Abstract
Tuberculosis is a chronic infectious disease that is transmitted by cough-propelled droplets that carry the etiologic bacterium, Mycobacterium tuberculosis. Although currently available drugs kill most isolates of M. tuberculosis, strains resistant to each of these have emerged, and multiply resistant strains are increasingly widespread. The growing problem of drug resistance combined with a global incidence of seven million new cases per year underscore the urgent need for new antituberculosis therapies. The recent publication of the complete sequence of the M. tuberculosis genome has made possible, for the first time, a comprehensive genomic approach to the biology of this organism and to the drug discovery process. We used a DNA microarray containing 97% of the ORFs predicted from this sequence to monitor changes in M. tuberculosis gene expression in response to the antituberculous drug isoniazid. Here we show that isoniazid induced several genes that encode proteins physiologically relevant to the drug’s mode of action, including an operonic cluster of five genes encoding type II fatty acid synthase enzymes and fbpC, which encodes trehalose dimycolyl transferase. Other genes, not apparently within directly affected biosynthetic pathways, also were induced. These genes, efpA, fadE23, fadE24, and ahpC, likely mediate processes that are linked to the toxic consequences of the drug. Insights gained from this approach may define new drug targets and suggest new methods for identifying compounds that inhibit those targets.
The transcription of genes encoding components of a multienzyme pathway often is regulated in response to the availability of cofactors, precursors, intermediates, or products of the same pathway. Compounds, including drugs, that selectively inhibit a pathway enzyme and thereby cause an accumulation of precursors and a depletion of products can be expected to selectively induce changes in the transcription of genes coding for enzymes that comprise the affected pathway, especially before a more generalized stress response ensues. The resulting gene expression profile would not only serve as a kind of signature of the inhibitor used, but would, in the case of inhibitors whose modes of action are unknown, incriminate the affected pathway and perhaps the specific targeted enzyme within the pathway. The realization of this idea requires that the expression of each gene in the genome be interrogated simultaneously in the presence and absence of inhibitor. The increasing availability of complete genome sequences provides one of the two preconditions for this approach, and the development of microarrays containing representatives of each of the genes provides the other. In the study described below, we have tested this method for pathway characterization. For this purpose, we fabricated a microarray containing DNA fragments corresponding to nearly every predicted ORF of the recently completed and annotated Mycobacterium tuberculosis genomic sequence of strain H37Rv (1). Here we describe the use of this array to generate the transcriptional response profile of M. tuberculosis during its exposure to isoniazid (INH), a drug that blocks the mycolic acid biosynthetic pathway.
We chose first to study the transcriptional response to INH because it is given to more tuberculosis patients than any other drug, and because among the arsenal of antituberculosis drugs, INH is the agent to which resistance emerges most frequently (2, 3). Additionally, our understanding of the intracellular consequences of INH activity on the biosynthesis of cell-wall lipids and the genes responsible for lipid metabolism has improved substantially in recent years. Therefore, a model system based on the effects of INH provides a conceptual framework for interpreting the transcriptional responses that we would detect by the microarray method and allows us to compare these results with published observations of genes and proteins that are known to be INH-induced. In turn, because INH is lethal to sensitive strains, the identification of enzymes that heretofore were not known to be within INH-inhibited pathways could lead to the selection of additional drug targets. The elucidation of the role of these enzymes in the production of mycolic acids, as discussed below, also could lead to a deeper understanding of the biosynthetic and regulatory processes involved in the production of this characteristic class of lipids.
INH selectively interrupts the synthesis of mycolic acids, the major component of the waxy, outer lipid envelope of mycobacteria (4). Mycolic acids are branched, β-hydroxy fatty acids composed of an intermediate-length (C24-C26), saturated alpha chain, and a longer (>C50) meromycolate chain that contains characteristic functional moieties. A convergence of elegant genetic and biochemical evidence has shown that INH blocks a type II fatty acid synthase (FAS-II) complex that is required for full-length extension of the meromycolate chain (5–7). Although the precise mechanism of INH-mediated killing remains unresolved, INH is thought to target three FAS-II complex proteins by binding to NADH in the pocket of the enoyl-acyl carrier protein (ACP) reductase, InhA, and by forming a covalent, ternary complex between the β-ketoacyl-ACP synthase, KasA, and an acyl carrier protein, AcpM (6, 7). As a consequence of INH activity, mature mycolates are not produced and become progressively depleted (4). At the same time, and in accordance with the pathway inhibition notion of INH action, an intracellular pool of saturated fatty acids (C24-C26) accumulates that presumably reflects the point at which meromycolate synthesis is interrupted by INH (8, 9). The accumulation of these fatty acid pathway precursors is associated with the increased production of AcpM and KasA (7) and implies that the induction of their genes is the consequence of a regulatory feedback mechanism that senses the imbalance of mycolic acid biosynthetic intermediates, which accumulate, and full-length mycolates, which are depleted. We reasoned that the INH-induced increase in KasA and AcpM, if mediated at the transcriptional level, should be reflected by parallel changes in the abundance of the corresponding transcripts. Further, we predicted that INH also would induce genes coding for other related components of the mycolic acid biosynthetic pathway. If so, a microarray containing DNA targets for nearly every ORF in the genome would detect the induction of these genes and others for which no a priori prediction was made.
