Abstract
Expression of Ldhc begins with the onset of meiosis in male germ cells and continues throughout spermatogenesis. Transcriptional regulatory mechanisms, especially in primary spermatocytes, are poorly described because of the lack of a reliable cell culture system. We constructed mouse transgenics and transfected germ cells in situ to study expression of the testis-specific isozyme of lactate dehydrogenase (LDH). From previous work, we determined that a 100-bp Ldhc core promoter contained potential cis regulatory elements, including a palindrome (−21 to +10), GC box (−70 to −65), and cAMP-responsive element (CRE) sites (−53 to −49, −39 to −35). We provide here the demonstration of a functional role for these sequences by expression of mutated transgenes in vivo. Our results reveal for the first time that mutation of the GC box does not abolish promoter activity, which remains testis-specific. Mutation of GC box or CRE sites resulted in a 73% and 74% reduction in promoter activity, respectively, in a transient transfection of germ cells in vivo by electroporation; the combination of GC box and CRE site mutations eliminates promoter activity. Therefore, we conclude that simultaneous occupancy of the GC box and CRE sites in the core promoter is necessary for full expression of Ldhc in the testis.
Keywords: CRE, electroporation, gamete biology, GC box, gene regulation, Ldhc, mutation, promoter, spermatogenesis, testis, transgenic mice
INTRODUCTION
Spermatogenesis is characterized by genes that are active exclusively in testis, but the mechanisms governing their expression and tissue specificity are largely unknown. The Ldhc gene encodes the important metabolic enzyme lactate dehydrogenase (LDH-C4), which is abundant in germ cells from pachytene spermatocytes to spermatozoa [1]. Previously, we demonstrated that a 100-bp core promoter is sufficient to drive robust tissue-specific expression of Ldhc in transgenic mice [2]. Our original analyses of the promoter were accomplished by in vitro transcription assays; we observed that mutation of a 31-bp palindromic sequence (PAL) in the Ldhc core promoter abolished transcription [3]. In addition, the 100-bp core promoter region contains GC box and cAMP-responsive element (CRE) binding sequences identified in front of a TATA box site. SP1/SP3 consensus sequences and an adjacent CRE binding site are the only regions conserved between the very different murine and human Ldhc promoter sequences. This observation implies important roles for these sites in regulating Ldhc expression. The murine SP1/SP3 transcription factor binding site was confirmed by footprinting and in vitro transfections [4, 5]. Bonny et al. [6] reported an SP1 consensus sequence (GC box) as part of the human Ldhc promoter. The quantitative contribution of SP1/SP3 to promoter activity and tissue specificity of gene expression is unknown. Due to lack of an appropriate germ cell line, two in vivo approaches were used in this research to characterize the 100-bp core promoter: transgenic mice and in vivo electroporation. In vivo DNA electroporation is a novel approach for transient transfection of spermatogenic cells in the live animal. This technique has been successfully applied to germ cell-specific promoter study [7, 8], RNA interference effects in germ cells [9], and the rescue of spermatogenesis in infertile mutant mice [10]. This approach does not address expression of the promoter constructs in somatic tissues. Here we show the effects of transgenic constructs with mutations of PAL or the GC box on promoter activity in vivo, and the quantitative contribution of GC box and CRE sites to promoter activity using in vivo transient transfection of germ cells by electroporation. We report that mutations in PAL and the GC box do not abolish promoter activity; rather, the capacity for efficient testis-specific expression of the transgenes is retained. Our novel finding in these studies is that there is an apparent functional redundancy between GC box and CRE sites, so mutation of one site decreases promoter activity, but mutating both abolishes promoter activity.
MATERIALS AND METHODS
Generation of Transgenic Mice
DNA constructs for generating transgenic animals are shown in Figure 1. The mouse Ldhc promoter from position −425 to +10 was cloned into pNASSβ (Clontech, Palo Alto, CA). To construct PALmu, the promoter fragment was amplified by PCR using sense primer Ldhc-425EcoRI (5’-CAT GAA TTC GTC TAC AGA GTT CCA GGA CG-3’) and antisense primer LdhcMuPalEcoRI/Re (5’-TAG AAT TCG GAT AAC TGT TGG GCT GCA GAG CCA ACA GTT ATA ACG GAA AAG G-3’); the palindrome mutation was introduced into the PCR fragment by the antisense primer, with an EcoRI site included in both primers for cloning. SPmu was prepared by reamplification of two PCR fragments that overlapped at one end and contained the GC box mutation. The primer set for the 5’ fragment was Ldhc-425EcoRI and LdhcMUSP1/Re (5’-ACG CAA GCC CAA ACC ACT GCC TGC TCA GGT ACC-3’); the primer set for the 3’ fragment was LdhcMUSP1/For (5’ -CAG TGG TTT GGG CTT GCG TGC TGA CGT TG-3’) and LdhcEcoRI/Re (5’-TAG AAT TCG GAT AAC TGT TGG GTC CAG GAG CCA ACA GTT A-3’). By mixing these two purified, overlapping PCR fragments, the final mutated fragment can be amplified with Ldhc-425EcoRI primer and LdhcEcoRI/Re primer. After restriction digestion with EcoRI this PCR product was cloned into the MCS region of the pNASSβ vector. An internal KpnI site is in the Ldhc promoter (position −88) and a unique HindIII site is located in the pNASSβ vector (Fig. 1). KpnI/HindIII double digestion of the recombinant DNA releases a fragment with the Ldhc promoter and the bacterial β-galactosidase reporter gene (LacZ). This fragment was purified on an agarose gel by a QIAquick Gel Extraction Kit (Qiagen, Valencia, CA). Constructs were validated by DNA sequencing. Transgenic animals were generated at the Northwestern University Transgenic Core Facility by pronuclear injection and screened by PCR and Southern blot. Mutations in the reporter construct were confirmed by sequencing the PCR product amplified from transgenic animal tail DNA. A 2.3-kb β-galactosidase gene DNA fragment was cut by SacI from the vector pNASSβ (12 to 2274) and used as a Southern blot probe and a transgene copy number standard as described previously [11].
FIG. 1.
