Abstract
Bacterial pathogens of both animals and plants use type III secretion machines to inject virulence proteins into host cells. Although many components of the secretion machinery are conserved among different bacterial species, the substrates for their type III pathways are not. The Yersinia type III machinery recognizes some secretion substrates via a signal that is encoded within the first 15 codons of yop mRNA. These signals can be altered by frameshift mutations without affecting secretion of the encoded polypeptides, suggesting a mechanism whereby translation of yop mRNA is coupled to the translocation of newly synthesized polypeptide. We report that the type III machinery of Erwinia chrysanthemi cloned in Escherichia coli recognizes the secretion signals of yopE and yopQ. Pseudomonas syringae AvrB and AvrPto, two proteins exported by the recombinant Erwinia machine, can also be secreted by the Yersinia type III pathway. Mapping AvrPto sequences sufficient for the secretion of reporter fusions in Yersinia revealed the presence of an mRNA secretion signal. We propose that 11 conserved components of type III secretion machines may recognize signals that couple mRNA translation to polypeptide secretion.
A search for factors that are necessary for the pathogenicity of Gram-negative microbes has identified many gene clusters that are closely related among different bacterial species (1). Several of these genetic loci encode type III secretion machines for the translocation of polypeptides across the bacterial double membrane envelope (1). Some mammalian pathogens such as Yersinia, Salmonella, Escherichia coli, Pseudomonas, and Shigella use type III machines for the injection of virulence factors into the cytosol of eukaryotic cells (2–7). A similar strategy is thought to be used by several plant pathogens; however, a direct demonstration of their protein injection has not yet been achieved (7). Salmonella typhimurium and perhaps other Gram-negative bacteria harbor two gene clusters that each specifies a type III machine (2). Mutants that abolish the function of individual type III machines arrest pathogenicity at distinct steps during Salmonella infection, indicating that protein secretion is needed for the continued successful interaction between the pathogen and its host (8).
Type III machines of pathogenic Yersinia species are encoded on a virulence plasmid and comprise 21 different ysc genes (Yop secretion) (9). Mutations in any one of the ysc genes abolish the type III secretion of 14 different Yop proteins (Yersinia outer proteins) by mutant Yersinia spp. (10). Yop proteins do not display amino acid sequence or physical similarity (9). Previous work revealed the existence of two independent type III export pathways for Yop proteins (11–13). The first pathway recognizes a secretion signal encoded in the first 15 codons of yop mRNA (13). This sequence can be altered by frameshift mutation without loss of function, suggesting that mRNA may provide a signal that leads to the coupling of yop translation and type III secretion of newly synthesized polypeptide (13–15). The second type III export pathway requires the presence of a bacterial Syc chaperone (specific Yop chaperone) (12, 16–18). For example, SycE binds as a homodimer to YopE amino acids 15–100 and maintains the polypeptide in a soluble state (19). The SycE2:YopE complex interacts with the type III machine, thereby leading to the displacement of SycE and to the targeting of YopE into the cytosol of eukaryotic cells (19, 20).
Although all type III machines are believed to catalyze similar reactions, i.e., the translocation of polypeptides across the bacterial envelope or their injection into the cytosol of eukaryotic cells, only 9 of the 21 ysc genes are conserved among all known type III machinery components of mammalian and plant pathogens (1). Can the secretion substrates of one type III machine be recognized by the export apparatus of another pathogen? Rosqvist et al. (21) reported that Yersinia YopE can be secreted by Salmonella in the presence, but not in the absence, of SycE chaperone (21). Similarly, Shigella IpaB could be secreted by Yersinia in the presence, but not in the absence, of IpgC chaperone (21). These results suggested that the secretion chaperone-mediated export pathway may be functional in many, if not all, Gram-negative bacteria. Because mRNA signals are functional even in the absence of secretion chaperones, this result also implies that type III machines cannot recognize RNA signals of heterologous secretion substrates. However, another study reported the secretion of Pseudomonas aeruginosa ExoS by Yersinia pseudotuberculosis, which occurred without the concomitant expression of a presumed ExoS chaperone (22). Thus, the mechanism by which type III machines recognize heterologous substrates has not yet been resolved.