Methods
Preparation of DNA Microarrays.
PCR primers were designed to amplify internal fragments of each predicted ORF described in the annotated genomic sequence of M. tuberculosis, strain H37Rv (1). Primers with equivalent predicted melting temperatures were selected by using primer 3 software [Steve Rozen and Helen J. Skaletsky (1996, 1997), code available at http://www-genome.wi.mit.edu/genome_software/other/primer3.html]; batch processing and data formatting were performed by a script written by Hugh Salamon (Stanford University). Primers were synthesized by Stanford University’s Protein and Nucleic Acid facility by using a 96-well, multiplex oligonucleotide synthesizer (10). The ORF-specific, double-stranded DNA fragments were produced by standard PCR amplification methods in 96-well plate format. All the PCR products were evaluated by agarose gel electrophoresis. Reactions with no product, multiple bands, or bands of the incorrect size were considered PCR failures and annotated as such in our database. Amplicon lengths ranged from 200 to 1,000 bp (580-bp average).
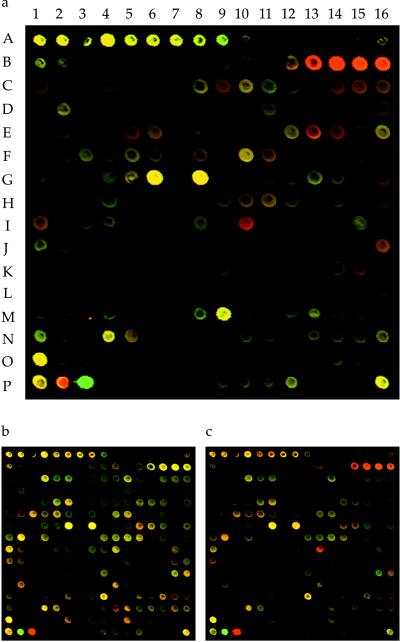
The microarrays were printed on poly-l-lysine-coated glass microscope slides as previously described by using a robotic spotting device fitted with stainless steel printing tips that enabled production of replicate arrays for each printing session (11, 12). During the course of these studies, three series of microarrays were constructed from independently amplified DNA fragment stocks. The most complete version contained validated fragments representing 3,834 (97%) of the predicted 3,924 ORFs. The results of this array series are presented in Table 1, and the two less complex microarrays confirmed the results of the larger version. The results presented in Fig. 1 are from a mini-array containing 203 different ORFs that were selected to contain a high proportion (29%) of genes encoding lipogenic or lipolytic enzymes.
Table 1.
Genes induced by INH or ethionamide treatment of an INH-sensitive strain
| Classification | ORF* | Gene | Proposed function | Fluorescent ratio†
|
Ref.* | |
|---|---|---|---|---|---|---|
| INH | ETH | |||||
| FAS-II | Rv2243 | fabD | Malonyl CoA-malony-ACP transacylase | 2.8 | 3.1 | |
| Rv2244 | acpM | Acyl carrier protein | 3.8 | 3.7 | 7 | |
| Rv2245 | kasA | β-ketoacyl synthase | 2.8 | 3.4 | 7 | |
| Rv2246 | kasB | β-ketoacyl synthase | 2.4 | 2.7 | ||
| Rv2247 | accD6 | Acetyl-CoA carboxylase (β chain) | 2.5 | 2.0 | ||
| Mycolate transfer | Rv0129c | fbpC | Trehalose dimycolyl transferase | 2.1 | 2.2 | 19 |
| Fatty acid β-oxidation | Rv3139 | fadE24 | Fatty-acyl-CoA dehydrogenase | 2.3 | 2.4 | |
| Rv3140 | fadE23 | Fatty-acyl-CoA dehydrogenase | 2.6 | 2.5 | ||
| Detoxification | Rv2428 | ahpC | Alkyl hydroperoxide reductase (subunit) | 2.9 | 2.5 | |
| Transport | Rv2846c | efpA | Efflux protein | 2.5 | 2.1 | |
| Unknown | Rv1592c | 3.0 | 2.4 | |||
| Rv1772 | 2.0 | 2.1 | ||||
| Rv0341 | 2.0 | 2.5 | 22 | |||
| Rv0342 | 2.6 | 2.1 | 22 | |||
ETH, ethionamide.
ORF nomenclature is based on ref. 1. Citations for individual genes refer to reports of INH-induced expression of specific genes or their protein products.
Average fluorescent ratio (drug-treated/untreated) resulting from the hybridization experiments of 2- to 6-hr drug exposures.
Figure 1.
