Abstract
A pseudoknot formed by a long-range interaction in the mRNA of the initiation factor 3 (IF3) operon is involved in the translational repression of the gene encoding ribosomal protein L35 by another ribosomal protein, L20. The nucleotides forming the 5′ strand of the key stem of the pseudoknot are located within the gene for IF3, whereas those forming the 3′ strand are located 280 nt downstream, immediately upstream of the Shine–Dalgarno sequence of the gene for L35. Here we show that premature termination of IF3 translation at a nonsense codon introduced upstream of the pseudoknot results in a substantial enhancement of L20-mediated repression of L35 expression. Conversely, an increase of IF3 translation decreases repression. These results, in addition to an analysis of the effect of mutations in sequences forming the pseudoknot, indicate that IF3 translation decreases L20-mediated repression of L35 expression. We propose that ribosomes translating IF3 disrupt the pseudoknot and thereby attenuate repression. The result is a novel type of translational coupling, where unfolding of the pseudoknot by ribosomes translating IF3 does not increase expression of L35 directly, but alleviates its repression by L20.
Keywords: pseudoknot, RNA folding, ribosome, translational control, feedback regulation
In phages and bacteria, genes corresponding to related functions are often grouped in large transcription units, known as operons. Coregulation of the individual genes within these operons often occurs at the level of initiation or termination of transcription. However, control of gene expression can also occur at the level of translation and the regulatory event is, in general, targeted to the first gene of the operon and propagated to the downstream gene by translational coupling. Translational coupling is where an event-altering translation of one gene also affects translation of the downstream gene without affecting its transcription. Translational coupling usually depends on the unfolding of an inhibitory secondary structure in the mRNA, which sequesters the ribosome binding site (RBS) of the distal gene. In general, ribosomes translating the upstream cistron unfold the inhibitory secondary structure, thus making the sequestered RBS accessible to incoming ribosomes (refs. 1–3 and references therein). In the case of the coupled expression of the RNA phage MS2 coat protein and lysis genes, the mechanism is slightly different; ribosomes that have terminated translation of the upstream coat protein gene first disrupt the secondary structure that sequesters the RBS of the lysis gene and then initiate translation of that gene (4). Finally, a more sophisticated mechanism involves stalling of the ribosome at a specific position of an upstream leader peptide sequence and the consequent alteration of an adjacent mRNA secondary structure resulting in the exposure of the RBS of the downstream gene. This mechanism, known as “translation attenuation,” governs the regulation of antibiotics-inducible genes in Gram-positive bacteria (5–7). Translational coupling can also rely on a mechanism that does not involve the disruption of an inhibitory secondary structure. In the case of coupling between genes V and VII of the single-stranded DNA phage f1, it has been proposed that gene V “hands over” ribosomes to the inherently defective RBS of the downstream gene VII (8).
This study describes a novel form of translational coupling that occurs in the initiation factor 3 (IF3) operon. This operon, also referred to as the infC–rpmI–rplT operon (9) or the IF3–L35–L20 operon (10), contains the infC, rpmI, and rplT genes encoding translation IF3 and the two ribosomal proteins, L35 and L20. The operon can also be extended to the upstream gene, thrS, encoding threonyl–tRNA synthetase. This set of genes is transcribed from four promoters (Fig. 1 and refs. 9 and 11). The first (pthrS) is located immediately upstream of thrS. The second (p0) and third (p0′) promoters are located within thrS, and the fourth (p1) within infC. The major promoters are pthrS and p0′. The operon also contains two leaky transcriptional terminators, t1, located at the end of infC, and t2, located at the end of the operon. Expression of the IF3 operon is regulated by two different control loops acting at the translational level. First, IF3 represses the expression of its own gene (12). Second, L20 directly represses the expression of rpmI, and indirectly that of its own gene, rplT, via translational coupling with rpmI (3). We have previously shown that a pseudoknot, formed by a long-range RNA–RNA base pairing interaction, strongly enhances L20-mediated repression of rpmI expression (10). The nucleotides forming the 5′ strand of the key stem of the pseudoknot are located within infC, whereas those forming the 3′ strand are located about 280 nt downstream, just upstream of the Shine–Dalgarno (SD) sequence of rpmI (Fig. 2). However, we have also shown that point mutations in the key stem do not fully abolish repression (10), suggesting that L20 might bind to other sites in the translational operator as well. In this work, we investigate the effect of infC translation on L20-mediated repression of rpmI expression.
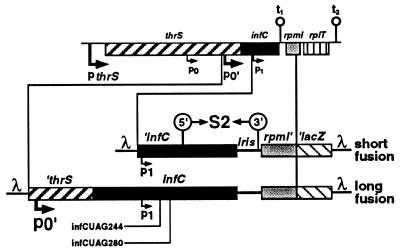
Figure 1.
