor eIF2α was rapidly, but transiently, increased after CLA treatment, potentially attenuating translation, kinases other than PERK could be responsible (52).
Interestingly, we observed that BiP and phosphorylated eIF2α increased with time in control cells. This phenomenon may be attributable to nutrient deprivation, because the levels of BiP (also named GRP78) and p-eIF2α are sensitive to glucose and nutrients (30, 53, 54). Cells consume nutrients during culture, and without a medium change, there may be nutrient deprivation at the later time points. This would be more pronounced in the control group, in which there were more viable cells than in the CLA treatment group. Importantly, this apparent stress did not lead to apoptosis, as indicated by the lack of PARP cleavage (Fig. 1), CHOP induction (Fig. 3), or annexin V-PI staining (20).
How does t10,c12-CLA induce ER stress?
We propose a mechanism that involves the production of reactive oxygen species, which are known to be able to induce an ER stress response (55). O'Shea, Stanton, and Devery (56) reported an increase in lipid peroxidation, measured using malondialdehyde as an end point, in MCF-7 cells treated with a mixed CLA isomer preparation (approximately equal amounts of c9,t11-CLA and t10,c12-CLA). Furthermore, Albright et al. (57) demonstrated an increase in 4-hydroxy-2-nonenal in both MCF-10A and MCF-7 cells treated with mixed CLA isomers. An increase in lipid peroxidation was also observed in healthy men and women consuming a mixed CLA isomer supplement, as evaluated by an increase in urinary 8-iso-prostaglandin F2α and 15-keto-dihydro-prostaglandin F2α (58). In the TM4t mammary tumor cell line used in the current studies, we found that the free radical scavengers α-tocopherol and hypotaurine blocked apoptosis induced by t10,c12-CLA (Y-C. Hsu and M. Ip, unpublished data), supporting a role for peroxidation products in CLA-induced apoptosis. We speculate that reactive oxygen species may alter the redox potential or induce the formation of aldehydic products which can conjugate with amino acids within proteins, thus altering their stability. ER stress would be induced in either scenario.
CLA and apoptosis
t10,c12-CLA, or a mixture of CLA isomers, has been shown to induce the apoptosis of a number of nonmammary as well as mammary tumor cell lines in vitro and to increase the apoptosis of preneoplastic cells in vivo (19, 59, 60). The induction of apoptosis may be a major factor contributing to the chemopreventive activity of this CLA isomer. We previously demonstrated that the intrinsic mitochondrial pathway played an important role in CLA-induced apoptosis (20). The current study significantly extends this observation and reports, for the first time, that t10,c12-CLA induces an atypical ER stress response, using certain components of the ER stress response to induce apoptosis. Interestingly, this effect of CLA may be selective for preneoplastic or neoplastic cells, because many normal cells are resistant to the apoptotic effects of CLA. We have shown, for example, that whereas dietary CLA induces the apoptosis of preneoplastic mammary epithelial cells in vivo, undifferentiated normal rat or mouse mammary epithelial cells are resistant (60, 61). The mechanism of this resistance is not yet known; however, it is intriguing that when normal mammary epithelial cells differentiate and become active in the synthesis and secretion of large amounts of milk proteins and lipid (as would happen in lactation) and are probably under ER stress as a consequence, they are highly susceptible to the apoptosis-inducing effect of CLA (25).
In summary, although c9,t11- and t10,c12-CLA are equally efficacious in some (16, 17), although not all (61), in vivo mammary cancer models, their mechanisms of action appear to differ. Ongoing studies in our laboratory are focused on understanding the pathways through which each of the isomers exhibits its chemopreventive activity.
Acknowledgments
The authors thank Drs. Mary Ann Warren, Jiping Zhang, Erica Berleth, Gokul Das, Patricia Masso-Welch, and Ke Zu as well as Barbara Lisafeld, Sue Shoemaker, and Sibel McGee for helpful discussion throughout these studies. The authors are grateful to Dr. Jennifer Black for assisting us with electron microscopy and Dr. Daniel Medina (Baylor College of Medicine, Houston, TX) for providing us with the TM4t mammary tumor cell line.