Recently, a cluster of hrp (hypersensitive response and pathogenicity) and hrc (hypersensitive response and conserved) genes encoding the Erwinia chrysanthemi type III machine was cloned in E. coli, thereby allowing the secretion of Pseudomonas syringae export substrates AvrB and AvrPto (23). We asked whether the Erwinia type III machine recognizes Yersinia substrates and report the secretion of YopE and YopQ. The Yersinia type III machine exported the P. syringae secretion substrates AvrB and AvrPto. The secretion signals of AvrB and AvrPto were mapped to the first 15 codons. The AvrPto signal could be frameshifted without loss of secretion, indicating that type III machines of Yersinia and Erwinia can recognize heterologous secretion substrates by their mRNA signals.
Materials and Methods
Bacterial Strains and Plasmids.
The strains and plasmids used in this study are listed in Table 1. E. coli strains were routinely grown in LB medium at 37°C for the isolation of plasmids and at 30°C in Luria medium for protein-secretion assays. The following concentrations were used for antibiotics: ampicillin (Ap), 25 μg/ml; chloramphenicol (Cm), 10 μg/ml; gentamycin (Gm), 10 μg/ml; kanamycin (km), 50 μg/ml; spectinomycin (Sp), 25 μg/ml. Npt fusions containing 15 codons and the frameshift-mutant derivatives were constructed as described (13). To construct the fusion of the first 44 codons of avrB to npt, DNA fragments were PCR-amplified with Taq DNA polymerase using primers AvrB-3 (5′-AACATATGGGCTGCGTCTCGTCAA-3′) and AvrB-4 (5′-AAGGTACCTAAGCATTGATCATAGACCTC-3′) and pAvrB-Flag as the template. The PCR product was then digested with NdeI and KpnI and ligated into pDA219 that had been cut with the same enzymes and purified using QiaQuik gel-purification columns (Qiagen, Chatsworth, CA). Plasmids carrying full-length yopE and yopQ were constructed by digestion of pDA36 and pDA243, respectively, by NdeI and KpnI followed by ligation into the same sites of pFlag-CTC such that the resulting proteins carried a C-terminal Flag epitope tag.
Table 1.
Bacterial strains and plasmids used in this work
| Designation | Relevant characteristics and use | Ref. |
|---|---|---|
| Y. enterocolitica | ||
| W22703 | 0:9 serotype, Nalr, wild-type isolate | 35 |
| E. coli | ||
| DH5α | supE44 ΔlacU169 (Φ80lacZΔM15)hsdR17 recA1 endA1 gyrA96 thi1 relA1 | 36 |
| P90C | araΔ(gpt-lac)5 | 37 |
| Plasmids | ||
| pDA219 | For construction of Npt fusions driven by the tac promoter, Cmr | 14 |
| pDA288 | pDA219 carrying avrB codons 1–15 fused to npt | This work |
| pDA289 | pDA219 carrying avrPto codons 1–15 fused to npt | This work |
| pDA291 | pDA219 carrying avrB codons 1–44 fused to npt | This work |
| pDA294 | pDA289 (−1 frameshift mutation) | This work |
| pDA295 | pDA289 (+1 frameshift mutation) | This work |
| pDA296 | pDA289 (−2 frameshift mutation) | This work |
| pDA297 | pDA289 (+2 frameshift mutation) | This work |
| pFlag-CTC | For construction of C-terminal fusions to Flag peptide, Apr | Kodak |
| pYopE–Flag | pFlag-CTC carrying yopE | This work |
| pYopQ–Flag | pFlag–CTC carrying yopQ | This work |
| pML123 | Broad host range expression vector, Gmr | 38 |
| pAvrB–Flag2 | pML123 carrying avrB Flag-tagged | 23 |
| pAvrPto–Flag | pML123 carrying avrPto Flag-tagged | 23 |
| pDA–AvrB15 | pDA219 carrying avrB with codons 2–15 deleted | This work |
| pDA–AvrPto10 | pDA219 carrying avrPto with codons 2–10 deleted | This work |
| pCPP2156 | pCPP19 carrying E. Chrysanthemi hrp cluster, Spr | 23 |
| pCPP2368 | pCPP2156:Tn5Cm that has HR− phenotype, Spr, Cmr | 23 |
Secretion Assays.