Schematic representation of the translational fusions. The region of the Escherichia coli chromosome region containing the thrS, infC, rpmI, and rplT genes is schematized at the top of the figure. The pthrS, p0, p0′, and p1 promoters are indicated by bent arrows and the transcriptional terminators, t1 and t2, by hairpins. The structures of the short and long rpmI′–′lacZ fusions are shown underneath. A prime on one side of a gene means that it is truncated on this side. iris is an acronym for infC–rpmI intergenic spacer. λ sequences are also indicated. Amber mutations infCUAG244 and infCUAG280 are shown. Locations of the two strands forming stem S2 of the pseudoknot (see Fig. 2) are indicated by 5′ and 3′ enclosed in circles.
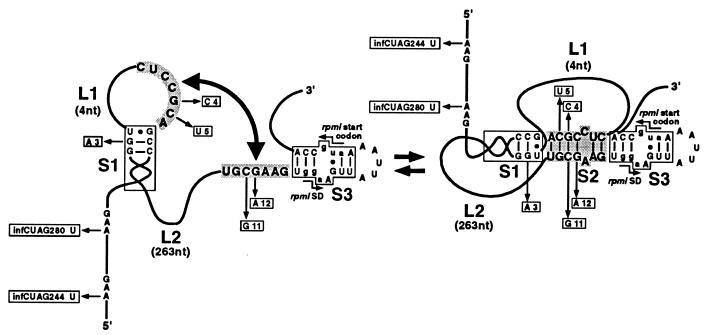
Figure 2.
Schematic representation of the long-range interaction required for formation of the pseudoknot involved in L20-mediated repression of rpmI expression. Stems S1 and S3 are represented by open boxes. L1 and L2 schematize the connecting loops of the pseudoknot. Their lengths are shown in parentheses. Nucleotides forming the two strands of stem S2 of the pseudoknot are shaded. The 5′ strand of stem S2 is sequence CUCCGCA from positions infC331 to infC337 and the 3′ strand is sequence UGCGAAG from positions iris 76 to iris 82. Mutations located in the sequences forming the pseudoknot (except mutations 7 and 8, which are located in stem S1) are within open boxes together with the corresponding base substitutions. The SD sequence and translation initiation codon of rpmI are indicated. Nucleotides forming the two sequences are in lowercase letters.
MATERIALS AND METHODS
Bacterial Strains, Plasmids, and General Techniques.
The E. coli K12 strains used in this work are: IBPC5311 [thi-1, argE3, ΔlacX74, galK2?, mtl-1, xyl-5, tsx-29?, rpsL, recA1, pps] (13), XAc [ara, argEam, Δ(lac-pro), nalA, rpoB, thi] (a generous gift from L. A. Isaksson, Department of Microbiology, University of Stockholm), and the amber-suppressor XA106 strain [ara, argEam, Δ(lac-pro), nalA, rpoB, supP] (14). Throughout this study, pBR322 (15) was used as a control plasmid and pBL6, a derivative of pBR322 carrying rplT, as an L20-overproducing plasmid (3). General bacteriological techniques were as described (16). General cloning techniques were as in refs. 17 and 18.
Mutagenesis.
All mutations used in this study have been described (10, 19). Mutation MUTp1, a p1 promoter-down mutation (changes the −10 box from TATAAT to AATCAT), was introduced into M13mp18MQ21ΔNB and M13mp18MIRΔNB (3) by site-directed mutagenesis using an oligonucleotide as described (20) to give M13mp18MQ21(MUTp1)ΔNB and M13mp18MIR(MUTp1)ΔNB, respectively. Mutation MUTp0′, a p0′ promoter-down mutation (changes the −35 box from TTGAGA to TTAAGA), was isolated after a screen for decreased expression of infC (M. Graffe, unpublished work). All mutations were sequenced using the dideoxy chain termination method (21).
λ Bacteriophage Constructions, Lysogenization, β-Galactosidase (β-Gal) Measurements.
Recombinant bacteriophages λMQ21ΔNB and λMIRΔNB, which contain p1-′infC–rpmI′–′lacZ and p0′-′thrS–infC–rpmI′–′lacZ translational fusions, respectively, have been described (3). A prime on one side of a gene means that it is truncated on this side. Hereafter, the two fusions and their corresponding derivatives will be referred to as the short and the long fusion, respectively. In both fusions, lacZ was fused in phase with the first 157 nt of rpmI. Derivatives from these fusions were constructed by first transferring all the mutations into M13mp18MQ21ΔNB and M13mp18MIRΔNB (3) or their derivatives with the appropriate restriction enzymes and subsequent cloning into λ as described (3, 10). Lysogenization, monolysogen screenings, growth conditions of plasmid-carrying monolysogens, and β-gal measurements were as described (3, 13, 22).