INH-induced mRNA expression profiles monitored by microarray hybridization analysis. (a) A subarray containing spots corresponding to 203 different M. tuberculosis ORFs illustrates the INH-induced gene response profile. Included were genes thought to encode components of the mycolic acid pathway. The expression response to a 4-hr treatment with INH is shown as a pseudocolored composite image. The two channels were pseudocolored according to the fluorescence intensity, either red (INH treated) or green (INH untreated), and overlaid to give the images shown. Yellow shades are derived from the combination of red and green, indicating relatively equivalent expression levels. Predominantly red spots at positions B13–16 correspond to genes of the FAS-II gene cluster (Rv2244–7). The spots at coordinates (P2–3) are positive controls of DNA prelabeled with Cy3 (P2) or Cy5 (P3). Reference spots contained M. tuberculosis whole-genomic DNA (A1, P1, and P16) and ribosomal DNA (A4–10). (b) INH treatment of an INH-resistant, ethionamide-sensitive strain (4309A) revealed little change in relative mRNA levels. (c) Treatment of the same strain with ethionamide produced a response pattern that was similar to the pattern produced by treatment of the INH-sensitive strain with INH (a).
Growth and Drug Treatment of M. tuberculosis Strains.
The INHs, ethionamides M. tuberculosis strain 1254 is a recent clinical isolate from San Francisco kindly provided by Peter Small (Stanford University). The INHr, ethionamides strain, 96A4309A, was kindly provided by Ed Desmond of the California Department of Health Services, Microbial Diseases Laboratory, Berkeley. Drug sensitivity testing was performed on each strain by Specialty Laboratories (Santa Monica, CA), and catalase activity was assessed by a semiquantitative catalase test (13). Single-colony isolates of each strain were cultivated in Middlebrook 7H9 medium (Difco; supplemented with 0.5% BSA, 0.2% glycerol, 0.2% dextrose, 14.5 mM NaCl, and 0.05% Tween-80) (14).
Cultures for experimental treatment were initiated by diluting a frozen stock inoculum 1:200 into fresh 7H9 media in vented, screw-cap, tissue culture flasks and grown to early log phase (0.15–0.3 OD600) with shaking (80 rpm) in a 5% CO2 atmosphere at 37°C. Drug treatment was begun by adding filtered stock solutions of INH (1 mg/ml, Sigma) or ethionamide (25 mg/ml, Sigma) to achieve the following final concentrations: 0.2 μg/ml or 1 μg/ml for INH and 5 μg/ml or 20 μg/ml for ethionamide. Upon completion of the predetermined duration of drug treatment, the bacteria were harvested by centrifugation, and the pellets were rapidly frozen in crushed dry ice and stored at −80°C for RNA isolation.
RNA Isolation.
Bacterial lysis and isolation of crude total RNA were achieved by a modification of methods using hot phenol and agitation with glass beads (15, 16). Briefly, frozen bacterial pellets were quickly resuspended in 0.4 ml of 70°C lysis buffer (20 mM sodium acetate, pH 5.2/0.5% SDS/1 mM EDTA) and transferred to 2-ml screw-cap tubes (fitted with rubber O-rings) containing 0.8 g glass beads and 1 ml of acidified phenol/chloroform (6:4, pH 5.2) preheated to 70°C. This mixture was agitated three times (30 sec at 5,000 rpm, enclosed in a Mini-Beadbeater homogenizer, BioSpec Products, Bartlesville, OK) over the course of an 8-min incubation at 65°C. The aqueous lysate subsequently was extracted three times in acidified phenol/chloroform/isoamyl alcohol (25:24:1) and precipitated in isopropanol. These crude RNA fractions typically contained substantial amounts of genomic DNA, so they were repurified by using a total RNA kit according to the manufacturer’s instructions (Qiagen, Valencia, CA). Remaining traces of DNA were digested by treating 30 μg RNA with 2 units DNase I (Boerhinger Mannheim) for 10 min at 37°C, followed by DNaseI inactivation at 70°C for 5 min. RNA samples were ethanol-precipitated, washed once in 70% ethanol, and redissolved in water to 1 μg/μl. Examination of the purified total RNA by gel electrophoresis revealed prominent 23S and 16S ribosomal bands, indicating that the pool was not significantly degraded.
Preparation of Labeled cDNA.
Fluorescently labeled cDNA copies of the total RNA pool were prepared by direct incorporation of fluorescent nucleotide analogs during a first-strand reverse transcription (RT) reaction. Each 40-μl labeling reaction consisted of 8 μg total RNA, 8 ng in vitro-derived transcripts as internal standards, 2 nmol random primers, 0.5 mM each of dATP, dCTP, and dGTP, 0.2 mM dTTP, 10 mM DTT, 0.3 units reverse transcriptase (Superscript II, Stratagene) in 1× reaction buffer provided by the manufacturer and 2 nmol of either Cy3-dUTP or Cy5-dUTP (Amersham Pharmacia). The RNA and primers were preheated to 70°C for 5 min and snap-cooled in ice water before adding the remaining reaction components. The RT reaction proceeded 10 min at 25°C followed by 2 hr at 42°C. Buffer exchange, purification, and concentration of the cDNA products was accomplished by three cycles of diluting the reaction mixture in 0.45 ml TE buffer (10 mM Tris, pH 8/1 mM EDTA) and filtering through Microcon-30 microconcentrators (Amicon).