Yersinia enterocolitica strains were grown overnight in tryptic soy broth at 26°C. Cultures were diluted 1:50 into 20 ml of fresh medium containing 5 mM EGTA and grown for 2 h at 26°C (OD600 = 0.2) before induction for type III secretion by a temperature switch to 37°C and incubation for 3 h (OD600 = 0.6). Where necessary, the tac promoter was induced by the addition of 1 mM isopropyl-β-d-thiogalactoside (IPTG) 30 min after the temperature shift. Twenty milliliters of induced culture were centrifuged at 7,000 × g for 15 min, and 15 ml of supernatant was removed and precipitated with 5% trichloroacetic acid (TCA). The remainder of the supernatant was discarded, and the pellet was suspended in 800 μl of water. A 600-μl aliquot of this suspension was precipitated with 600 μl of ice-cold 10% TCA. All TCA precipitates were centrifuged for 15 min at 17,000 × g and washed for 10 min on ice with acetone. Pellets were collected by centrifugation at 17,000 × g for 10 min and air-dried. Proteins were suspended in 150 μl of 0.5 M Tris⋅HCl/4% SDS (pH 8.0) and added to an equal volume of sample buffer. Proteins separated by SDS/PAGE were transferred to Immobilon P membrane (Millipore) and identified by their binding to polyclonal antibody followed by chemiluminescent detection. A dilution series of purified protein antigen was also immunoblotted to ensure that signal intensity corresponded to antigen concentration. Where indicated, the signals were quantified by densitometric scanning of the developed x-ray films.
E. coli colonies were inoculated into 5 ml of Luria medium containing the appropriate antibiotics and grown overnight at 30°C. Cells were washed twice in Luria medium, diluted to OD600 = 0.2, and grown at 30°C until OD600 = 0.6. When the OD600 reached 0.35, the culture was induced for the expression of type III substrates by adding IPTG to 1 mM. Cultures were centrifuged first for 15 min at 4,300 × g, and the pellet was saved. The supernatant was again centrifuged for 40 min at 17,500 × g to remove residual cells and precipitated with ice cold TCA to a final concentration of 20%. After incubation on ice for 2–4 h, the precipitate was collected by centrifugation for 40 min at 17,300 × g. The sediment was washed twice with cold acetone, air-dried, and solubilized by boiling in SDS sample buffer. The cell sediment was diluted into 4 ml of water, and 100-μl aliquots were mixed with sample buffer before boiling. Samples were separated by SDS/PAGE before immunoblotting with specific antisera. A dilution series of purified protein antigen was also immunoblotted to ensure that signal intensity corresponded to antigen concentration. Where indicated, the signals were quantified by densitometric scanning of the developed x-ray films.
Results
Secretion of YopE and YopQ by E. coli Expressing the Erwinia Type III Machine.
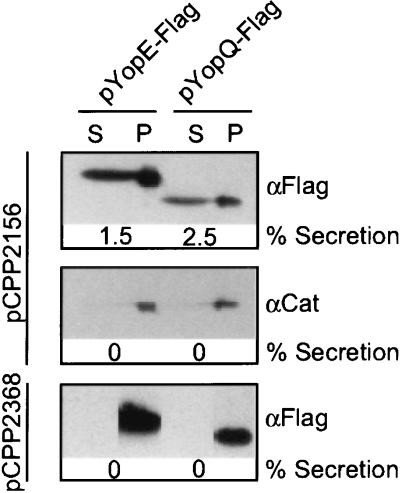
Type III secretion of YopQ occurs solely via an mRNA signal, whereas YopE secretion can be initiated by two independent type III pathways (12, 14). One pathway recognizes the signal located in the first 15 codons of yopE mRNA (13). The second pathway absolutely requires binding of SycE to YopE amino acid residues 15–100 (12). Thus, in the absence of SycE chaperone, YopE can only be exported via the RNA signal, similar to the type III secretion of YopQ. We asked whether the type III machine of Erwinia can recognize Yersinia RNA signals and secrete YopE and YopQ. Y. enterocolitica yopE and yopQ were expressed by the IPTG-inducible tac promoter in E. coli DH5α (pCPP2156), containing the type III secretion machine of Erwinia. A Flag epitope tag was appended to the ORF of both yopE and yopQ, thereby permitting detection of polypeptides via peptide specific antibody. Both YopE–Flag and YopQ–Flag were secreted into the culture supernatant of E. coli expressing Erwinia type III genes (Fig. 1). Chloramphenicol acetyltransferase, a resident of the bacterial cytoplasm, was detected only in the sediment of the fractionated cultures (Fig. 1). An isogenic E. coli mutant DH5α (pCPP2368), carrying a transposon insertion in the Erwinia hrc cluster (23), failed to secrete either YopE–Flag or YopQ–Flag. Together, these results suggest that the mRNA secretion signals of YopE and YopQ are recognized by the Erwinia type III machine.
Figure 1.