RESULTS
Translation of infC Decreases L20-Mediated Repression of rpmI Expression.
Regulation of rpmI expression by L20 was studied using rpmI′–′lacZ translational fusions cloned in bacteriophage λ and integrated into the E. coli chromosome at attλ. Throughout this study, we used two types of rpmI′–′lacZ fusions, which differ in the size of the region located upstream of rpmI. In the shorter of the two fusions, transcription starts at the minor p1 promoter within infC coding sequences, whereas transcription of the longer fusion starts mainly at the p0′ promoter, upstream of infC (Fig. 1). Consequently, infC is translated in the long fusion, whereas in the short fusion, it is not. Both fusions contain all of the sequences required for regulation of rpmI expression (9). The short fusion synthesized 110 Miller units of β-gal in the presence of pBR322, as a control plasmid, and 1.8 units in the presence of pBL6, a pBR322 derivative that carries rplT expressed from the lac promoter and that overproduces L20. The corresponding β-gal activities with the long fusion were 1800 units and 160 units. The effect of L20 on the fusions is given as “repression factor,” which is the ratio of the β-gal activity in the presence of pBR322 to that in the presence of pBL6. Thus, L20-mediated repression of rpmI expression was 62-fold with the short fusion, but only 11-fold with the long fusion. The effect of L20 on rpmI expression was measured with pulse-labeling experiments followed by SDS/PAGE. Steady-state measurements of β-gal activity and immediate effects of L20 induction on the synthesis of the L35–β-gal hybrid protein gave equivalent results (unpublished work). Only the results of the steady-state measurements will be given below.
Premature Termination of infC Translation Increases the Level of L20-Mediated Repression of rpmI Expression.
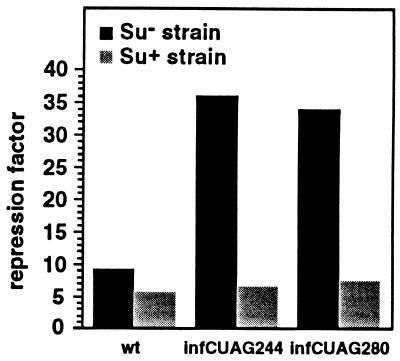
The difference in L20-mediated repression between the short and long fusion could be explained if ribosomes translating infC were to unfold the pseudoknot and/or eject L20 from its binding site. If this is true, one would predict that by preventing ribosomes from traversing the region of infC, which constitutes part of the translational operator, one should be able to restore control to the levels seen in the short fusion. Premature termination of infC translation upstream of the sequences forming stem S1 of the pseudoknot was achieved by introducing an amber codon at positions infC244 or infC280 in the long fusion (mutations infCUAG244 and infCUAG280, respectively, see Figs. 1 and 2). The effect of these amber codons was studied in both the presence (Su+) and absence (Su−) of the supP amber suppressor mutation. Neither of the amber codons showed significant polar effects, since β-gal activities of the respective lysogens were 1739 and 1664 units vs. 2445 for the wild-type (wt) lysogen in the Su− strain. However, premature termination of infC translation in the Su− strain increased control about 4-fold; repression factors were 36.0 and 34.0 for mutations infCUAG244 and infCUAG280, respectively, compared with 9.2 for the wild type (Fig. 3). In contrast, when infC translation was restored in the Su+ strain, repression factors were slightly but reproducibly higher than that obtained with the wild type (6.8 and 7.4, respectively, compared with 5.7 for the wt) (Fig. 3). This slight difference is most probably explained by the fact that suppression, although very efficient (see below), is not 100% efficient. Premature termination and restoration of infC translation in the Su− and Su+ strains were verified by immunoblotting experiments using anti-IF3 antibodies (data not shown). These experiments showed that the intensity of the truncated form of IF3 generated by either infCUAG244 or infCUAG280 mutations, increased 8- to 9-fold when the host was changed from supP to wild type, indicating that suppression was very efficient. This experiment confirms a prediction of our hypothesis, that decreased translation of infC increases L20-mediated repression of rpmI expression.
Figure 3.
Effect of premature termination of infC translation on L20-mediated repression of rpmI expression. Amber mutations infCUAG244 and infCUAG280 were independently introduced in the long fusion at positions infC244 and infC280, respectively. The effect of each mutation, compared with the wild type, was measured using the (Su−) XAc and (Su+) XA106 strains as hosts. In XAc, β-gal activities (in Miller units) with the wild type, the amber mutations UAG244 and UAG280, are 2152, 1850, and 1525 in the presence of the control plasmid pBR322 and 236, 51, and 45 in the presence of the L20-overproducing plasmid pBL6. In XA106, the corresponding activities are 2173, 2386, and 1838 in the presence of the control plasmid and 370, 349, and 247 in the presence of the L20-overproducing plasmid.