Microarray Hybridization and Data Analysis.
The two cDNA pools to be compared were mixed and applied to the array in a hybridization mixture containing 3.5× SSC, 0.3% SDS, and 10 μg yeast tRNA. Hybridization took place under a glass coverslip in a humidified slide chamber submerged in a 63°C water bath for 4–6 hr. The slides were washed, dried, and scanned for fluorescence by using a laser-activated, confocal scanner (12). Average signal intensity and local background measurements were obtained for each spot on the array by using custom-written software (Michael Eisen, scanalyze, 1998, available at http://rana.stanford.edu).
Local background was subtracted from the value of each spot on the array. Spots were considered negative and eliminated from further analysis if the values for both channels were less than a threshold value defined as one SD above the average value of the negative control spots that contained non-M. tuberculosis DNA. The two channels were normalized with respect to the median values for the remaining set of M. tuberculosis DNA spots. The Cy5/Cy3 fluorescence ratios and log10-transformed ratios were calculated from the normalized values.
Estimates of the assay sensitivity were made for the experiments shown in Fig. 1 by adding known masses of four different non-M. tuberculosis, in vitro-derived transcripts to the labeling reaction. The amount of these artificial transcripts added per 8 μg of total M. tuberculosis RNA ranged from 0.01 to 10 pg, corresponding to approximately 0.3–300 transcripts per cell (assuming the mass of total RNA in a bacterium is 5.8 × 10−14 g (17). Thus, when the lower detection limit was defined as two SDs above the mean local background of all spots in the experiment, the sensitivity was estimated to be 3–30 transcripts per cell.
For selection of drug-induced genes, the z value [(ratio − mean)/SD] was calculated for each spot and assigned a positive value for induced genes and a negative value for repressed genes. Spots were considered induced and reported in Table 1 if the sum of the z values, obtained for the 40-min, 2-hr, and 6 hr-treated samples within a given time course, was greater than six for two independent time courses.
RT-PCR.
The RT step was primed randomly and then subdivided into the various PCR steps containing the appropriate primer pairs for amplification of the target genes. Each 30-μl RT reaction contained 2 μg purified total RNA, 300 ng random primers, 200 mM each of dATP, dCTP, dGTP, and dTTP, 10 mM DTT, and 20 units Moloney murine leukemia virus reverse transcriptase (Boerhinger Mannheim), in a 1× buffer supplied by the manufacturer. The RNA and primers were preheated to 70°C for 5 min and snap-cooled in ice water before adding the remaining components; the RT reactions proceeded 10 min at 25°C and 1 hr at 42°C. To assess the contribution of contaminating DNA templates in the RNA samples, RNase-treated controls were performed in a parallel manner with an addition to the RT reaction of 15 pg RNaseA (Qiagen) that had been preheated for 15 min at 65°C to eliminate residual DNase activity. Positive controls for the PCR step were performed by adding M. tuberculosis genomic DNA as template to two RT reactions that were either treated or untreated with RNaseA.
The PCR phase contained two primer pairs in each reaction to allow coamplification and the internal comparison of two target genes. The 70-μl reactions contained 1 μl of template nucleic acid from the appropriate RT reaction, 1 unit Taq polymerase (Sigma), and 25 pmol each of four ORF-specific primers in the 1× PCR mixture described above. Each reaction proceeded through 26 cycles of the following temperature profile: 94°C, 40 sec; 52°C, 45 sec; 72°C, 45 sec. A PCR negative control reaction, containing the appropriate primer pairs but no added template, was performed with each experiment. Amplified products were analyzed by electrophoresis in 2% agarose gels, and images of the ethidium-bromide-stained bands were obtained with Polaroid film or a digital camera (AlphaImager 2000, Alpha Innotech, San Leandro, CA). Primers used to amplify inhA were 5′-ACTCGTCGATCGCGTTTCACAT-3′ and 5′-GTCGGCGAGATGATGTCACCC-3′; for fas they were 5′-CATGACCTACCTGCAGTGGCTG-3′ and 5′-AACGTCACGAAAAAGGGCACAT-3′; for kasA they were 5′-TCGGACGCCTTTCATATGGTG-3′ and 5′-TCGGGTGTCTCGTAGTTCAGGG-3′; for efpA they were 5′-ACCATCTCGTCGGTGCTGTG-3′ and 5′-CGGAACAAGTGGAACGGC-3′; for ahpC they were 5′-CACTCGCAGTACCTCATCGACGT-3′ and 5′-GCTAACCATTGGCGATCAATTCC-3′.