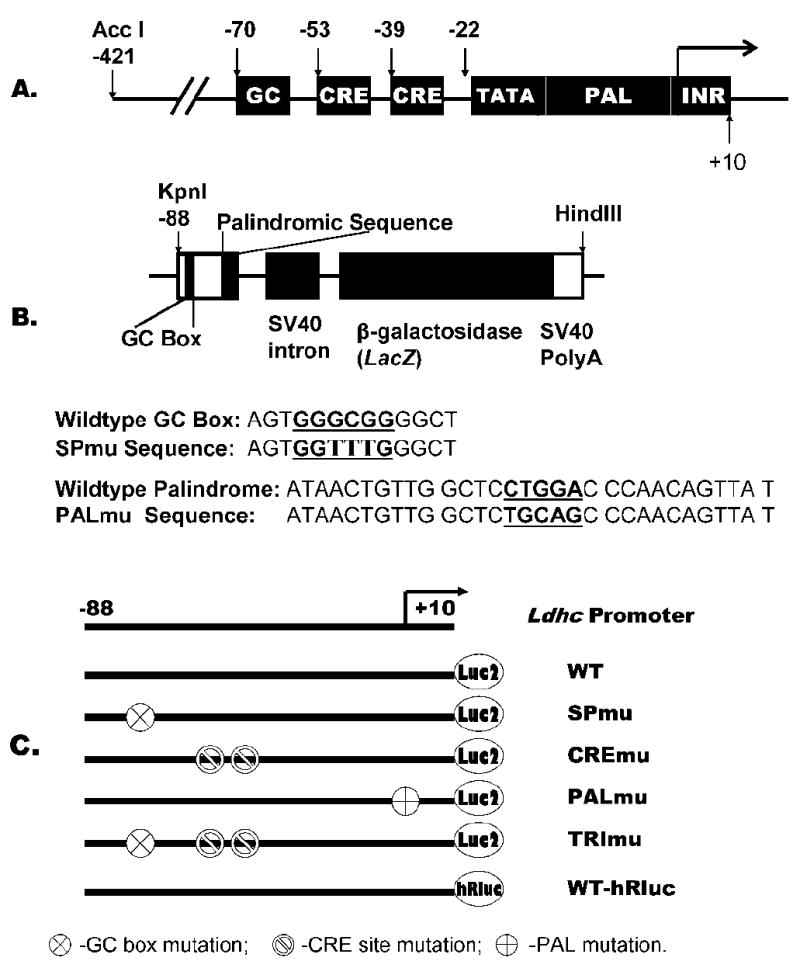
Schematic diagrams illustrating the Ldhc core promoter sequence and DNA constructs. A) The 100-bp core promoter containing a GC box, 2 CRE sites, TATA box, palindromic sequence, and transcription initiation element (INR). B) The transgene construct depicting the Ldhc core promoter sequence from −88 to +12 cloned into pNASSβ for LacZ reporter gene expression, mutation sites for SPmu and PALmu constructs were underlined. C) Constructs used for in vivo electroporation; WT-hRluc was used as a transfection control vector co-injected with other test vectors.
β-Galactosidase Staining and Activity Assay
Testis β-galactosidase (β-gal) activity staining basically followed the procedure previously described [12, 13]. Briefly, mice were killed, and testes were excised and fixed for 2 h at 4°C in PBS-buffered 2% paraformaldehyde containing 0.2% glutaraldehyde, 0.01% sodium deoxycholate, and 0.02% NP-40. Then the testis was washed briefly in PBS, and kept in staining buffer at room temperature overnight. The staining stock solution contained 100 mM sodium phosphate (pH7.3) with 1.3 mM MgCl2, 3 mM K3Fe(CN)6, and 3mM K4Fe(CN)6, to which was added 50 μl 2% X-gal per ml. β-galactosidase activity in fresh tissues was measured using the Galacto-light plus kit BL100P (Applied Biosystems, Bedford, MA). The protocol for chemiluminescent detection of β-galactosidase activity was described previously [13]. Tissues were homogenized, centrifuged, and heated at 48°C for 1 h to destroy endogenous β-galactosidase activity. After centrifugation, the supernatant was stored at −70°C until activity was measured. Protein concentration was determined with the BioRad Protein Assay reagent (Bio-Rad Laboratories, Hercules, CA).
Immunohistochemistry Staining
Whole testes were incubated in Bouin fixative from 4 h to overnight after puncturing the testis capsule in several places to facilitate fixative permeation. Tissue was embedded in paraffin, and 5 μm sections were cut. The sections were deparaffinized and rehydrated with xylene, ethanol (100%, 95%, 70%, 50% for 3–5 min for each step), and then in PBS for 10 min. Slides were treated with Peroxo-Block (Zymed Laboratories, San Francisco, CA) to eliminate endogenous peroxidase. Then endogenous Avidin/Biotin blocking solution (Zymed Laboratories) was added to eliminate background caused by endogenous avidin- or biotin-like substances. A Histostain Plus Rabbit Primary (DAB) kit (Zymed Laboratories) was used for immunohistochemistry (IHC) staining according to the manufacturer’s instructions, except that the tissue sections were blocked for 1 h and incubated with primary antibody for 1 h at room temperature. Rabbit polyclonal anti-β-gal was purchased from MP Biomedicals (Irvine, CA) and used at a 1:600 dilution; goat anti-rabbit secondary antibody was purchased from Sigma-Aldrich (St. Louis, MO) and used at a 1:30 000 dilution. Slides were counterstained with hematoxylin and mounted in Histomount (Zymed Laboratories).