Increased Translation of infC Decreases the Level of L20-Mediated Repression of rpmI Expression.
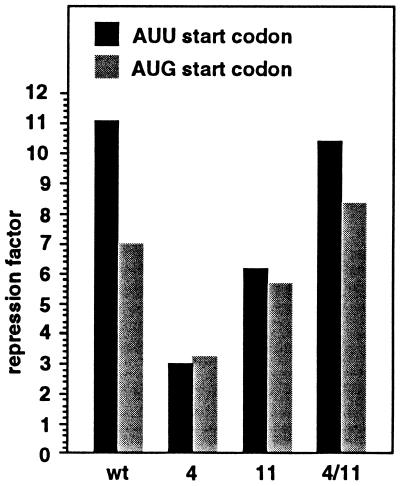
To take this analysis one step further, we investigated the effect of increased translation of infC on rpmI control by mutating the start codon of infC on the long fusion. Changing the AUU start codon to the more efficient AUG codon was previously shown to increase the expression of infC 10-fold (19, 23). To avoid the deleterious effect of IF3 overproduction on the cells we combined the AUG mutant codon with the MUTp0′ mutation, which decreases expression from the p0′ promoter by about 4-fold (data not shown). Because of this decrease in the strength of the p0′ promoter, expression from the p1 promoter is no longer negligible as in the wt. To keep p1 promoter much weaker than p0′, we also lowered the activity of the former promoter with the MUTp1 mutation. This mutation decreases the expression from the p1 promoter by about 8-fold (data not shown). Replacing the AUU start codon with AUG decreased the repression factor from 11.2 to 7.0 (first two columns, Fig. 4), consistent with the hypothesis that translation of infC modulates L20-mediated control. We believe that translating ribosomes transiently unfold the pseudoknot and/or eject L20, which, in turn, causes a decrease in control and an increase in rpmI expression. Although the expression of infC is increased 10-fold, control is not completely abolished, suggesting that the pseudoknot can re-form and permit L20 binding between two translating ribosomes. Below we will present evidence that the pseudoknot can re-form when infC is highly translated.
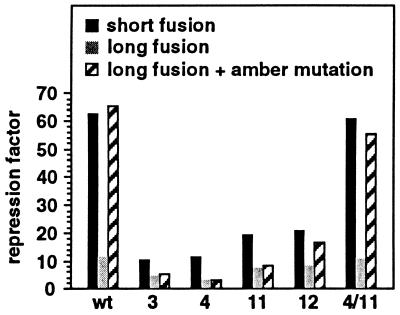
Figure 4.
Effect of increased infC translation on L20-mediated repression of rpmI expression. The effect of varying initiation frequency of infC translation was measured on the long fusion containing both the MUTp0′ and MUTp1 promoter-down mutations. The effect was measured on the long fusion without additional mutation (wild type), with each of the two compensatory mutations in stem S2 of the pseudoknot (4 and 11) and with both mutations combined (4/11). Positions of the mutations are shown in Fig. 2. The host strain used in this experiment is IBPC5311. Normal initiation frequency was provided by the wild-type AUU start codon, whereas increased frequency was provided by the mutant AUG start codon. With the wild-type start codon, β-gal activities (in Miller units) measured with wild type, mutations 4, 11, and 4/11 are 108, 151, 145, and 83.2 in the presence of the control plasmid pBR322 and 9.7, 50.5, 23.3, and 8 in the presence of the L20-overproducing plasmid pBL6. With the mutant start codon, the corresponding activities are 127, 183, 163, and 122 in the presence of the control plasmid and 18.2, 56, 28.8, and 14.6 in the presence of the L20-overproducing plasmid.
Mutations That Destabilize the Pseudoknot Reduce the Effect of infC Translation on L20-Mediated Repression of rpmI Expression.
We had previously shown that point mutations that destabilize either stem S1 or stem S2 of the pseudoknot decrease control of the short fusion (10). If the effect of infC translation on control is caused by temporary disruption of the pseudoknot, one would predict that mutations destabilizing stems S1 or S2 would reduce this effect. Therefore, we assessed the effect of various cis-acting mutations in the long fusion in the presence or absence of the amber mutation infCUAG244 in a Su− background. Because we expected that in the long fusion containing the amber codon mutations in the pseudoknot would have a similar effect to the same mutations in the short fusion, we included the latter fusion in our study (Fig. 5). When translation of infC proceeded through the wild-type pseudoknot in the long fusion, the repression factor was reduced about 6- to 7-fold compared with that measured with both the short fusion and the long fusion, where infC translation is interrupted upstream of the pseudoknot (Fig. 5, three first columns). On the other hand, when the pseudoknot was destabilized or disrupted by mutations 3, 4, 11, and 12 (see Fig. 2), reduction of control is at most 2-fold compared with that measured with the long fusion, where infC translation is interrupted (Fig. 5). As expected, when mutations 4 and 11 (see Fig. 2) were combined to restore base pairing in stem S2, the effect of infC translation on control was reestablished (Fig. 5, last three columns). Therefore, our results indicate that the effect of infC translation on control relies on the presence of the pseudoknot.