Abbreviations
ATF, activating transcription factor
CHOP, C/EBP-homologous protein
CLA, conjugated linoleic acid
eIF, eukaryotic initiation factor
ER, endoplasmic reticulum
ERSE, endoplasmic reticulum stress response element
GRP78, 78 kDa glucose-regulated protein
IRE1, inositol-requiring enzyme 1
MTT, thiazolyl blue tetrazolium bromide
PARP, poly(ADP-ribose) polymerase
PERK, double-stranded RNA-dependent protein kinase (PKR)-like endoplasmic reticulum-resident kinase
siRNA, small interfering RNA
t10,c12, trans-10,cis-12
XBP1, X-box binding protein 1
Published, JLR Papers in Press, February 8, 2008.
Footnotes
This work was supported by National Institutes of Health Grant CA-61763 (C.I., M.M.I.), by predoctoral Department of Defense Grant W81XWH-06-1-0288 (L.O.), by an Avon-American Association for Cancer Research Scholar Award (X.M.), and by the shared resources of the National Cancer Institute Roswell Park Cancer Center Support Grant CA-16056.
References
- 1.Schroder M., and R. J. Kaufman. 2005. The mammalian unfolded protein response. Annu. Rev. Biochem. 74 739–789. [DOI] [PubMed] [Google Scholar]
- 2.Szegezdi E., S. E. Logue, A. M. Gorman, and A. Samali. 2006. Mediators of endoplasmic reticulum stress-induced apoptosis. EMBO Rep. 7 880–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang K., and R. J. Kaufman. 2006. The unfolded protein response: a stress signaling pathway critical for health and disease. Neurology. 66 (Suppl. 1): S102–S109. [DOI] [PubMed] [Google Scholar]
- 4.Harding H. P., Y. Zhang, A. Bertolotti, H. Zeng, and D. Ron. 2000. Perk is essential for translational regulation and cell survival during the unfolded protein response. Mol. Cell. 5 897–904. [DOI] [PubMed] [Google Scholar]
- 5.Ye J., R. B. Rawson, R. Komuro, X. Chen, U. P. Dave, R. Prywes, M. S. Brown, and J. L. Goldstein. 2000. ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol. Cell. 6 1355–1364. [DOI] [PubMed] [Google Scholar]
- 6.Yoshida H., T. Matsui, A. Yamamoto, T. Okada, and K. Mori. 2001. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 107 881–891. [DOI] [PubMed] [Google Scholar]
- 7.Lee K., W. Tirasophon, X. Shen, M. Michalak, R. Prywes, T. Okada, H. Yoshida, K. Mori, and R. J. Kaufman. 2002. IRE1-mediated unconventional mRNA splicing and S2P-mediated ATF6 cleavage merge to regulate XBP1 in signaling the unfolded protein response. Genes Dev. 16 452–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yoshida H., T. Okada, K. Haze, H. Yanagi, T. Yura, M. Negishi, and K. Mori. 2000. ATF6 activated by proteolysis binds in the presence of NF-Y (CBF) directly to the cis-acting element responsible for the mammalian unfolded protein response. Mol. Cell. Biol. 20 6755–6767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yoshida H., K. Haze, H. Yanagi, T. Yura, and K. Mori. 1998. Identification of the cis-acting endoplasmic reticulum stress response element responsible for transcriptional induction of mammalian glucose-regulated proteins. Involvement of basic leucine zipper transcription factors. J. Biol. Chem. 273 33741–33749. [DOI] [PubMed] [Google Scholar]
- 10.Yoshida H., T. Matsui, N. Hosokawa, R. J. Kaufman, K. Nagata, and K. Mori. 2003. A time-dependent phase shift in the mammalian unfolded protein response. Dev. Cell. 4 265–271. [DOI] [PubMed] [Google Scholar]