The Erwinia type III machine secretes Yersinia Yops. E. coli DH5α (pCPP2156) expressing type III genes of E. chrysanthemi was transformed with plasmids that expressed either the yopE or the yopQ gene under control of the IPTG-inducible tac promoter. Cultures were induced for expression of Yops and centrifuged. Bacteria in the sedimented pellet (P) were separated from the culture medium in the supernatant (S). Supernatant samples were concentrated 60-fold to facilitate quantitation by SDS/PAGE. Proteins were analyzed by immunoblotting by using peptide antibody against the Flag epitope that had been appended to C-terminal yopE and yopQ sequences. The cytoplasmic protein, chloramphenicol acetyl transferase, supplied on an additional plasmid, was not secreted in these strains. As a control for type III secretion, E. coli DH5α (pCPP2368) carrying a transposon insertion in the Erwinia hrc gene cluster did not secrete YopE–Flag andYopQ–Flag. Yersinia YopQ and YopE harbor an mRNA signal within the first 15 codons that functions to couple mRNA translation and type III secretion of these polypeptides.
We wished to map the secretion signals for the recombinant Erwinia type III machine, and examined yopE, yopQ, and avr (see below) fusions to the neomycin phosphotransferase reporter (npt) gene. However, expression of npt fusions in E. coli DH5α carrying pCPP2156 and pCPP2368 proved to be toxic and caused leakage of cytoplasmic proteins into the supernatant of centrifuged cultures. Furthermore, our genetic characterization of export signals was hindered by the low efficiency of type III secretion in E. coli DH5α (pCPP2156). We routinely observed only 1–4% secretion for most secretion substrates, indicating that the cloned type III pathway of Erwinia was not fully induced under the conditions used. Because of these obstacles, we pursued the molecular characterization of secretion signals in Y. enterocolitica, a pathogen that secretes large amounts of proteins via the type III pathway (9).
Secretion of AvrB and AvrPto by the Y. enterocolitica Type III Machine.
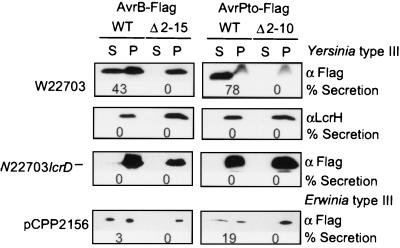
The Erwinia type III machine recognizes and secretes P. syringae export substrates, AvrB and AvrPto (23) (Fig. 2). We wondered whether Pseudomonas proteins can also be secreted by the mammalian pathogen Y. enterocolitica ,and we expressed AvrB–Flag as well as AvrPto–Flag under control of the constitutively expressed npt promoter in strain W22703 (Fig. 2). Forty-three percent of AvrB–Flag and 78% of AvrPto–Flag were found in the extracellular medium of Y. enterocolitica cultures. No type III secretion of AvrB–Flag and AvrPto–Flag was observed in a Yersinia mutant unable to express LcrD, a component of the type III machine that is located in the cytoplasmic membrane (Fig. 2). As a control, the cytoplasmic protein LcrH was found in the bacterial sediment but not in the culture supernatant of all Yersinia samples examined (Fig. 2).
Figure 2.
Yersinia type III machines secrete Pseudomonas secretion substrates. Y. enterocolitica W22703 was transformed with plasmids carrying the avrB and avrPto genes under control of the constitutively expressed npt promoter. Yersinia cultures were induced for type III secretion by temperature shift to 37°C and chelation of calcium from the culture medium. Proteins were detected by immunoblotting with antibody against the Flag epitope, which had been appended to C-terminal avrB and avrPto sequences. Yersinia efficiently recognized the secretion signals of the Pseudomonas proteins, as 43% of AvrB and 78% of AvrPto were secreted, whereas the Yersinia cytoplasmic protein, LcrH, was not secreted. As a control for type III secretion, Yersinia carrying a null mutation in lcrD were unable to secrete AvrB or AvrPto. Deletion of the first 15 or 10 codons of AvrB or AvrPto, respectively, abolished secretion. The secretion of AvrB and AvrPto by the Erwinia type III machine cloned in E. coli was analyzed by concentrating culture supernatants 7.5 (AvrPto) and 80 fold (AvrB).
The Secretion Signals of AvrB and AvrPto.