Figure 5.
Comparison of the effect of mutations on L20-mediated repression of rpmI expression with or without interruption of infC translation. The effect of mutations was measured on the wild-type- and the amber mutation infCUAG244-containing long fusions using the (Su−) IBPC5311 strain as a host. Positions of the mutations are shown in Fig. 2. Columns denoted by wt correspond to the wild-type pseudoknot. With the short fusion, β-gal activities (in Miller units) measured with wild type, mutations 3, 4, 11, 12, and 4/11 are taken from ref. 10 and are as follows: 110, 156, 503, 309, 292, 284, and 122 in the presence of the control plasmid pBR322 and 1.8, 14.9, 54.6, 27.2, 15.3, 12.9, and 2 in the presence of the L20-overproducing plasmid pBL6. With the long fusion, the corresponding activities are 1818, 2795, 5415, 2881, 3611, 4092, and 1752 in the presence of the control plasmid and 159, 625, 475, 934, 471, 505, and 156 in the presence of the L20-overproducing plasmid. With the long fusion with the amber mutation at position infC244 the corresponding activities are 1460, 2004, 3444, 2739, 3070, 3827, and 1294 in the presence of the control plasmid and 22.4, 375, 190, 924, 364, 229, and 23.8 in the presence of the L20-overproducing plasmid.
The Regulatory Pseudoknot also Forms in the Native Long Fusion.
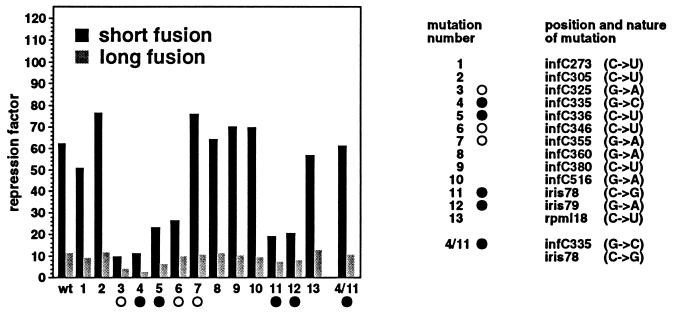
In a previous work (10), we have shown that mutations that strongly affect the control on the short fusion were found exclusively in stems S1 or S2 (see Fig. 2) of the pseudoknot. Mutations located upstream (Fig. 6, mutations 1 and 2) or downstream (Fig. 6, mutation 13) of the pseudoknot and those that fall between stems S1 and S2 (Fig. 6, mutations 8–10) had no or little effect on control. The effect of this same set of mutations and that of mutations located in the pseudoknot (Fig. 6, mutations 3–7, 11, and 12 in stems S1 and S2) on control were analyzed using the long fusion. The results show that mutations 3–6, 11, and 12 in stems S1 and S2 of the pseudoknot, which decrease control on the short fusion, also affect control on the long fusion, albeit to a lesser extent (Fig. 6). In addition, mutations that had no effect on control with the short fusion are also without effect on the long fusion. These results suggest that repression of rpmI expression by L20 occurs similarly in the long and the short fusion. The fact that mutations affecting control on the long fusion are all located in the pseudoknot indicates that this structure plays a role in control of the long fusion as well. Since infC is translated on the long fusion, this result raises the question of whether the pseudoknot is able to re-form between translating ribosomes. This question was addressed by analyzing the effect of mutations 4 and 11, which disrupt the same G-C base pair in stem S2 of the pseudoknot (see Fig. 2). Both mutations decreased control of the long fusion; repression factors were 3.1 and 7.7 for mutations 4 and 11, respectively, compared with 11.4 for the wild type (Fig. 6). Because mutations 4 and 11 in combination restore base pairing in stem S2 of the pseudoknot and totally reestablish control (repression factor 11.2; Fig. 6), we conclude that the pseudoknot also forms on the native long fusion. Apparently, the long-range interaction (stem S2) can still form, although infC is translated. This means that with an AUU initiation codon, the frequency of initiation, i.e., the distance between two translating ribosomes, is sufficient for the pseudoknot to regularly re-form on the translated mRNA. This even seems to be true when the translation initiation frequency is increased by a change of the infC start codon from AUU to AUG. Our results indicate that with an AUG start codon, combination of mutation 4 with mutation 11 substantially increases control, since the repression factor raises from 3.3 and 5.9 for mutations 4 and 11 separately to 8.4 for the combination (Fig. 4). Therefore, we conclude that the pseudoknot is able to re-form between two translating ribosomes even when the frequency of infC translation initiation is increased 10-fold with respect to the normal frequency. The repression ratio with mutant 4 is the same with both AUU and AUG initiation codons (Fig. 4), as if residual repression with the mutant is independent of infC translation. This might be explained by some residual effect of L20 on rpmI expression that is pseudoknot independent (see Introduction) and thus not suspected to be sensitive to infC translation.