Plasmid Constructs for Electroporation
A construct (Ldhc-425/+29-pGL4) was created by PCR using primer pair Ldhc-425For (5’-CAT AGA TCT GCC TGG TCT ACA GAG TTC CAG GAC G-3’) and Ldhc-20Re (5’-CAT AAG CTT GCA CAG GTA AGA AAC CAG GAT AAC TGT TG-3’) and mouse genomic DNA. The PCR product was cloned into the BglII/HindIII site of vector pGL4-luc2 (Promega, Madison, WI). This construct was confirmed by sequencing and used as template for following PCR reactions except as otherwise specified. The Ldhc promoter for the electroporation constructs all started from position −88 and ended at +12. Transfection control construct (WT-hRluc): PCR with primer set Ldhc-88For (5’-CAT AGA TCT GGT ACC TGA GCA GGC AGT G-3’) and Ldhc+1Re (5’-CAT AAG CTT GGA TAA CTG TTG GGT CCA GGA GCC AAC AG-3’) was cut by BglII/HindIII and cloned into Renilla luciferase vector pGL4.70[hRluc] (Promega). All the following PCR products were cut by BglII and HindIII and cloned into firefly luciferase vector pGL4.10[luc2] (Promega): Wildtype construct (WT): PCR with primer set Ldhc-88For and Ldhc+1Re; SPmu construct (SPmu): PCR with primers Ldhc-88For, Ldhc+1Re and template pNASSβ(Spmu) (plasmid from transgene construct, above); PALmu construct (PALmu): PCR with primers Ldhc-88For and PALmu/Re (5’-CAT AAG CTT GGA TAA CTG TTG GGC TGC AGA GCC AAC AGT TAT AAC GGA AAA GG-3’) and template pNASSβ(PALmu) (described above); CREmu construct (CREmu): PCR1 by primers Ldhc-425For and CREmu/Re (5’-GGA AAA AAA CAA AGT CAA AAA AAG CAC GCA AGC CCC GCC CAC-3’); and PCR2 by primers TRImu/For (5’-GCG TGC TTT TTT TGA CTT TGT TTT TTT CCT TTT CCG TTA TAA CTG TTG GC-3’) and Ldhc+32/Re (5’-CAC ACC TTA CTG CTG ACT CCG-3’). PCR products from PCR1 and PCR2 were diluted 200 times and mixed as a template for final PCR with primers Ldhc-88For and Ldhc+1Re; TRImu construct (TRImu): PCR1 by primers Ldhc-425For, TRImu/Re (5’-GGA AAA AAA CAA AGT CAA AAA AAG CAC GCA AGC CCA AAC CAC-3’) and template pNASSβ(SPmu); and PCR2 by primers TRImu/For, Ldhc+32/Re, and template pGL4 (Ldhc-425/+500). PCR products from PCR1 and PCR2 were diluted 200 times and combined together as a template for final PCR with primer Ldhc-88For and Ldhc+1Re.
In Vivo Electroporation
A mixture consisting of 17 μg test plasmid (Fig. 1) and 1.7 μg control plasmid (WT-hRluc) was injected via the efferent ducts into the surgically exposed testis of a 5-wk-old CD1 mouse (Harlan, Indianapolis, IN) [14]. Injection pipettes were prepared using Borosilicate glass capillaries (WP Instruments [WPI], Sarasota, FL), a micropipette puller, and MicroBeveler 48000 (WPI). Micropipette insertion into the rete testis was done manually and the DNA solution was delivered by UltraMicroPump II (WPI). The square wave electroporator CUY21EDIT (NEPA GENE) and Tweezertrodes 520 (BTX Instrument Division of Genetronics, San Diego, CA) were used for electroporation according to Somboonthum et al. [8]. Briefly, electric pulses were applied eight times in four different directions at 40 V for 50 ms at intervals of 950 ms. Testes were harvested 3 days after electroporation for luciferase activity analysis. Luciferase reporter activity was measured by a Dual-Luciferase Reporter Assay System (Promega). Transfection efficiency was normalized using cotransfected Renilla luciferase activity. For each construct four testes were used.
Electrophoretic Mobility Shift Assay
The gel shift assay kit supplied by Promega was used for this procedure according to the manufacturer with the following modifications. The probes were labeled with [γ-32p] ATP. The reaction product was purified on a G-50 micro column (Amersham Pharmacia, Piscataway, NJ), and labeling efficiency was monitored with a Beckman LS 6500 Scintillation Counter. Electrophoretic mobility shift assay (EMSA) on a 5% 0.75-mm thick acrylamide gel was performed in the cold room until the dye front reached the bottom of the gel. The following oligonucleotides were used for the EMSA: GC box Ldhc-SP (5’-AGC AGG CAG TGG GCG GGG CTT G-3’); mutated GC box Ldhc-SPmu (5’-AGC AGG CAG TGG TTT GGG CTT G-3’); competitor SP1 consensus (5’-ATT CGA TCG GGG CGG GGC GAGC); CRE sites, Ldhc-CRE (5’-TGC GTG CTG ACG TTG ACT TTG TGA CGT TCC-3’); mutated Ldhc CRE sites, Ldhc-CREmu1 (5’-TGC GTG CTT TTT TTG ACT TTG TTT TTT TCC-3’); and canonical CRE sequence (5’-CTA GCT CTC TGA CGT CAG GCA ATC TCT-3’) used as competitor. SP1 (sc-59), SP3 (sc-644) and CRE modulator(CREM; sc-440) antibodies for supershifts were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA), and antibody recognizing CREB was obtained from Rockland Immunochemicals (Gilbertsville, PA).
The use of animals in this study was approved by the Northwestern University Institutional Animal Care and Use Committee within the guidelines for humane care and treatment of rodents.
RESULTS
The 100-bp Ldhc core promoter was mutated in the hinge region of the PAL sequence as in the in vitro transcription assay [3] and designated PALmu (Fig. 1). The construct with a mutation in the GC box was designated SPmu (Fig. 1). According to Yang and Thomas [4], this consensus sequence binds transcription factor SP1 and may play an important role in transcription and possibly tissue specificity of Ldhc expression. Six transgenic founder lines were obtained for PALmu, with four of them positive for β-gal expression. Three SPmu lines showed β-gal expression (Table 1). The founder lines that did not produce pups were sacrificed for tissue harvest and reporter expression assays.
TABLE 1.
Analysis of transgenic founders and offspring.
| β-Gal reporter expressionb |
|||
|---|---|---|---|
| Founder linea | Founder | Pups | Transgene copy no. |
| PALmu | |||
| 323 (M) | 4 Positive | 5 | |
| 321 (M) | Positive | ||
| 327 (M) | Positive | ||
| 330 (F) | 1 Positive | 24 | |
| SPmu | |||
| 230 (M) | Positive | ||
| 260 (M) | 1 Positive | 4 | |
| 272 (M) | 3 Positive | 5 | |
M, Male; F, Female.
Positive β-gal expression determined by visible X-gal staining in testis tubules; significant level of β-gal activity in testis compared to background level in somatic tissues; and by IHC staining of testis paraffin section by β-gal antibody.