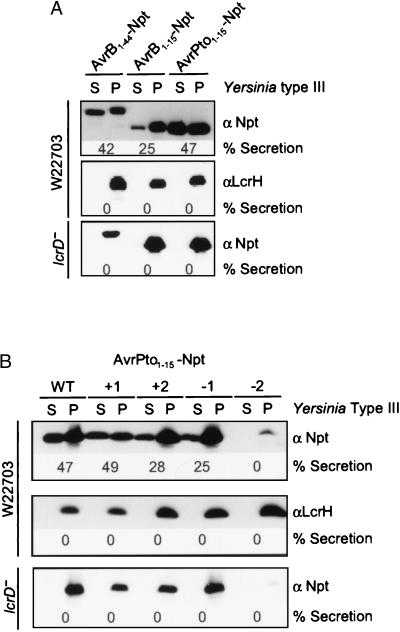
To determine whether the P. syringae secretion substrates harbored mRNA signals similar to those of Yersinia Yops, we mapped and characterized the secretion signals of the Pseudomonas proteins. Fusion of the first 15 codons of avrPto to npt were sufficient to promote secretion of the hybrid Npt protein into the medium of Yersinia cultures (47% secretion) (Fig. 3A). Fusion of the first 15 codons of avrB promoted secretion of 25% of the Npt reporter. We also examined larger fusions and observed that the first 44 codons of avrB improved the efficiency of signal recognition as 42% of the Npt hybrid was located in the culture supernatant, similar to the secretion of full-length AvrB–Flag.
Figure 3.
Pseudomonas AvrPto is secreted by the type III machine via an mRNA secretion signal. (A) The secretion signal encoded by the first 15 codons is not only necessary but also sufficient for the secretion of reporter proteins. The first 15 codons of avrB and avrPto were fused to neomycin phosphotransferase (Npt) and secretion was measured in Y. enterocolitica W22703 with specific antibody to Npt. Both the first 15 codons of avrB and avrPto are sufficient to cause 25% and 47% secretion, respectively, of reporter fusions into the extracellular medium of Yersinia cultures. A fusion of the first 44 codons of avrB to npt caused an increase in secretion of the hybrid protein to 42%. (B) The secretion signal encoded within the first 15 codons of avrPto is recognized at the level of mRNA sequence, because it can tolerate frameshift mutations. Frameshift mutations were generated by nucleotide insertions (+1, +2) or deletions (−1, −2) immediately after the AUG translational start. Frameshifts were suppressed by reciprocal deletions and insertions at the fusion site with npt. The +1, +2, and −1 frameshift mutations did not interfere with Yersinia type III secretion because 49, 28 and 25% of the polypeptides were found in the culture supernatant. The −2 frameshift mutation abolished mRNA expression of the npt reporter.
To examine whether these signals were absolutely necessary for the type III secretion of AvrB and AvrPto, we performed deletions of the first 10 or 15 codons of Pseudomonas mRNAs and expressed them under control of the IPTG-inducible tac promoter. Deletion of codons 2–15 of avrB mRNA, as well as deletion of codons 2–10 of avrPto mRNA, abolished all type III export of the mutant polypeptides by both the cloned Erwinia and the Yersinia type III machines (Fig. 2). To examine whether the Pseudomonas signals were encoded in the mRNA sequence, we focused on the secretion signal of AvrPto because it caused more secretion than the AvrB signal. Frameshift mutations were constructed by either inserting or deleting nucleotides immediately following the AUG translational start of avrPto mRNA. Each frameshift was corrected by reciprocal mutation at the fusion site with npt. The +1 frame shift mutation did not affect the efficiency of secretion, as 49% of the mutant protein was found in the culture supernatant (Fig. 3B). The −1 and +2 frameshift mutations of avrPto permitted 25% and 28% secretion of the mutant polypeptides into the culture medium of Yersinia. The −2 frame shift severely reduced expression of the mutant polypeptide, small amounts of which were detected in the bacterial cytoplasm but not in the culture medium. Together, these results revealed that the signals necessary and sufficient for the secretion of AvrB and AvrPto are located in the first 15 codons. Furthermore, the AvrPto signal is encoded in the mRNA sequence, because it can be altered by some frameshift mutations without loss of secretion.