Results and Discussion
To monitor the INH-induced gene response profile at an early stage in the inhibitory effect of the drug, we treated log-phase liquid cultures with INH and collected bacteria at defined times throughout the first 8 hr of treatment. Total RNA was isolated from each sample, and a fluorescently labeled cDNA replica of each was prepared by a randomly primed, RT reaction that incorporated one of two fluorescent nucleotide analogues into the cDNA. The RNA isolated from cells collected at the beginning of treatment (t = 0) was the reference sample to which each of the samples isolated from later time points were compared. The t = 0 sample was labeled with one fluorochrome (typically Cy3), and an equivalent mass of RNA from each of the later time points was labeled with the other (typically Cy5). For each time point, the two differentially labeled cDNA samples were mixed and cohybridized to the array. The resulting signal intensities from each fluorophore were compared for each ORF represented on the microarray, thus allowing us to monitor the relative changes in mRNA abundance as a consequence of INH treatment.
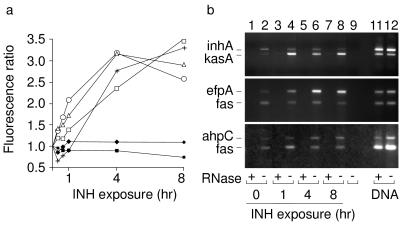
A highly induced set of genes belong to an operon-like cluster of five genes that encode polypeptide components of the FAS-II complex [denoted as Rv2243–47 by Cole et al. (1)] including AcpM (Rv2244) and KasA (Rv2245) (Fig. 1a, Table 1). Thus, microarray results corroborated biochemically derived observations and highlighted genes directly involved in the processes inhibited by INH. The induction of each of these genes was detected as early as 20 min after treatment (Fig. 2a), indicating that interruption of the FAS-II cycle is rapidly sensed and responded to at a transcriptional level within a fraction of a generation time (24–36 hr) of M. tuberculosis.
Figure 2.
Temporal profile of INH-induced expression of selected genes. (a) Microarray-based determinations of selected gene responses resulting from 20- min, 40-min, 1-hr, 4-hr, and 8-hr treatments with INH. The results are depicted as ratios of the fluorescence intensities of labeled cDNAs from bacteria exposed to INH for the indicated times compared with the zero time point. Ratio values for inhA (■), fas (♦), kasA (○), fadE24 (▵), ahpC (+), and efpA (□) derived from microarray hybridization experiments are plotted with respect to the duration of INH exposure. (b) The change in abundance of selected transcripts in total RNA was estimated by RT-PCR samples prepared from organisms exposed to INH for 0, 1, 4, and 8 hr. Primers were designed to amplify internal regions of kasA (318 bp), efpA (521 bp), and ahpC (484 bp) and the internal reference genes inhA (432 bp) and fas (306 bp). RNA samples isolated from bacteria treated with INH for the indicated times (lanes 1–8) were reverse-transcribed and PCR-amplified with primers specific to kasA and inhA (Top), efpA and fas (Middle), and ahpC and fas (Bottom). The pairwise comparison of two genes for each experiment, using amplified fragments of similar size, allowed direct comparison of the corresponding transcripts. To assess RNA-specific amplification, a duplicate RNA sample for each time point was digested with RNase (lanes 1, 3, 5, and 7). Positive controls for the PCR step contained M. tuberculosis genomic DNA as template and were either treated (lane 11) or untreated (lane 12) with RNase. A PCR negative control reaction (lane 9), containing the appropriate primer pairs but no added template, was performed with each experiment.
fbpC also was induced. It encodes an abundant, exported antigen that possesses trehalose-dimycolyl transferase activity that likely mediates a terminal step of mycolate maturation by esterifying mycolates to specific carbohydrates in the cell wall (18). FbpC, also known as antigen 85-C, is one of three highly related proteins known collectively as the antigen 85 complex, so its induction is consistent with the report that antigen 85 proteins are induced by INH (19). As illustrated by the proposed mycolic acid biosynthesis pathway depicted in Fig. 3, the mycolyl transferase step is subsequent to the point of INH inhibition, and thus fbpC is presumably up-regulated by a feedback mechanism that senses the depletion of mature mycolates.
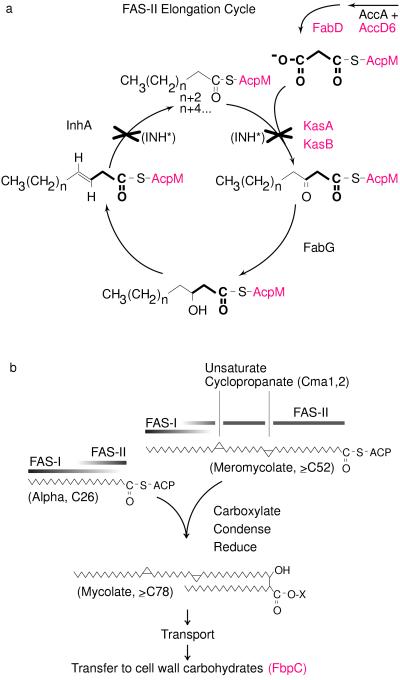
Figure 3.