We detected activity of the transgene reporter by X-gal staining of the intact testis, β-gal activity in tissue extracts, and IHC of paraffin-embedded sections. Two or all three methods were applied to animals that had been confirmed on Southern blots to contain the transgene. X-gal staining of the testis clearly indicated that both the PALmu and SPmu transgenes were expressed (Fig. 2). A stage-specific pattern of β-gal expression was observed in seminiferous tubules in which only alternating segments were stained. This is consistent with IHC detection of β-gal in spermatocytes, but not later stages of spermatogenesis. For each test animal, the testis, brain, heart, kidney, liver, and spleen were homogenized, and soluble fractions were used for β-gal activity measurement (Fig. 3). In the PALmu founder line 323, β-gal activity in testes was two orders of magnitude higher compared to the background amounts in all somatic tissue samples (Fig. 3A). Other PALmu founder lines showed lower activity readings but a similar expression pattern. In SPmu founder line 272, the three offspring examined for β-gal activity had significantly higher levels in testis (P < 0.05), whereas only background activity was detected in somatic tissues (Fig. 3B). The same pattern was observed with the other two SPmu founder lines. This β-gal activity distribution indicates that the Ldhc core promoter with either mutation retains activity for efficient testis-specific expression of Ldhc. Reporter gene expression was limited to pachytene primary spermatocytes (Fig. 4) [2, 15], though LDH-C4 is always detectable from meiosis to postmeiotic stages.
FIG. 2.
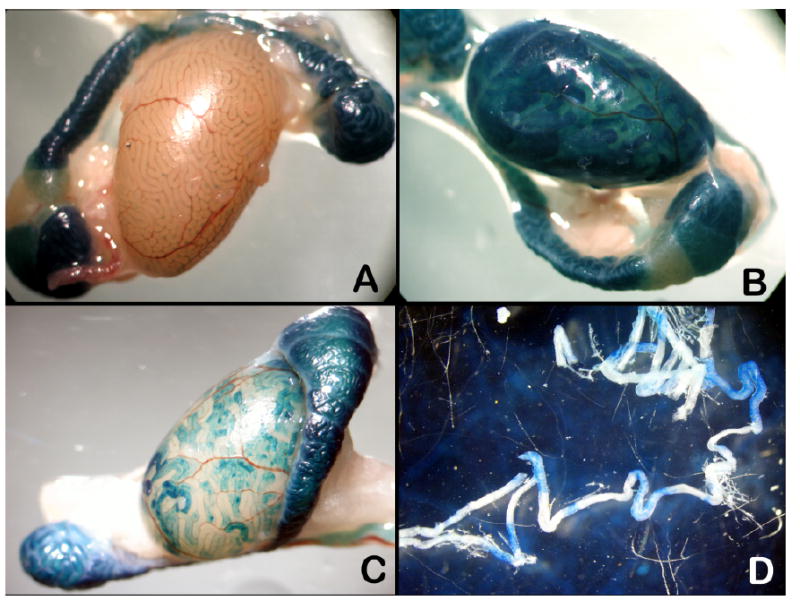
X-gal stained transgenic testis. A) PALmu transgenic littermate identified as negative by PCR. B) PALmu transgenics. C) SPmu testis. D) Stage-specific staining pattern in seminiferous tubules of a PALmu mouse; epididymal staining is from endogenous β-gal. Original magnification ×10.
FIG. 3.
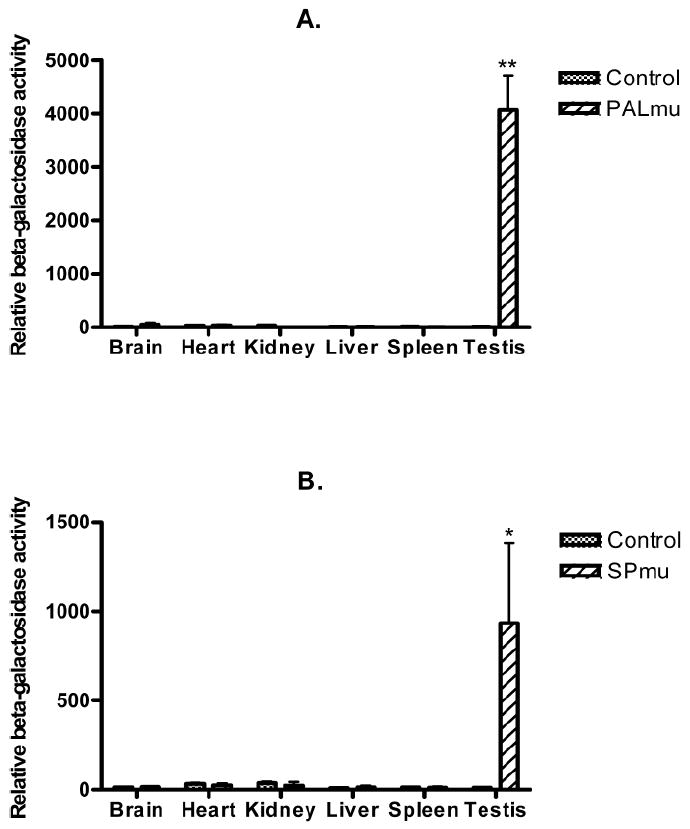
β-galactosidase activity analyses. Average β-gal activity from founder lines PALmu323 and SPmu272 were illustrated with nontransgenic control animals. All readings calibrated as RLU (relative light unit) per μg protein. One-way ANOVA and Dunnett’s Multiple Comparsion Test were performed between testis and somatic tissues. Bar represents means ± SEM, n = 3. *P < 0.05, **P < 0.01 indicates significant difference.
FIG. 4.