- 11.Oyadomari S., and M. Mori. 2004. Roles of CHOP/GADD153 in endoplasmic reticulum stress. Cell Death Differ. 11 381–389. [DOI] [PubMed] [Google Scholar]
- 12.Nakagawa T., H. Zhu, N. Morishima, E. Li, J. Xu, B. A. Yankner, and J. Yuan. 2000. Caspase-12 mediates endoplasmic-reticulum-specific apoptosis and cytotoxicity by amyloid-beta. Nature. 403 98–103. [DOI] [PubMed] [Google Scholar]
- 13.Gaullier J. M., J. Halse, H. O. Hoivik, K. Hoye, C. Syvertsen, M. Nurminiemi, C. Hassfeld, A. Einerhand, M. O'Shea, and O. Gudmundsen. 2007. Six months supplementation with conjugated linoleic acid induces regional-specific fat mass decreases in overweight and obese. Br. J. Nutr. 97 550–560. [DOI] [PubMed] [Google Scholar]
- 14.Syvertsen C., J. Halse, H. O. Hoivik, J. M. Gaullier, M. Nurminiemi, K. Kristiansen, A. Einerhand, M. O'Shea, and O. Gudmundsen. 2007. The effect of 6 months supplementation with conjugated linoleic acid on insulin resistance in overweight and obese. Int. J. Obes. (Lond). 31 1148–1154. [DOI] [PubMed] [Google Scholar]
- 15.Ip M. M., P. A. Masso-Welch, and C. Ip. 2003. Prevention of mammary cancer with conjugated linoleic acid: role of the stroma and the epithelium. J. Mammary Gland Biol. Neoplasia. 8 101–116. [DOI] [PubMed] [Google Scholar]
- 16.Ip C., Y. Dong, M. M. Ip, S. Banni, G. Carta, E. Angioni, E. Murro, S. Spada, M. P. Melis, and A. Saebo. 2002. Conjugated linoleic acid isomers and mammary cancer prevention. Nutr. Cancer. 43 52–58. [DOI] [PubMed] [Google Scholar]
- 17.Hubbard N. E., D. Lim, and K. L. Erickson. 2003. Effect of separate conjugated linoleic acid isomers on murine mammary tumorigenesis. Cancer Lett. 190. 13–19. [DOI] [PubMed]
- 18.Masso-Welch P. A., D. Zangani, C. Ip, M. M. Vaughan, S. F. Shoemaker, S. O. McGee, and M. M. Ip. 2004. Isomers of conjugated linoleic acid differ in their effects on angiogenesis and survival of mouse mammary adipose vasculature. J. Nutr. 134 299–307. [DOI] [PubMed] [Google Scholar]
- 19.Ip C., M. M. Ip, T. Loftus, S. Shoemaker, and W. Shea-Eaton. 2000. Induction of apoptosis by conjugated linoleic acid in cultured mammary tumor cells and premalignant lesions of the rat mammary gland. Cancer Epidemiol. Biomarkers Prev. 9 689–696. [PubMed] [Google Scholar]
- 20.Ou L., C. Ip, B. Lisafeld, and M. M. Ip. 2007. Conjugated linoleic acid induces apoptosis of murine mammary tumor cells via Bcl-2 loss. Biochem. Biophys. Res. Commun. 356 1044–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen-Levy Z., J. Nourse, and M. L. Cleary. 1989. The bcl-2 candidate proto-oncogene product is a 24-kilodalton integral-membrane protein highly expressed in lymphoid cell lines and lymphomas carrying the t(14;18) translocation. Mol. Cell. Biol. 9 701–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu W., A. Cowie, G. W. Wasfy, L. Z. Penn, B. Leber, and D. W. Andrews. 1996. Bcl-2 mutants with restricted subcellular location reveal spatially distinct pathways for apoptosis in different cell types. EMBO J. 15 4130–4141. [PMC free article] [PubMed] [Google Scholar]