Roles of INH-induced genes in the context of a proposed pathway for mycolic acid biosynthesis. (a) Mycolic acids of M. tuberculosis are elongated by the multienzyme, AcpM-dependent FAS-II complex (7, 23, 24). By analogy with the Escherichia coli FAS-II system (25), the enzymatic steps required for the addition of a two-carbon subunit, provided by malonyl-AcpM (highlighted in bold), are shown together with some of the proteins that contribute to the process. Activated INH (INH*) is thought to inhibit FAS-II by binding to NADH in the pocket of InhA and by forming a covalent, ternary complex between KasA and AcpM (6, 7). The operon-like cluster of genes encoding AccD6, FabD, KasA, KasB, and AcpM (red) all were induced by INH (Table 1), perhaps as a consequence of the depleted meromycolate pool. In contrast, neither fabG nor inhA, which are organized into a two-gene cluster at a separate position on the chromosome, was induced by INH (Fig. 2). (b) A proposed mycolate synthesis pathway shows the predicted roles of FAS-II, described above, and FAS-I, which elongates short-chain fatty-acyl-CoA esters (23, 26). The precise fatty acyl intermediates and ACP-charging enzymes that mediate the FAS-I to FAS-II transition have not been characterized. Meromycolate chains can be functionalized by several different moieties (27). For clarity, however, only the cyclopropyl-containing meromycolate and the enzymes that introduce the distal (Cma1) and proximal (Cma2) moieties are shown (28). The mycolyl transferase (FbpC) is one of several exported proteins predicted to catalyze the transesterification of mycolates to carbohydrate moieties in the cell wall (18). Microarray hybridization experiments showed that fbpC (red) also was induced by INH treatment.
Several other INH-induced genes encode proteins that have not been biochemically characterized, but which are homologous with proteins of known function: two adjacent acyl-CoA dehydrogenases, FadE23 and FadE24 (Rv3139–40), an efflux protein, EfpA (Rv2846c), and a subunit of the alkyl-hydroperoxide reductase, AhpC (Rv2428) (Fig. 2a and Table 1). The acyl-CoA dehydrogenases likely participate in the degradation of fatty acids by β-oxidation into acetyl-CoA subunits. Although this activity is not directly related to mycolic acid biosynthesis, it may reflect a response designed to recycle the fatty acids that accumulate as a result of INH treatment. The induction of each gene in the five-gene FAS-II cluster and each gene of the two-gene fadE23–24 cluster also demonstrates how microarray analysis helps reveal the presence and boundaries of bacterial operons. Although the results do not prove the existence of a multigenic transcript, the position, orientation, and parallel regulation of these genes is consistent with the definition of an operon.
EfpA is predicted to be a proton-energized transporter with 14 transmembrane domains, but the molecules it translocates have not been defined (20). The INH-mediated induction of efpA naturally raises the possibility that the protein transports molecules relevant to mycolic acid production, such as nascent mycolates or synthesis precursors. If EfpA mediates an essential mycolate biosynthetic function, then it may be an appropriate novel drug target, especially because the gene is present only within the pathogenic members of the mycobacterial genus. An alternative explanation for efpA induction is based on the understanding that some efflux pumps provide important mechanisms for expelling drugs and other toxic compounds from the intracellular environment (21). Thus, INH may have triggered a similar mechanism that is mediated in M. tuberculosis by the induction of EfpA. However, because M. tuberculosis is exquisitely sensitive to INH, it seems unlikely that wild-type EfpA effectively reduces intracellular INH levels, and no drug-resistant strains of M. tuberculosis have yet been identified with efpA mutations that could explain the emergence of resistance (20). Further investigation is warranted to elucidate EfpA function and the molecules that signal its induction.
The induction of ahpC and Rv0341–2 (Table 1) were delayed slightly compared with the genes described above (Fig. 1), indicating that they may be responding to a secondary, toxic consequence of INH activity. This interpretation is supported by the knowledge that alkyl hydroperoxide reductases help detoxify the cell by reducing specific classes of reactive oxygen species. A previous report that transcripts for Rv0341–2 are induced by INH (22) is consistent with the results reported here, and it helps validate the ability of microarray hybridization analysis to identify differences between two pools of total RNA.
To further test the validity of the microarray results, we performed RT-PCR to compare the abundance of kasA, efpA, and ahpC transcripts from INH-treated and untreated bacteria (Fig. 2). As internal controls we analyzed the relative abundance of inhA (Rv1484) and fas (Rv2524c) transcripts by this technique because the array results (Fig. 2a) showed that their expression was not changed by INH exposure. Because their encoded products participate directly in fatty acid biosynthesis (fas encodes the type I FAS, FAS-I), their inclusion in this assay allowed us to test the specificity of the INH response within a group of functionally related enzymes. INH-induced expression of kasA, efpA, and ahpC was confirmed by RT-PCR; the RT-PCR assay also corroborated the microarray-based observation that the expression of inhA and fas was not changed by INH treatment (Fig. 2b).