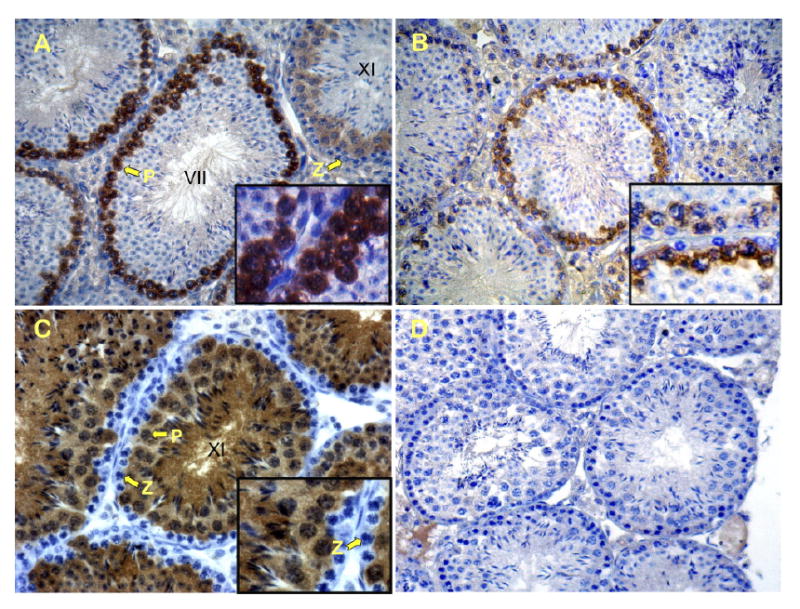
IHC staining of testis sections from PALmu and SPmu transgenic mice. A) Anti-β-gal staining of PALmu transgenic testis section. B) Anti-β-gal staining in SPmu testis sections. C and D) Nontransgenic sections stained with anti-LDHC and anti-β-gal antibodies respectively. Arrows in A and C indicate zygotene (Z) and pachytene (P) spermatocytes. Reaction product is visible only in pachytene and later stages of spermatogenesis (see insets, magnification ×1200). Original magnification ×400.
In vivo electroporation was first tested using GFP reporter vector pAcGFP-N1 from Clontech (Fig. 5). Plasmid DNA solution with 10% trypan blue dye was microinjected into testis seminiferous tubules through the efferent duct. About 50% of surface tubules were filled with plasmid DNA solution. Twenty four hours after electroporation, robust GFP expression was observed in these seminiferous tubules; both frozen sections and paraffin-embedded sections demonstrated efficient plasmid DNA transfection of testicular cells. Five different promoter constructs, along with a Renilla luciferase control vector (WT-hRluc), were injected into seminiferous tubules to examine relative promoter activity (Fig. 1C and Fig. 6). PALmu did not affect promoter activity, but SPmu and CREmu led to 73% and 74% reduction, respectively. The double mutation designated TRImu abolished promoter activity completely. Specific DNA-protein interactions on the GC box and CRE sites were demonstrated by a gel mobility shift assay (EMSA) using testis nuclear extract (Fig. 7). Multiple DNA-protein bands were observed for the Ldhc-SP oligo probe. Fifty-fold cold probe (Ldhc-SP) was able to compete with all the bands; however, addition of 50-fold mutated probe (Ldhc-SPmu) did not change the gel-shift band pattern. A consensus SP1 sequence can also compete with the shifted bands. Supershifts were observed with SP1 or SP3 antibody for the Ldhc-SP site. Similar results were obtained with Ldhc-CRE oligo with distinctive gel-shift bands observed; either 50-fold of cold probe or consensus CRE oligo sequence was able to compete with the binding of testis nuclear proteins. No competition was observed by the oligo with the CRE site mutated. Supershift was detected with the CREB or the CREM antibody (Fig. 7).
FIG. 5.
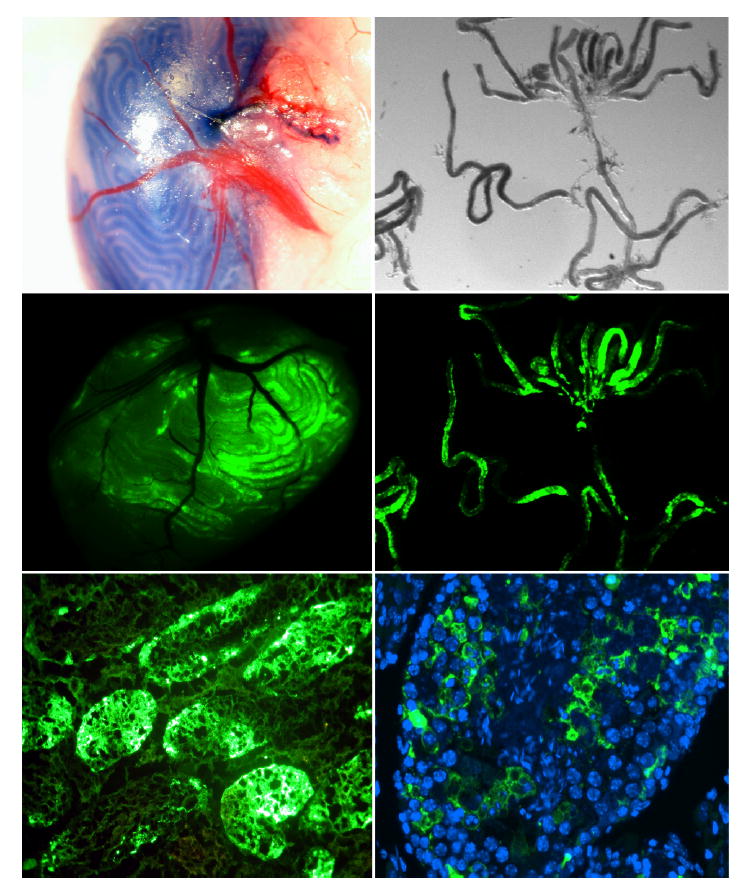
In vivo electroporation. Microinjection of pAcGFP-N1 into seminiferous tubules via the efferent duct, followed by electroporation of surgically exposed testis. Diagram shows GFP expression 24 h after electroporation. Top: Bright field view of seminiferous tubules injected with DNA solution containing 10% trypan blue tracking dye. Middle: Fluorescence microscopy of testis (left) and seminiferous tubules (right). Bottom: Fluorescence in frozen section (left) and paraffin-embedded section (right) showing GFP expression in transfected germ cells. Original magnification ×10 for top and middle panel; original magnification ×100 and ×400 for bottom panel left and right, respectively.
FIG. 6.