Figure 6.
Effect of the mutations on L20-mediated repression of rpmI expression. Locations of the mutations and the corresponding base substitutions are shown on the right. Mutations indicated by open circles and solid circles are located in stems S1 and S2 of the pseudoknot, respectively (see Fig. 2). The host strain used in this experiment is IBPC5311. β-gal activities (in Miller units) measured with the short and the long fusion for wild type, mutations 3, 4, 11, 12, and 4/11 are indicated in the legend to Fig. 5. β-gal activities (in Miller units) measured with the short fusion for mutations 1, 2, 5–10, and 13 are taken from ref. 10 and are as follows: 95.1, 111, 341, 152, 89.9, 135, 112, 95.1, and 53.4 in the presence of the control plasmid pBR322 and 1.9, 1.4, 14.4, 5.7, 1.2, 2.1, 1.6, 1.4, and 0.9 in the presence of the L20-overproducing plasmid pBL6. The corresponding activities measured with the long fusion are 1603, 1698, 3896, 1572, 1489, 1883, 1877, 1340, and 1590 in the presence of the control plasmid and 177, 144, 620, 154, 140, 163, 186, 138, and 123 in the presence of the L20-overproducing plasmid.
DISCUSSION
The Long-Range Pseudoknot as a Coupling Device.
In the IF3 operon, expression mainly comes from promoters upstream of infC, p0′ in particular. However, the two last cistrons of the operon are also expressed from the minor p1 promoter, which is internal to infC. The rpmI′–′lacZ long fusion, starting at p0′, carries the translation initiation signals of infC, whereas the short fusion, starting at p1, does not. In this work, we investigate the effect of infC translation on the regulation of rpmI expression. We first show that L20-mediated repression of rpmI expression is much less efficient in the long fusion, where infC is translated, than in the short, where no translation occurs. That the lower efficiency of repression on the long fusion is due to translation of infC is clear since: (i) the efficiency of repression is increased by premature termination of infC translation upstream of the pseudoknot; (ii) the efficiency of repression is decreased when infC translation is increased using a more efficient start codon. The simplest explanation is that ribosomes translating IF3 disrupt the pseudoknot, which relies on sequences internal to infC. This disruption most probably perturbs the folding of the whole translational operator, thus decreasing L20 binding and rpmI repression. We know from this and previous work (10) that mutations that destabilize the pseudoknot strongly decrease rpmI repression on the short fusion. In the presence of such mutations, we expected translation of infC to have a weaker effect on control, because the pseudoknot has been already destabilized or even disrupted. Here we show this to be true. Furthermore, the effect of infC translation was restored when base pairing was reestablished in stem S2 of the pseudoknot.
These results indicate that translation of infC modulates L20-mediated repression of rpmI expression. Thus, expression of infC and rpmI, and consequently that of rplT (see Introduction), are coupled at the translational level. The melting of an RNA secondary structure by translating ribosomes is common to most translational coupling systems (see Introduction). However, the coupling mechanism described here is different in the sense that translating ribosomes, instead of unfolding an inhibitory structure that masks the RBS of rpmI, unfold a pseudoknot that enhances the repression of the expression of that gene.
As outlined previously, IF3 negatively autoregulates infC expression (12). Therefore, should the cellular concentration of IF3 increase, translation of infC will decrease and consequently, L20-mediated repression of rpmI expression will increase because of the coupling mechanism described in this work. In other words, the coupling between infC and rpmI provides a connection between the two separate regulatory loops of the IF3 operon (see Introduction).
Pseudoknot-Dependent Regulation of rpmI Expression and RNA Folding.