- 23.Xie Q., V. I. Khaoustov, C. C. Chung, J. Sohn, B. Krishnan, D. E. Lewis, and B. Yoffe. 2002. Effect of tauroursodeoxycholic acid on endoplasmic reticulum stress-induced caspase-12 activation. Hepatology. 36 592–601. [DOI] [PubMed] [Google Scholar]
- 24.Kerkela R., L. Grazette, R. Yacobi, C. Iliescu, R. Patten, C. Beahm, B. Walters, S. Shevtsov, S. Pesant, F. J. Clubb, et al. 2006. Cardiotoxicity of the cancer therapeutic agent imatinib mesylate. Nat. Med. 12 908–916. [DOI] [PubMed] [Google Scholar]
- 25.Ip M. M., P. A. Masso-Welch, S. F. Shoemaker, W. K. Shea-Eaton, and C. Ip. 1999. Conjugated linoleic acid inhibits proliferation and induces apoptosis of normal rat mammary epithelial cells in primary culture. Exp. Cell Res. 250 22–34. [DOI] [PubMed] [Google Scholar]
- 26.Duriez P. J., and G. M. Shah. 1997. Cleavage of poly(ADP-ribose) polymerase: a sensitive parameter to study cell death. Biochem. Cell Biol. 75 337–349. [PubMed] [Google Scholar]
- 27.Benavides A., D. Pastor, P. Santos, P. Tranque, and S. Calvo. 2005. CHOP plays a pivotal role in the astrocyte death induced by oxygen and glucose deprivation. Glia. 52 261–275. [DOI] [PubMed] [Google Scholar]
- 28.McCullough K. D., J. L. Martindale, L. O. Klotz, T. Y. Aw, and N. J. Holbrook. 2001. Gadd153 sensitizes cells to endoplasmic reticulum stress by down-regulating Bcl2 and perturbing the cellular redox state. Mol. Cell. Biol. 21 1249–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Calfon M., H. Zeng, F. Urano, J. H. Till, S. R. Hubbard, H. P. Harding, S. G. Clark, and D. Ron. 2002. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 415 92–96. [DOI] [PubMed] [Google Scholar]
- 30.Harding H. P., I. Novoa, Y. Zhang, H. Zeng, R. Wek, M. Schapira, and D. Ron. 2000. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol. Cell. 6 1099–1108. [DOI] [PubMed] [Google Scholar]
- 31.Maytin E. V., M. Ubeda, J. C. Lin, and J. F. Habener. 2001. Stress-inducible transcription factor CHOP/gadd153 induces apoptosis in mammalian cells via p38 kinase-dependent and -independent mechanisms. Exp. Cell Res. 267 193–204. [DOI] [PubMed] [Google Scholar]
- 32.Tajiri S., S. Yano, M. Morioka, J. Kuratsu, M. Mori, and T. Gotoh. 2006. CHOP is involved in neuronal apoptosis induced by neurotrophic factor deprivation. FEBS Lett. 580 3462–3468. [DOI] [PubMed] [Google Scholar]
- 33.Kim D. G., K. R. You, M. J. Liu, Y. K. Choi, and Y. S. Won. 2002. GADD153-mediated anticancer effects of N-(4-hydroxyphenyl)retinamide on human hepatoma cells. J. Biol. Chem. 277 38930–38938. [DOI] [PubMed] [Google Scholar]
- 34.Powolny A., K. Takahashi, R. G. Hopkins, and G. Loo. 2003. Induction of GADD gene expression by phenethylisothiocyanate in human colon adenocarcinoma cells. J. Cell. Biochem. 90 1128–1139. [DOI] [PubMed] [Google Scholar]
- 35.Scott D. W., and G. Loo. 2004. Curcumin-induced GADD153 gene up-regulation in human colon cancer cells. Carcinogenesis. 25 2155–2164. [DOI] [PubMed] [Google Scholar]
- 36.Wu Y., H. Zhang, Y. Dong, Y. M. Park, and C. Ip. 2005. Endoplasmic reticulum stress signal mediators are targets of selenium action. Cancer Res. 65 9073–9079. [DOI] [PubMed] [Google Scholar]