Discussion
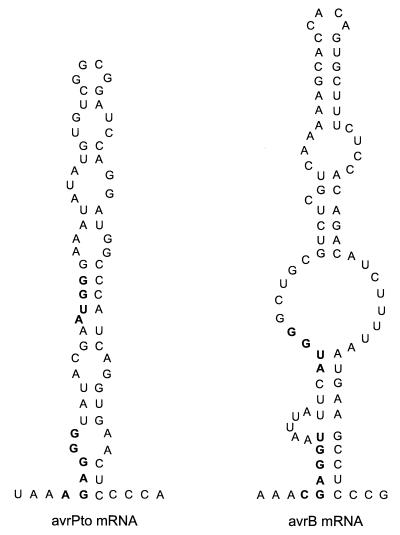
When cloned in E. coli, the type III gene cluster of E. chrysanthemi catalyzes the secretion of heterologous substrates from either P. syringae (23) or Y. enterocolitica. Characterization of the nature of the secretion signal of Yersinia Yops and Pseudomonas AvrPto revealed signals within the first 15 codons of the respective mRNAs. These signals can tolerate some frameshift mutations and still provide for the secretion of the mutant polypeptide chains, suggesting that the mRNA of secretion substrates is recognized by the type III pathways of Erwinia, Yersinia, and Pseudomonas. A computational analysis (24) of the mRNA sequences of avrPto and avrB revealed secondary structures similar to those previously described for Yersinia yopE and yopN (Fig. 4). The Shine–Dalgarno-binding site for 16S ribosomal RNA as well as the AUG translational start site, two elements required for translational initiation (25), are engaged in a hyphenated stem–loop structure. Codons 2–4 of avrPto and avrB are positioned within a loop and may function as recognition signals for the type III pathway (13).
Figure 4.
Predicted RNA structures of the AvrPto and AvrB secretion signals. RNA sequences were subjected to folding analysis by using the Zuker program (24). The displayed structures show areas encompassing the ribosome-binding sites (bold), including the Shine–Dalgarno sequence (−13 to −8), start codon (+1 to +4), and downstream sequence of avrB and avrPto secretion signals. ΔG value (Gibbs energy) for avrB is −19.9 kCal (1 Cal = 4.18 J) and that of avrPto is −17.0 kCal.
The cloned E. chrysanthemi type III machine secretes both Avr and Yop proteins much less efficiently than either Pseudomonas or Yersinia spp. (9, 26). We think it is likely that the type III secretion pathway of the recombinant Erwinia machine is not properly induced, either because a specific signal is not provided in our experiments or because a regulatory gene(s) is absent from the recombinant E. coli strain (26). For example, Yersinia type III secretion is largely repressed by the addition of Ca2+ ions to the media or by temperatures below 30°C (27). We have tested whether Erwinia type III secretion is regulated by similar factors but could not detect conditions that improved secretion efficiency (data not shown).
Partial sequencing of the cloned E. chrysanthemi type III machine combined with its overall sequence and organizational similarity with the known Erwinia amylovora type III system suggests that 20 genes encode the E. chrysanthemi secretion machine (23), 11 of which are conserved in the Yersinia type III system (yscCDJLNQRSTU and lcrD) (9). Moreover, nine of these genes (yscCJNQRSTU and lcrD) are found in every type III system characterized to date, while two genes, yscL (hrpE) and yscD (hrpQ), are shared by only some of them (1). Thus, it is likely that type III machines comprising all 11 components will be able to recognize mRNA secretion signals of type III substrates. This hypothesis is in agreement with previous work on the recognition of substrates by heterologous organisms. For example, the Salmonella SPI-1 gene cluster lacks 1 of the 11 genes (yscL) (2) and is unable to recognize the RNA secretion signal of YopE (21). On the other hand, P. aeruginosa encodes all 11 of these genes (4), and substrates can be secreted without chaperones, suggesting RNA signaling (22).
Ten of the eleven conserved type III genes are homologous to genes that are required for assembly of the flagellar basal-body hook complex and for export of flagellin (yscDJLNQRSTU and lcrD) (1). The one gene not found in the flagellar system is yscC, an outer-membrane secretin of type III machines (28). Homologous secretins are also required for the type II secretion of proteins from the periplasm of Gram-negative bacteria across the outer membrane (29). Outer-membrane secretins are not needed to promote flagellar assembly (30). This type III pathway functions to polymerize translocated flagellin into a proteinaceous filament rather than releasing a secreted polypeptide into the extracellular milieu (31). We find the conservation of genes between type III secretion machines and the flagellar assembly complex compelling and speculate that the latter may also recognize mRNA signals to export polypeptides across the bacterial envelope.
If 11 components compose the core of type III machines, why does E. chrysanthemi use 9 additional hrp genes and Yersinia 10 additional ysc genes to direct protein secretion and translocation into host cells? This complexity is probably due to the large number of secretion substrates, the unique obstacles associated with protein translocation across host-surface barriers, and the differing destinations of the proteins that travel the pathway. For example, Yersinia secretes 14 different Yops, which are directed to distinct locations during infection (9). One group of Yops is injected into the cytosol of eukaryotic host cells (type III targeting) (20). A second group is secreted into the extracellular milieu, while yet another class of Yops remains associated with the bacterial surface and may be involved in regulating type III targeting and secretion (32). Future work will need to unravel all levels of substrate specificity of different Yops during infection. A similarly complex situation exists for some plant pathogens, which use type III machines to assemble a pilus-like structure (33) that is proposed to deliver Hrp proteins across the thick cell wall of plants into the target cell cytosol (34).