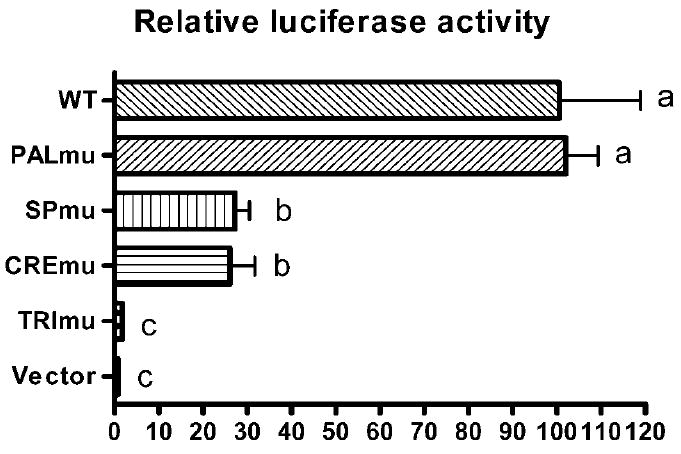
Analysis of Ldhc promoter activity by in vivo electroporation. Horizontal axis represents the percentage of luciferase activity for each mutated promoter sequence compared to the wildtype promoter. Ldhc promoter sequences extended from −88 to +12 in all constructs. WT, wildtype promoter; PALmu, PAL sequence mutation; SPmu, GC box mutation; CREmu, both CRE sites mutated; TRImu, combined mutation of GC box and both CRE sites. One-way ANOVA and two-tailed, unpaired t-test between groups were performed. Bars with different letters are significantly different (P < 0.01).
FIG. 7.
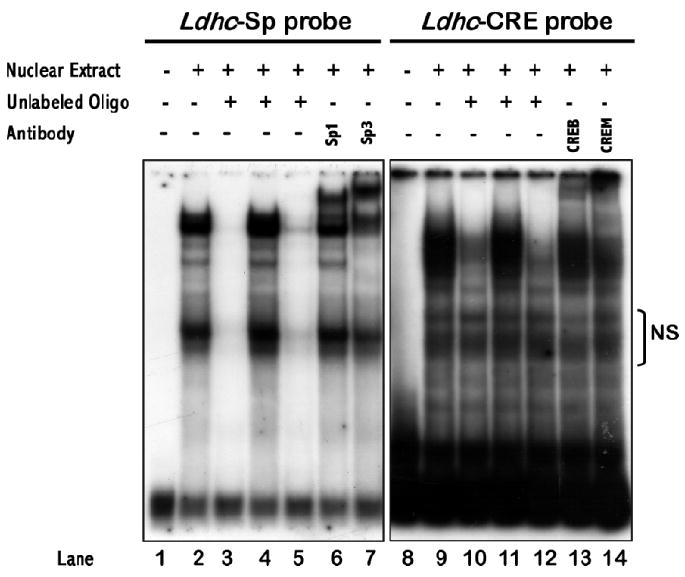
Gel-shift assay demonstrating specific interaction of Ldhc promoter oligos containing GC box (Ldhc-SP) or CRE sequences (Ldhc-CRE) with testis nuclear protein extract (TN). Lanes 1 to 7, oligo Ldhc-SP used as probe; lane 1: Ldhc-SP probe; lane 2: TN + probe; lanes 3–5: TN + probe + 50-fold non-radioactive Ldhc-SP, Ldhc-SPmu, and SP1 consensus, respectively; lanes 6–7: TN + probe + antibody. Lanes 8–14, oligo Ldhc-CRE used as probe; lane 8: Ldhc-CRE; lane 9: TN + probe; lanes 10–12: TN + probe + 50-fold non-radioactive Ldhc-CRE, Ldhc-CREmu1, and CRE consensus, respectively; lanes 13–14: TN + probe + antibody. NS, Nonspecific.
DISCUSSION
LDH-C4 is one of the most abundant proteins in mouse testis at a concentration even higher than β-actin (Liang and Goldberg, unpublished observation). But expression of the gene is strictly limited to meiotic and postmeiotic germ cells and does not occur in any somatic tissues. That makes Ldhc an ideal model gene for studying the regulation of tissue-specific expression.
Previously the 100-bp Ldhc promoter (−88 to +10) was defined by its ability to drive testis-specific expression in an in vitro transcription assay [3] and with generation of transgenic animals [2]. Mutation of PAL (−21 to +10) abolished promoter activity [3]. To determine whether PAL contains regulatory elements, we constructed a transgene mutation similar to that described by Zhou and Goldberg [3]. Of the four transgenic founder lines produced (Table 1), line 323 yielded the most male transgenic pups. In this line the β-gal reporter activity in the testis is two to three orders of magnitude higher than in somatic tissues (Fig. 3A), demonstrating clearly that PAL mutation does not change the testis-specific expression pattern; therefore, PAL is not responsible for tissue specificity of Ldhc expression. The discrepancy between the cell-free in vitro transcription assay and this result could be due to the special secondary structure of the PAL sequence. Our in vivo electroporation assay also indicated that the PAL mutation did not affect promoter activity. Based on these observations, we conclude that PAL is not involved in Ldhc regulation.
By DNase I treatment of the Ldhc proximal promoter (−524/+38) sequence from isolated spermatocytes, Yang and Thomas [4] identified a single footprint over the GC box motif that contained three DNase I hypersensitive sites located upstream from the CCAT box (−117/−114). These results suggest that Ldhc expression involves enhancer-assisted, SP1-mediated transcriptional activation. Therefore, we hypothesized that SP1/SP3 were essential for Ldhc expression. Surprisingly, the Ldhc core promoter with a mutated GC box remained transcriptionally active and retained tissue specificity (Figs. 2-4). However, the average transgene reporter activity in line Spmu 272 is only one fourth that of PALmu 323. Our in vivo electroporation assay indicated that the GC box mutation caused a 73% reduction in total promoter activity (Fig. 6). These results suggest that SP1/SP3, though not essential for Ldhc basal and tissue-specific activity, nevertheless enhance its expression.
There is strong evidence that the cAMP signaling pathway is a major mechanism for regulating gene expression at different stages of spermatogenesis [16]. Two bZIP transcription factor family members are the CREB and the CREM, which bind to the CRE in a target gene to activate its expression. For example, Ldha contains a CRE site (−48/−41) in the proximal promoter and is regulated by cAMP [17].
Our EMSA results described here strongly support CRE-CREM/CREB DNA-protein interactions. The functional assays indicate that the CRE sequence is an essential element for Ldhc gene activation. The CRE site is conserved as well in the human Ldhc promoter. Because the LDH-C4 catalyzes the reversible oxidation of lactate, we can suggest that FSH-induced cAMP signaling plays an important role in regulating glycolysis during spermatogenesis. There is abundant evidence that spermatozoa rely on aerobic glycolysis to transduce the energy required for motility and fertilization. For example, targeted disruption of the testis-specific glyceraldehyde phosphate dehydrogenase (GAPDS) gene [18], a key glycolytic enzyme, inhibits sperm motility and fertilizing capacity. A similar phenotype has been observed in our laboratory by targeted disruption of Ldhc (Odet and Goldberg et al., unpublished results).