During folding of an RNA molecule, formation of secondary structure motifs likely precedes that of tertiary interactions (24–29). It is known that formation of a simple secondary structure like a hairpin helix is very rapid, occurring in less than 1 msec in vitro (24). In contrast, formation of a short-range RNA–RNA tertiary interaction like a local pseudoknot has been shown to take tenths of a second (30). Recent experiments have demonstrated that a long-range tertiary structure like the pseudoknot, which constitutes the P3–P7 domain of the Tetrahymena ribozyme, takes tens of seconds to minutes to form in vitro (29, 31). Although the two regions forming stem S2 of the pseudoknot involved in rpmI control are separated even more (≈280 nt) than those forming the key stem of the P3–P7 domain (≈170 nt), it is unlikely that stem S2 takes tens of seconds to form in vivo for several reasons. The first is based on the reasonable assumption that rpmI mRNA, a message encoding a ribosomal protein, is a well-translated mRNA. Logically, translation of rpmI should initiate at a frequency of at least once every 3 sec corresponding to that of lacZ (32), a well-translated gene. If the time between two initiation events is 3 sec or less, a folding time of the order of tens of seconds for stem S2 would be too slow to permit regulation of rpmI translation. The folding time is therefore either shorter or on the same time scale than the time between two initiation events at the RBS of rpmI, i.e., in the second range. The second reason to believe that the folding time of stem S2 is faster than tens of seconds in vivo is directly based on our experimental data. When we mutated the inefficient AUU start codon of infC into an AUG codon, we provided the translation initiation region of infC with features typical of well-translated genes like lacZ (an efficient SD sequence and an AUG start codon). Our data indicate that, even when infC starts with an AUG codon, compensatory mutations that restore base pairing in stem S2 of the pseudoknot increase control when compared with single mutations. This result means that refolding can occur between translating ribosomes, which are only a few seconds apart. Therefore, in our case, the folding time of a long-range pseudoknot is faster than expected if we consider the in vitro studies performed on the Tetrahymena ribozyme. This result is in agreement with the fact that in vivo folding of the Tetrahymena ribozyme is known to occur 20- to 50-fold more rapidly in vivo than in vitro (33). This acceleration is apparently not due to species-specific proteins (34), but rather to some general factors such as the so-called RNA chaperones that may be involved in the folding of a wide range of RNAs (35).
Although tools are at hand to measure folding times in vitro (29), they are often missing in vivo. We believe that the pseudoknot-dependent coupling mechanism described here could be an adequate system for in vivo measurements of folding times. The fact that compensatory mutations exist that simultaneously restore base pairing in stem S2 of the pseudoknot and reestablish control of both fusions is a good indication that the pseudoknot really forms in vivo. Because the translation initiation frequency of infC can be increased both in cis and in trans, we expect to be able to find the translation initiation frequency at which compensatory mutations in stem S2 can no longer reestablish rpmI control, to measure the minimal time necessary for the pseudoknot to form in vivo.
Acknowledgments
We thank M. H. de Smit for very helpful comments. We are indebted to C. Condon for critical reading of the manuscript. This work was supported by grants from the Centre National de la Recherche Scientifique (UPR 9073); the Ministère de l’Education Nationale, de l’Enseignement Supérieur et de la Recherche (ACCSV n° 9506049); and from European Economic Community Human Capital and Mobility Network.
ABBREVIATIONS
- RBS
ribosome binding site
- SD
Shine–Dalgarno
- IF3
initiation factor 3
- β-gal
β-galactosidase
References
- 1.de Smit M H, van Duin J. Prog Nucleic Acid Res Mol Biol. 1990;38:1–35. doi: 10.1016/s0079-6603(08)60707-2. [DOI] [PubMed] [Google Scholar]
- 2.Zengel J M, Lindahl L. Prog Nucleic Acid Res Mol Biol. 1994;47:331–369. doi: 10.1016/s0079-6603(08)60256-1. [DOI] [PubMed] [Google Scholar]