The physiological relevance and specificity of the microarray-determined transcriptional response to INH was explored further by testing the response of an INH-resistant strain. INH is a prodrug that requires intracellular oxidation by the mycobacterial catalase/peroxidase, KatG, to form the bioactive species. Consequently, resistance to INH emerges most frequently because of mutations that inactivate KatG. Ethionamide is structurally related to INH and used therapeutically as a second-line antibiotic, but it does not have the same requirement for KatG-mediated activation. Because INH and ethionamide both inhibit mycolate biosynthesis and are believed to target the same FAS-II enzymes, we predicted that ethionamide would generate a similar response profile to that of INH. We further predicted that a catalase-negative, INH-resistant strain would not exhibit INH-dependent gene expression, whereas when treated with ethionamide, the same strain would respond with the response profile that is characteristic of FAS-II inhibition.
To test the first prediction, we evaluated the response of the INH-sensitive strain to ethionamide by using the same microarray hybridization system and experimental design described above. The results showed that ethionamide treatment induces the same genes that were induced by INH (Table 1). The parallel nature of the INH and ethionamide responses suggests that the expression profiles revealed by microarray hybridization provide a characteristic signature that is specific for the cellular process that is affected by the compound. This conclusion is further supported by the observation that the INH and ethionamide response profiles were distinct from the profiles obtained from bacteria exposed in a similar manner to a variety of different toxic compounds, including hydrogen peroxide, ethanol, and aminoglycoside antibiotics (not shown). To test the second prediction, we used microarray hybridization to assay a catalase-negative, INH-resistant strain for its transcriptional response to INH and ethionamide. The results revealed no significant changes in gene expression upon INH exposure, indicating that the prodrug form of INH is indeed inactive, both generally and with respect to the FAS-II system. In contrast, the ethionamide-induced gene response profile resembled that of the INH-sensitive strain, displaying the characteristic signature of FAS-II inhibition (Fig. 1 b and c). Taken together these results also show that the characteristic INH/ethionamide response is the result of intracellular conditions associated with the drug’s mode of action and not an unrelated response to the physiologically inactive prodrug.
Our understanding of the molecular consequences of INH on mycolate biosynthesis provides confidence that many of the INH-responsive genes encode proteins that are relevant to the drug’s mode of action. Because DNA microarray hybridization analysis allowed us to monitor most of the genetic repertoire of the organism, we were able to identify genes potentially involved in mycolic acid metabolism, whose role in this process had not been previously recognized. Because the affected enzymatic pathway contains proven drug targets, it is reasonable to propose that other proteins operating in the same pathway also might be appropriate targets for new drug development. The results of this study also demonstrate that the transcriptional responses of a microorganism provide information about the physiological state of the bacterium. Accordingly, using this approach, it may soon be possible to predict the mode of action of a novel compound based on a physiologically derived interpretation of its expression response to that compound. Furthermore, drug-responsive promoters identified by the experimental approach described here may serve as sensors of the intracellular conditions that are characteristic of the drug’s activity. This feature may prove valuable in designing screens to identify novel compounds that exert similar effects on the cell.
Acknowledgments
We are grateful for the important contributions of M. Eisen, T. Garnier. H. Salamon, and P. Small. This work was supported by National Institutes of Health Grants AI35969 and AI44826, the Carlsberg Foundation, and the Danish Natural Science Research Council (H.-H.K.).