Our mutational analysis in which Ldhc promoter activity is abolished is a significant finding that allows us to propose a mechanism for the tissue-specific expression of Ldhc and possibly other genes that contain these cis elements and show testis-restricted activation. We suggest that transcription factor availability and/or methylation can effectively block gene expression. The GC box (GGGCGG) and CRE (TGACGT) sequences contain CpG dinucleotides that are targets for methylation. Kroft et al. [19] reported that these sites are indeed hypermethylated in somatic tissue, but hypomethylated in testis; methylation of an Ldhc promoter (−425/+10) renders it inactive in an in vitro transfection assay. Repressor ICER, inhibitor of CREM/CREB, may be involved in suppressing Ldhc promoter activity in somatic cells. Because an animal with a mutated GC box transgene still retains a testis-specific expression of reporter activity, it is possible that transcription factor CREM coordinated with a testis-specific coactivator is responsible for tissue specificity. It is well established that Activator of CREM (ACT) in testis interacts with CREM and regulates postmeiotic germ cell-specific genes [20]. Coactivator ACT is only detected in spermatids, but Ldhc expression starts in pachytene spermatocytes; therefore, we suggest that a coactivator like FHL4 (Four and a half LIM domain) may function in the early stages of spermatogenesis [21]. FHL4 is a testis-specific coactivator that shares a high degree of homology with ACT; its expression can be detected first in spermatocytes.
Our results with mutated sequences also demonstrate that a long-range distal enhancer is not required for testis specificity, and the likely random chromosomal integration of transgenes eliminates the possibility that tissue specificity is due to different chromatin contexts in germ cells and somatic cells. Although the Ldhc core promoter is relatively short, there are numerous examples of other similarly structured testis-specific core promoter sequences. These include the 91-bp angiotensin-converting enzyme (Ace) promoter [22], a 330-bp calmegin promoter [23], the 87-bp β4-galactosyltransferase-I promoter [24], the 193-bp Gsg2 (haspin) promoter [25], a 248-bp Hils1 promoter [26], the 165-bp Hst70/Hsp70.2 promoter [27], the 187-bp Pdha2 promoter [28], the 116-bp proenkephalin promoter [29], the 187-bp protamine 1 promoter [30], and the 294-bp Sp10 promoter [31]. These sequences control either spermatid-specific or testis-specific gene expression (including both meiotic and postmeiotic cells). Common cis elements are not found by comparison of these promoter sequences, but a GC box is present in 6 of 11 gene promoters and a CRE site in 7 of 11 gene promoters. Transcription repression by protein-DNA interaction of these sites is likely to be a major mechanism for silencing testis-specific genes in somatic tissues. Further study of the Ldhc promoter should provide answers to regulatory mechanisms for testis specific genes.
Acknowledgments
We thank Dr. Lynn Doglio and the Northwestern University Transgenic Core Facility for generation of transgenic mice. We are grateful to Dr. Derek McLean for instruction in seminiferous tubule microinjection. We also thank Chongwen Duan for technical help with immunohistochemistry.
Footnotes
Supported by NIH grant R01 HD005863-35. Core facilities used in this research were provided by the Lurie Cancer Center, Northwestern University.
References
- 1.Hintz M, Goldberg E. Immunohistochemical location of LDH-X during spermatogenesis in mouse testes. Dev Biol. 1977;57:375–384. doi: 10.1016/0012-1606(77)90222-6. [DOI] [PubMed] [Google Scholar]
- 2.Li S, Zhou W, Doglio L, Goldberg E. Transgenic mice demonstrate a testis-specific promoter for lactate dehydrogenase, Ldhc. J Biol Chem. 1998;273:31191–31194. doi: 10.1074/jbc.273.47.31191. [DOI] [PubMed] [Google Scholar]
- 3.Zhou W, Goldberg E. A dual-function palindromic sequence regulates testis-specific transcription of the mouse lactate dehydrogenase c gene in vitro. Biol Reprod. 1996;54:84–90. doi: 10.1095/biolreprod54.1.84. [DOI] [PubMed] [Google Scholar]
- 4.Yang J, Thomas K. Molecular and functional characterization of the promoter region of the mouse Ldhc gene: enhancer-assisted, SP1-mediated transcriptional activation. Nucleic Acids Res. 1997;25:2213–2220. doi: 10.1093/nar/25.11.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thomas K, Sung DY, Yang J, Johnson K, Thomas W, Millette C, McCarrey J, Breitberg A, Gibbs R, Walker W. Identification, characterization, and functional analysis of SP1 transcript variants expressed in germ cells during mouse spermatogenesis. Biol Reprod. 2005;72:898–907. doi: 10.1095/biolreprod.104.030528. [DOI] [PubMed] [Google Scholar]
- 6.Bonny C, Cooker LA, Goldberg E. Deoxyribonucleic acid-protein interactions and expression of the human testis-specific lactate dehydrogenase promoter: transcription factor SP1 plays a major role. Biol Reprod. 1998;58:754–759. doi: 10.1095/biolreprod58.3.754. [DOI] [PubMed] [Google Scholar]
- 7.Ike A, Ohta H, Onishi M, Iguchi N, Nishimune Y, Nozaki M. Transient expression analysis of the mouse ornithine decarboxylase antizyme haploid-specific promoter using in vivo electroporation. FEBS Letters. 2004;559:159–164. doi: 10.1016/S0014-5793(04)00065-1. [DOI] [PubMed] [Google Scholar]
- 8.Somboonthum P, Ohta H, Yamada S, Onishi M, Ike A, Nishimune Y, Nozaki M. cAMP-responsive element in TATA-less core promoter is essential for haploid-specific gene expression in mouse testis. Nucleic Acids Res. 2005;33:3401–3411. doi: 10.1093/nar/gki652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shoji M, Chuma S, Yoshida K, Morita T, Nakatsuji N. RNA interference during spermatogenesis in mice. Dev Biol. 2005;282:524–534. doi: 10.1016/j.ydbio.2005.03.030. [DOI] [PubMed] [Google Scholar]