- 3.Lesage P, Chiaruttini C, Dondon J, Graffe M, Milet M, Springer M. J Mol Biol. 1992;228:366–386. doi: 10.1016/0022-2836(92)90827-7. [DOI] [PubMed] [Google Scholar]
- 4.Adhin M R, van Duin J. J Mol Biol. 1990;213:811–818. doi: 10.1016/S0022-2836(05)80265-7. [DOI] [PubMed] [Google Scholar]
- 5.Lovett P S. J Bacteriol. 1990;172:1–6. doi: 10.1128/jb.172.1.1-6.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gu Z, Harrod R, Rogers E J, Lovett P S. Proc Natl Acad Sci USA. 1994;91:5612–5616. doi: 10.1073/pnas.91.12.5612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harrod R, Lovett P S. Proc Natl Acad Sci USA. 1995;92:8650–8654. doi: 10.1073/pnas.92.19.8650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ivey-Hoyle M, Steege D A. J Mol Biol. 1989;208:233–244. doi: 10.1016/0022-2836(89)90385-9. [DOI] [PubMed] [Google Scholar]
- 9.Lesage P, Truong H N, Graffe M, Dondon J, Springer M. J Mol Biol. 1990;213:465–475. doi: 10.1016/S0022-2836(05)80208-6. [DOI] [PubMed] [Google Scholar]
- 10.Chiaruttini C, Milet M, Springer M. EMBO J. 1996;15:4402–4413. [PMC free article] [PubMed] [Google Scholar]
- 11.Wertheimer S J, Klotsky R-A, Schwartz I. Gene. 1988;63:309–320. doi: 10.1016/0378-1119(88)90534-3. [DOI] [PubMed] [Google Scholar]
- 12.Butler J S, Springer M, Dondon J, Graffe M, Grunberg-Manago M. J Mol Biol. 1986;192:767–780. doi: 10.1016/0022-2836(86)90027-6. [DOI] [PubMed] [Google Scholar]
- 13.Springer M, Plumbridge J A, Butler J S, Graffe M, Dondon J, Mayaux J F, Fayat G, Lestienne P, Blanquet S, Grunberg-Manago M. J Mol Biol. 1985;185:93–104. doi: 10.1016/0022-2836(85)90185-8. [DOI] [PubMed] [Google Scholar]
- 14.Coulondre C, Miller J H. J Mol Biol. 1977;117:525–567. doi: 10.1016/0022-2836(77)90056-0. [DOI] [PubMed] [Google Scholar]
- 15.Bolivar F, Rodriguez R L, Greene P J, Heynecker H L, Boyer H W, Crosa J H, Falkow S. Gene. 1977;2:95–113. [PubMed] [Google Scholar]
- 16.Miller J H. Experiments in Molecular Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1972. [Google Scholar]
- 17.Davis R W, Botstein D, Roth J R. Advanced Bacterial Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1980. [Google Scholar]
- 18.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 19.Sacerdot C, Chiaruttini C, Engst K, Graffe M, Milet M, Mathy N, Dondon J, Springer M. Mol Microbiol. 1996;21:331–346. doi: 10.1046/j.1365-2958.1996.6361359.x. [DOI] [PubMed] [Google Scholar]
- 20.Kunkel T A, Roberts J D, Zakour R A. In: Recombinant DNA: Part E. Wu R, Grossman L, editors. Vol. 154. New York: Academic; 1987. pp. 367–382. [Google Scholar]
- 21.Sanger F, Nicklen S, Coulson A R. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Springer M, Graffe M, Butler J S, Grunberg-Manago M. Proc Natl Acad Sci USA. 1986;83:4384–4388. doi: 10.1073/pnas.83.12.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Butler J S, Springer M, Grunberg-Manago M. Proc Natl Acad Sci USA. 1987;84:4022–4025. doi: 10.1073/pnas.84.12.4022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Crothers D M, Cole P E, Hilbers C W, Shulman R G. J Mol Biol. 1974;87:63–88. doi: 10.1016/0022-2836(74)90560-9. [DOI] [PubMed] [Google Scholar]
- 25.Herschlag D. Biochemistry. 1992;31:1386–1399. doi: 10.1021/bi00120a015. [DOI] [PubMed] [Google Scholar]
- 26.Bevilacqua P C, Kierzek R, Johnson K A, Turner D H. Science. 1992;258:1355–1358. doi: 10.1126/science.1455230. [DOI] [PubMed] [Google Scholar]
- 27.Banerjee A R, Jaeger J A, Turner D H. Biochemistry. 1993;32:153–163. doi: 10.1021/bi00052a021. [DOI] [PubMed] [Google Scholar]
- 28.Jaeger L, Westhof E, Michel F. J Mol Biol. 1993;234:331–346. doi: 10.1006/jmbi.1993.1590. [DOI] [PubMed] [Google Scholar]
- 29.Zarrinkar P P, Williamson J R. Science. 1994;265:918–924. doi: 10.1126/science.8052848. [DOI] [PubMed] [Google Scholar]
- 30.Wyatt J R, Puglisi J D, Tinoco I., Jr J Mol Biol. 1990;214:455–470. doi: 10.1016/0022-2836(90)90193-P. [DOI] [PubMed] [Google Scholar]
- 31.Banerjee A R, Turner A R. Biochemistry. 1995;34:6504–6512. doi: 10.1021/bi00019a031. [DOI] [PubMed] [Google Scholar]
- 32.Sørensen M A, Pedersen S. J Mol Biol. 1991;222:265–280. doi: 10.1016/0022-2836(91)90211-n. [DOI] [PubMed] [Google Scholar]
- 33.Bass B L, Cech T R. Nature (London) 1984;308:820–826. doi: 10.1038/308820a0. [DOI] [PubMed] [Google Scholar]
- 34.Zhang F, Ramsey E S, Woodson S A. RNA. 1995;1:284–292. [PMC free article] [PubMed] [Google Scholar]
- 35.Herschlag D. J Biol Chem. 1995;270:20871–20874. doi: 10.1074/jbc.270.36.20871. [DOI] [PubMed] [Google Scholar]