- 37.Zu K., T. Bihani, A. Lin, Y. M. Park, K. Mori, and C. Ip. 2006. Enhanced selenium effect on growth arrest by BiP/GRP78 knockdown in p53-null human prostate cancer cells. Oncogene. 25 546–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim S. H., C. I. Hwang, Y. S. Juhnn, J. H. Lee, W. Y. Park, and Y. S. Song. 2007. GADD153 mediates celecoxib-induced apoptosis in cervical cancer cells. Carcinogenesis. 28 223–231. [DOI] [PubMed] [Google Scholar]
- 39.Ma Y., J. W. Brewer, J. A. Diehl, and L. M. Hendershot. 2002. Two distinct stress signaling pathways converge upon the CHOP promoter during the mammalian unfolded protein response. J. Mol. Biol. 318 1351–1365. [DOI] [PubMed] [Google Scholar]
- 40.Rutkowski D. T., and R. J. Kaufman. 2004. A trip to the ER: coping with stress. Trends Cell Biol. 14 20–28. [DOI] [PubMed] [Google Scholar]
- 41.Yamamoto K., T. Sato, T. Matsui, M. Sato, T. Okada, H. Yoshida, A. Harada, and K. Mori. 2007. Transcriptional induction of mammalian ER quality control proteins is mediated by single or combined action of ATF6alpha and XBP1. Dev. Cell. 13 365–376. [DOI] [PubMed] [Google Scholar]
- 42.Fawcett T. W., J. L. Martindale, K. Z. Guyton, T. Hai, and N. J. Holbrook. 1999. Complexes containing activating transcription factor (ATF)/cAMP-responsive-element-binding protein (CREB) interact with the CCAAT/enhancer-binding protein (C/EBP)-ATF composite site to regulate Gadd153 expression during the stress response. Biochem. J. 339 135–141. [PMC free article] [PubMed] [Google Scholar]
- 43.Luo S., P. Baumeister, S. Yang, S. F. Abcouwer, and A. S. Lee. 2003. Induction of Grp78/BiP by translational block: activation of the Grp78 promoter by ATF4 through an upstream ATF/CRE site independent of the endoplasmic reticulum stress elements. J. Biol. Chem. 278 37375–37385. [DOI] [PubMed] [Google Scholar]
- 44.Yamaguchi H., and H. G. Wang. 2004. CHOP is involved in endoplasmic reticulum stress-induced apoptosis by enhancing DR5 expression in human carcinoma cells. J. Biol. Chem. 279 45495–45502. [DOI] [PubMed] [Google Scholar]
- 45.Yoshida T., T. Shiraishi, S. Nakata, M. Horinaka, M. Wakada, Y. Mizutani, T. Miki, and T. Sakai. 2005. Proteasome inhibitor MG132 induces death receptor 5 through CCAAT/enhancer-binding protein homologous protein. Cancer Res. 65 5662–5667. [DOI] [PubMed] [Google Scholar]
- 46.Saleh M., J. C. Mathison, M. K. Wolinski, S. J. Bensinger, P. Fitzgerald, N. Droin, R. J. Ulevitch, D. R. Green, and D. W. Nicholson. 2006. Enhanced bacterial clearance and sepsis resistance in caspase-12-deficient mice. Nature. 440 1064–1068. [DOI] [PubMed] [Google Scholar]
- 47.Saleh M., J. P. Vaillancourt, R. K. Graham, M. Huyck, S. M. Srinivasula, E. S. Alnemri, M. H. Steinberg, V. Nolan, C. T. Baldwin, R. S. Hotchkiss, et al. 2004. Differential modulation of endotoxin responsiveness by human caspase-12 polymorphisms. Nature. 429 75–79. [DOI] [PubMed] [Google Scholar]
- 48.Hitomi J., T. Katayama, Y. Eguchi, T. Kudo, M. Taniguchi, Y. Koyama, T. Manabe, S. Yamagishi, Y. Bando, K. Imaizumi, et al. 2004. Involvement of caspase-4 in endoplasmic reticulum stress-induced apoptosis and Abeta-induced cell death. J. Cell Biol. 165 347–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reddy R. K., C. Mao, P. Baumeister, R. C. Austin, R. J. Kaufman, and A. S. Lee. 2003. Endoplasmic reticulum chaperone protein GRP78 protects cells from apoptosis induced by topoisomerase inhibitors: role of ATP binding site in suppression of caspase-7 activation. J. Biol. Chem. 278 20915–20924. [DOI] [PubMed] [Google Scholar]