- 10.Yomogida K, Yagura Y, Nishimune Y. Electroporated transgene rescued spermatogenesis in infertile mutant mice with a Sertoli cell defect. Biol Reprod. 2002;67:712–717. doi: 10.1095/biolreprod.101.001743. [DOI] [PubMed] [Google Scholar]
- 11.Camper SA. Research applications of transgenic mice. Biotechniques. 1987;5:638–650. [Google Scholar]
- 12.Langford KG, Shai SY, Howard TE, Kovac MJ, Overbeek PA, Bernstein KE. Transgenic mice demonstrate a testis-specific promoter for angiotensin-converting enzyme. J Biol Chem. 1991;266:15559–15562. [PubMed] [Google Scholar]
- 13.Shaper NL, Harduin-Lepers A, Shaper JH. Male germ cell expression of murine β4-galactosyltransferase. J Biol Chem. 1994;269:25165–25171. [PubMed] [Google Scholar]
- 14.Ogawa T, Arechaga JM, Avarbock MR, Brinster RL. Transplantation of testis germinal cells into mouse seminiferous tubules. Intl J Dev Biol. 1997;41:111–122. [PubMed] [Google Scholar]
- 15.Kroft TL, Li S, Doglio L, Goldberg E. A transgenic analysis of mouse lactate dehydrogenase c promoter activity in the testis. J Androl. 2003;24:843–852. doi: 10.1002/j.1939-4640.2003.tb03135.x. [DOI] [PubMed] [Google Scholar]
- 16.Don J, Stelzer G. The expanding family of CREB/CREM transcription factors that are involved with spermatogenesis. Mol Cell Endocrinol. 2002;187:115–124. doi: 10.1016/s0303-7207(01)00696-7. [DOI] [PubMed] [Google Scholar]
- 17.Short ML, Huang D, Milkowski DM, Short S, Kunstman K, Soong CJ, Chung KC, Jungmann RA. Analysis of the rat lactate dehydrogenase A subunit gene from promoter/regulatory region. Biochem J. 1994;304:391–398. doi: 10.1042/bj3040391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miki K, Qu W, Goulding EH, Willis WD, Bunch DO, Strader LF, Perreault SD, Eddy EM, O’Brien DA. Glyceraldehyde 3-phosphate dehydrogenase-S, a sperm-specific glycolytic enzyme, is required for sperm motility and male fertility. Proc Natl Acad Sci U S A. 2004;101:16501–16506. doi: 10.1073/pnas.0407708101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kroft T, Jethanandani P, McLean DJ, Goldberg E. Methylation of CpG dinucleotides alters binding and silences testis-specific transcription directed by the mouse lactate dehydrogenase c promoter. Biol Reprod. 2001;65:1522–1527. doi: 10.1095/biolreprod65.5.1522. [DOI] [PubMed] [Google Scholar]
- 20.Macho B, Brancorsini S, Fimia GM, Setou M, Hirokawa N, Sassone-Corsi P. CREM-dependent transcription in male germ cells controlled by a kinesin. Science. 2002;298:2388–2390. doi: 10.1126/science.1077265. [DOI] [PubMed] [Google Scholar]
- 21.Fimia GM, De Cesare D, Sassone-Corsi P. A family of LIM-only transcriptional coactivators: tissue-specific expression and selective activation of CREB and CREM. Mol Cell Biol. 2000;20:8613–8622. doi: 10.1128/mcb.20.22.8613-8622.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Howard T, Balogh R, Overbeek P, Bernstein KE. Sperm-specific expression of angiotensis-converting enzyme (ACE) is mediated by a 91-base-pair promoter containing a CRE-like element. Mol Cell Biol. 1993;13:18–27. doi: 10.1128/mcb.13.1.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Watanabe D, Okabe M, Hamajima N, Morita T, Nishina Y, Nishimune Y. Characterization of the testis-specific gene ‘calmegin’ promoter sequence and its activity defined by transgenic mouse experiments. FEBS Lett. 1995;368:509–512. doi: 10.1016/0014-5793(95)00729-s. [DOI] [PubMed] [Google Scholar]
- 24.Charron M, Shaper NL, Rajput B, Shaper JH. A novel 14-base-pair regulatory element is essential for in vivo expression of murine β4-galactosidase-I in late pachytene spermatocytes and round spermatids. Mol Cell Biol. 1999;19:5823–5832. doi: 10.1128/mcb.19.8.5823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tokuhiro K, Miyagawa Y, Yamda S, Hirose M, Ohta H, Nishimune Y, Tanaka H. The 193 base pair Gsg2 (Haspin) promoter region regulates germ-cell-specific expression bidirectionally and synthronously in mice. Biol Reprod. 2007;76:407–414. doi: 10.1095/biolreprod.106.055236. [DOI] [PubMed] [Google Scholar]
- 26.Iguchi N, Tanaka H, Yamada S, Nishimura H, Nishimune Y. Control of mouse Hils1 gene expression during spermatogenesis: identification of regulatory element by transgenic mouse. Biol Reprod. 2004;70:1239–1245. doi: 10.1095/biolreprod.103.024760. [DOI] [PubMed] [Google Scholar]
- 27.Scieglinska D, Vydra N, Krawczyk Z, Widlak W. Location of promoter elements necessary and sufficient to direct testis-specific expression of the Hst70/Hsp70.2 gene. Biochem J. 2004;379:739–747. doi: 10.1042/BJ20031842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iannello RC, Young J, Sumarsono S, Tymms MJ, Dahl HM, Gould J, Hedger M, Kola I. Regulation of Pdha2 expression is mediated by proximal promoter sequences and CpG methylation. Mol Cell Biol. 1997;17:612–619. doi: 10.1128/mcb.17.2.612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu F, Tokeson J, Persengiev SP, Ebert K, Kilpatrick DL. Novel repeat elements direct rat proenkephalin transcripton during spermatogenesis. J Biol Chem. 1997;272:5056–5062. doi: 10.1074/jbc.272.8.5056. [DOI] [PubMed] [Google Scholar]
- 30.Zambrowicz B, Palmiter RD. Testis-specific and ubiquitous proteins bind to functionally important regions of the mouse protamine 1 promoter. Biol Reprod. 1994;50:65–72. doi: 10.1095/biolreprod50.1.65. [DOI] [PubMed] [Google Scholar]
- 31.Reddi PP, Flickinger CJ, Herr JC. Round spermatid-specific transcription of the mouse Sp10 gene is mediated by a 294-pair proximal promoter. Biol Reprod. 1999;61:1256–1266. doi: 10.1095/biolreprod61.5.1256. [DOI] [PubMed] [Google Scholar]