Acknowledgments
We thank members of our laboratories for critical review of this manuscript. D.M.A. was supported by the Microbial Pathogenesis Training Grant AI 07323 from the Public Health Service to the Department of Microbiology and Immunology at UCLA School of Medicine. Work in the laboratory of A.C. was supported by Grants MCB-9631530 from the National Science Foundation and 97–35303-4488 (A.C.) and 98–35303-6662 (D.E.F.) from the National Research Initiative Competitive Grants Program/U.S. Department of Agriculture. Work in the laboratory of O.S. was supported by United States Public Health Service Grant AI 42797 from the National Institutes of Health–National Institute of Allergy and Infectious Diseases Branch.
Abbreviations
- cat
chloramphenicol acetyl-transferase
- hrp
hypersensitive response and pathogenicity
- hrc
hypersensitive response and conserved
- npt
neomycin phosphotransferase
- syc
specific Yop chaperone
- yop
Yersinia outer protein
- ysc
Yop secretion
- IPTG
isopropyl-β-d-thiogalactoside
References
- 1.Hueck C J. Microbiol Mol Biol Rev. 1998;62:379–433. doi: 10.1128/mmbr.62.2.379-433.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Collazo C M, Galan J E. Gene. 1997;192:51–59. doi: 10.1016/s0378-1119(96)00825-6. [DOI] [PubMed] [Google Scholar]
- 3.Cornelis G R, Wolf-Watz H. Mol Microbiol. 1997;23:861–867. doi: 10.1046/j.1365-2958.1997.2731623.x. [DOI] [PubMed] [Google Scholar]
- 4.Frank D W. Mol Microbiol. 1997;26:621–629. doi: 10.1046/j.1365-2958.1997.6251991.x. [DOI] [PubMed] [Google Scholar]
- 5.Straley S C, Skrzypek E, Plano G V, Bliska J B. Infect Immun. 1993;61:3105–3110. doi: 10.1128/iai.61.8.3105-3110.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaper J B. Trends Microbiol. 1998;6:169–172. doi: 10.1016/s0966-842x(98)01266-9. [DOI] [PubMed] [Google Scholar]
- 7.Alfano J R, Collmer A. J Bacteriol. 1997;179:5655–5662. doi: 10.1128/jb.179.18.5655-5662.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee V T, Schneewind O. Immunol Rev. 1999;168:241–255. doi: 10.1111/j.1600-065x.1999.tb01296.x. [DOI] [PubMed] [Google Scholar]
- 9.Cornelis G R, Boland A, Boyd A P, Geuijen C, Iriarte M, Neyt C, Sory M-P, Stainier I. Microbiol Mol Biol Rev. 1998;62:1315–1352. doi: 10.1128/mmbr.62.4.1315-1352.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Michiels T, Vanooteghem J-C, Lambert de Rouvroit C, China B, Gustin A, Boudry P, Cornelis G R. J Bacteriol. 1991;173:4994–5009. doi: 10.1128/jb.173.16.4994-5009.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sory M-P, Boland A, Lambermont I, Cornelis G R. Proc Natl Acad Sci USA. 1995;92:11998–12002. doi: 10.1073/pnas.92.26.11998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheng L W, Anderson D M, Schneewind O. Mol Microbiol. 1997;24:757–765. doi: 10.1046/j.1365-2958.1997.3831750.x. [DOI] [PubMed] [Google Scholar]
- 13.Anderson D M, Schneewind O. Science. 1997;278:1140–1143. doi: 10.1126/science.278.5340.1140. [DOI] [PubMed] [Google Scholar]
- 14.Anderson D M, Schneewind O. Mol Microbiol. 1999;31:1139–1148. doi: 10.1046/j.1365-2958.1999.01254.x. [DOI] [PubMed] [Google Scholar]
- 15.Anderson D M, Schneewind O. Curr Opin Microbiol. 1999;2:18–24. doi: 10.1016/s1369-5274(99)80003-4. [DOI] [PubMed] [Google Scholar]