Abbreviations
- INH
isoniazid
- FAS
fatty acid synthase
- RT
reverse transcription
References
- 1.Cole S T, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon S V, Eiglmeier K, Gas S, Barry C E, 3rd, et al. Nature (London) 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 2.W.H.O. Anti-Tuberculosis Drug Resistance in the World: The W.H.O./IUATLD Global Project on Anti-Tuberculosis Drug Resistance Surveillance. Geneva: W.H.O.; 1997. [Google Scholar]
- 3.Pablos-Mendez A, Raviglione M C, Laszlo A, Binkin N, Rieder H L, Bustreo F, Cohn D L, Lambregts-van Weezenbeek C S, Kim S J, Chaulet P, Nunn P. N Engl J Med. 1998;338:1641–1649. doi: 10.1056/NEJM199806043382301. [DOI] [PubMed] [Google Scholar]
- 4.Takayama K, Wang L, David H L. Antimicrob Agents Chemother. 1972;2:29–35. doi: 10.1128/aac.2.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Banerjee A, Dubnau E, Quemard A, Balasubramanian V, Um K S, Wilson T, Collins D, de Lisle G, Jacobs W R., Jr Science. 1994;263:227–230. doi: 10.1126/science.8284673. [DOI] [PubMed] [Google Scholar]
- 6.Rozwarski D A, Grant G A, Barton D H R, Jacobs W R, Jr, Sacchettini J C. Science. 1998;279:98–102. doi: 10.1126/science.279.5347.98. [DOI] [PubMed] [Google Scholar]
- 7.Mdluli K, Slayden R A, Zhu Y, Ramaswamy S, Pan X, Mead D, Crane D D, Musser J M, Barry C E., 3rd Science. 1998;280:1607–1610. doi: 10.1126/science.280.5369.1607. [DOI] [PubMed] [Google Scholar]
- 8.Mdluli K, Sherman D R, Hickey M J, Kreiswirth B N, Morris S, Stover C K, Barry C E., 3rd J Infect Dis. 1996;174:1085–1090. doi: 10.1093/infdis/174.5.1085. [DOI] [PubMed] [Google Scholar]
- 9.Takayama K, Schnoes H K, Armstrong E L, Boyle R W. J Lipid Res. 1975;16:308–317. [PubMed] [Google Scholar]
- 10.Lashkari D A, McCusker J H, Davis R W. Proc Natl Acad Sci USA. 1997;94:8945–8947. doi: 10.1073/pnas.94.17.8945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schena M, Shalon D, Davis R W, Brown P O. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 12.DeRisi J L, Iyer V R, Brown P O. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- 13.Kubica G P, Jones W, Abbott V D, Beam R E, Kilburn J O, Caper J O., Jr Am Rev Respir Dis. 1966;94:400–405. doi: 10.1164/arrd.1966.94.3.400. [DOI] [PubMed] [Google Scholar]
- 14.Jacobs W R, Jr, Kalpana G V, Cirillo J D, Pascopella L, Snapper S B, Udani R A, Jones W, Barletta R G, Bloom B R. Methods Enzymol. 1991;204:537–555. doi: 10.1016/0076-6879(91)04027-l. [DOI] [PubMed] [Google Scholar]
- 15.Mangan J A, Sole K M, Mitchison D A, Butcher P D. Nucleic Acids Res. 1997;25:675–676. doi: 10.1093/nar/25.3.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aiba H, Hanamura A, Yamano H. J Biol Chem. 1991;266:1721–1727. [PubMed] [Google Scholar]
- 17.Neidhardt F C, editor. Escherichia coli and Salmonella typhimurium Cellular and Molecular Biology. Vol. 1. Washington, DC: Am. Soc. Microbiol.; 1987. p. 4. [Google Scholar]
- 18.Belisle J T, Vissa V D, Sievert T, Takayama K, Brennan P J, Besra G S. Science. 1997;276:1420–1422. doi: 10.1126/science.276.5317.1420. [DOI] [PubMed] [Google Scholar]
- 19.Garbe T R, Hibler N S, Deretic V. Antimicrob Agents Chemother. 1996;40:1754–1756. doi: 10.1128/aac.40.7.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Doran J L, Pang Y, Mdluli K E, Moran A J, Victor T C, Stokes R W, Mahenthiralingam E, Kreiswirth B N, Butt J L, Baron G S, et al. Clin Diagn Lab Immunol. 1997;4:23–32. doi: 10.1128/cdli.4.1.23-32.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paulsen I T, Brown M H, Skurray R A. Microbiol Rev. 1996;60:575–608. doi: 10.1128/mr.60.4.575-608.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alland D, Kramnik I, Weisbrod T R, Otsubo L, Cerny R, Miller L P, Jacobs W R, Jr, Bloom B R. Proc Natl Acad Sci USA. 1998;95:13227–13232. doi: 10.1073/pnas.95.22.13227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Odriozola J M, Ramos J A, Bloch K. Biochim Biophys Acta. 1977;488:207–217. doi: 10.1016/0005-2760(77)90178-3. [DOI] [PubMed] [Google Scholar]
- 24.Quemard A, Sacchettini J C, Dessen A, Vilcheze C, Bittman R, Jacobs W R, Jr, Blanchard J S. Biochemistry. 1995;34:8235–8241. doi: 10.1021/bi00026a004. [DOI] [PubMed] [Google Scholar]
- 25.Magnuson K, Jackowski S, Rock C O, Cronan J E., Jr Microbiol Rev. 1993;57:522–542. doi: 10.1128/mr.57.3.522-542.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kolattukudy P E, Fernandes N D, Azad A K, Fitzmaurice A M, Sirakova T D. Mol Microbiol. 1997;24:263–270. doi: 10.1046/j.1365-2958.1997.3361705.x. [DOI] [PubMed] [Google Scholar]
- 27.Barry C E, 3rd, Lee R E, Mdluli K, Sampson A E, Schroeder B G, Slayden R A, Yuan Y. Prog Lipid Res. 1998;37:143–179. doi: 10.1016/s0163-7827(98)00008-3. [DOI] [PubMed] [Google Scholar]
- 28.George K M, Yuan Y, Sherman D R, Barry C E., 3rd J Biol Chem. 1995;270:27292–27298. doi: 10.1074/jbc.270.45.27292. [DOI] [PubMed] [Google Scholar]