- 50.Karaskov E., C. Scott, L. Zhang, T. Teodoro, M. Ravazzola, and A. Volchuk. 2006. Chronic palmitate but not oleate exposure induces endoplasmic reticulum stress, which may contribute to INS-1 pancreatic beta-cell apoptosis. Endocrinology. 147 3398–3407. [DOI] [PubMed] [Google Scholar]
- 51.Morris J. A., A. J. Dorner, C. A. Edwards, L. M. Hendershot, and R. J. Kaufman. 1997. Immunoglobulin binding protein (BiP) function is required to protect cells from endoplasmic reticulum stress but is not required for the secretion of selective proteins. J. Biol. Chem. 272 4327–4334. [DOI] [PubMed] [Google Scholar]
- 52.Dever T. E. 2002. Gene-specific regulation by general translation factors. Cell. 108 545–556. [DOI] [PubMed] [Google Scholar]
- 53.Kimball S. R. 2002. Regulation of global and specific mRNA translation by amino acids. J. Nutr. 132 883–886. [DOI] [PubMed] [Google Scholar]
- 54.Little E., M. Ramakrishnan, B. Roy, G. Gazit, and A. S. Lee. 1994. The glucose-regulated proteins (GRP78 and GRP94): functions, gene regulation, and applications. Crit. Rev. Eukaryot. Gene Expr. 4 1–18. [DOI] [PubMed] [Google Scholar]
- 55.Hanada S., M. Harada, H. Kumemura, O. M. Bishr, H. Koga, T. Kawaguchi, E. Taniguchi, T. Yoshida, T. Hisamoto, C. Yanagimoto, et al. 2007. Oxidative stress induces the endoplasmic reticulum stress and facilitates inclusion formation in cultured cells. J. Hepatol. 47 93–102. [DOI] [PubMed] [Google Scholar]
- 56.O'Shea M., C. Stanton, and R. Devery. 1999. Antioxidant enzyme defence responses of human MCF-7 and SW480 cancer cells to conjugated linoleic acid. Anticancer Res. 19 1953–1959. [PubMed] [Google Scholar]
- 57.Albright C. D., E. Klem, A. A. Shah, and P. Gallagher. 2005. Breast cancer cell-targeted oxidative stress: enhancement of cancer cell uptake of conjugated linoleic acid, activation of p53, and inhibition of proliferation. Exp. Mol. Pathol. 79 118–125. [DOI] [PubMed] [Google Scholar]
- 58.Basu S., A. Smedman, and B. Vessby. 2000. Conjugated linoleic acid induces lipid peroxidation in humans. FEBS Lett. 468 33–36. [DOI] [PubMed] [Google Scholar]
- 59.Lee S. H., K. Yamaguchi, J. S. Kim, T. E. Eling, S. Safe, Y. Park, and S. J. Baek. 2006. Conjugated linoleic acid stimulates an anti-tumorigenic protein NAG-1 in an isomer specific manner. Carcinogenesis. 27 972–981. [DOI] [PubMed] [Google Scholar]
- 60.Thompson H., Z. J. Zhu, S. Banni, K. Darcy, T. Loftus, and C. Ip. 1997. Morphological and biochemical status of the mammary gland as influenced by conjugated linoleic acid: implication for a reduction in mammary cancer risk. Cancer Res. 57 5067–5072. [PubMed] [Google Scholar]
- 61.Ip M. M., S. O. McGee, P. A. Masso-Welch, C. Ip, X. Meng, L. Ou, and S. F. Shoemaker. 2007. The t10,c12 isomer of conjugated linoleic acid stimulates mammary tumorigenesis in transgenic mice over-expressing erbB2 in the mammary epithelium. Carcinogenesis. 28 1269–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]