- 16.Wattiau P, Cornelis G R. Mol Microbiol. 1993;8:123–131. doi: 10.1111/j.1365-2958.1993.tb01209.x. [DOI] [PubMed] [Google Scholar]
- 17.Wattiau P, Bernier B, Deslee P, Michiels T, Cornelis G R. Proc Natl Acad Sci USA. 1994;91:10493–10497. doi: 10.1073/pnas.91.22.10493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wattiau P, Woestyn S, Cornelis G R. Mol Microbiol. 1996;20:255–262. doi: 10.1111/j.1365-2958.1996.tb02614.x. [DOI] [PubMed] [Google Scholar]
- 19.Cheng L W, Schneewind O. J Biol Chem. 1999;274:22102–22108. doi: 10.1074/jbc.274.31.22102. [DOI] [PubMed] [Google Scholar]
- 20.Lee V T, Anderson D M, Schneewind O. Mol Microbiol. 1998;28:593–601. doi: 10.1046/j.1365-2958.1998.00822.x. [DOI] [PubMed] [Google Scholar]
- 21.Rosqvist R, Hakansson S, Forsberg A, Wolf-Watz H. EMBO J. 1995;14:4187–4195. doi: 10.1002/j.1460-2075.1995.tb00092.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frithz-Lindsten E, Du Y, Rosqvist R, Forsber A. Mol Microbiol. 1997;25:1125–1139. doi: 10.1046/j.1365-2958.1997.5411905.x. [DOI] [PubMed] [Google Scholar]
- 23.Ham J H, Bauer D W, Fouts D E, Collmer A. Proc Natl Acad Sci USA. 1998;95:10206–10211. doi: 10.1073/pnas.95.17.10206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zuker M. Science. 1989;244:48–52. doi: 10.1126/science.2468181. [DOI] [PubMed] [Google Scholar]
- 25.Gold L, Pribnow D, Schneider T, Shinedling S, Singer B S, Stormo G. Annu Rev Microbiol. 1981;35:365–403. doi: 10.1146/annurev.mi.35.100181.002053. [DOI] [PubMed] [Google Scholar]
- 26.van Dijk, K., Fouts, D. E., Rehm, A. H., Hill, A. R., Collmer, A. & Alfano, J. R. (1999) J. Bacteriol.181, in press. [DOI] [PMC free article] [PubMed]
- 27.Straley S C, Plano G V, Skrzypek E, Haddix P L, Fields K A. Mol Microbiol. 1993;8:1005–1010. doi: 10.1111/j.1365-2958.1993.tb01644.x. [DOI] [PubMed] [Google Scholar]
- 28.Koster M, Bitter W, de Cock H, Allaoui A, Cornelis G R, Tommassen J. Mol Microbiol. 1997;26:789–797. doi: 10.1046/j.1365-2958.1997.6141981.x. [DOI] [PubMed] [Google Scholar]
- 29.Pugsley A P. Microbiol Rev. 1993;57:50–108. doi: 10.1128/mr.57.1.50-108.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Daefler S, Guilvout I, Hardie K R, Pugsley A P, Russel M. Mol Microbiol. 1997;24:465–475. doi: 10.1046/j.1365-2958.1997.3531727.x. [DOI] [PubMed] [Google Scholar]
- 31.Macnab R M. Annu Rev Genet. 1992;26:131–158. doi: 10.1146/annurev.ge.26.120192.001023. [DOI] [PubMed] [Google Scholar]
- 32.Lee V T, Schneewind O. Mol Microbiol. 1999;31:1619–1629. doi: 10.1046/j.1365-2958.1999.01270.x. [DOI] [PubMed] [Google Scholar]
- 33.Roine E, Wei W, Yuan J, Nurmiaho-Lassila E L, Kalkinnen N, Romantachuk M, He S Y. Proc Natl Acad Sci USA. 1997;94:3459–3464. doi: 10.1073/pnas.94.7.3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.He S Y, Huang H C, Collmer A. Cell. 1993;73:1255–1266. doi: 10.1016/0092-8674(93)90354-s. [DOI] [PubMed] [Google Scholar]
- 35.Cornelis G R, Colson C. J Gen Microbiol. 1975;87:285–291. doi: 10.1099/00221287-87-2-285. [DOI] [PubMed] [Google Scholar]
- 36.Hanahan D. J Mol Biol. 1983;166:557–572. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 37.Miller J H, Albertini A M. J Mol Biol. 1983;164:59–71. doi: 10.1016/0022-2836(83)90087-6. [DOI] [PubMed] [Google Scholar]
- 38.Labes M, Puhler A, Simon R. Gene. 1990;89:37–46. doi: 10.1016/0378-1119(90)90203-4. [DOI] [PubMed] [Google Scholar]