Abstract
A novel method of P-element mutagenesis is described for the isolation of mutants affecting the development of the Drosophila compound eye. It exploits the interaction between the Bride of Sevenless (Boss) ligand and the Sevenless (Sev) receptor tyrosine kinase that triggers the formation of the UV-sensitive photoreceptor neuron, R7. Transposition of a boss cDNA transgene, in an otherwise boss mutant background, was used as a “phenotypic trap” in live flies to identify enhancers expressed during a narrow time window in eye development. Using a rapid behavioral screen, more than 400,000 flies were tested for restoration of R7. Some 1,800 R7-containing flies were identified. Among these, 21 independent insertions with expression of the boss reporter gene in the R8 cell were identified by a external eye morphology and staining with an antibody against Boss. Among 900 lines with expression of the boss reporter gene in multiple cells assessed for homozygous mutant phenotypes, insertions in the marbles, glass, gap1, and fasciclin II genes were isolated. This phenotypic enhancer-trap facilitates (i) the isolation of enhancer-traps with a specific expression pattern, and (ii) the recovery of mutants disrupting development of specific tissues. Because the temporal and tissue specificity of the phenotypic trap is dependent on the choice of the marker used, this approach can be extended to other tissues and developmental stages.
Keywords: P-element mutagenesis, mutants, sevenless, bride of sevenless
The compound eye of Drosophila melanogaster contains an array of some 800 ommatidia or simple eyes. Each ommatidium comprises 8 photoreceptor neurons (R cells, designated R1–R8) and a set of accessory cells, including lens-secreting cone cells and pigment cells. The development of this structure has been described in detail at the level of individual identifiable cells (reviewed in refs. 1 and 2) and is readily amenable to classical and molecular genetic analysis. Studies over the past decade have utilized this system to gain insights into the cellular and molecular strategies regulating development (2).
Progress in unraveling the regulatory pathways controlling compound eye development has occurred, in large part, through the application of genetic screens based on chemical, irradiation, or P-element mutagenesis (3, 4). Whereas chemical mutagenesis results in relatively high frequency of randomly distributed mutations (3), these lesions do not provide a convenient molecular handle for identifying the disrupted gene. In contrast, although P elements provide an immediate aid to cloning the gene, insertional mutagenesis occurs at a lower frequency and is less random (4). A modified form of P-element mutagenesis, called the enhancer-trap, incorporates a histological detection system for assessing whether the P element has inserted into the vicinity of a gene of interest (4, 5). Altered P elements carry a lacZ gene fused to a minimal promoter, which can respond to nearby tissue-specific enhancers. Expression patterns are detected by β-galactosidase activity staining. The role of the nearby gene can then be assessed by analyzing flies homozygous for the P-element insertion or, more often, by generating mutant alleles through imprecise excision or local-hopping insertional mutagenesis (4). A considerable limitation in enhancer-trap mutagenesis, however, is that a histological screen is used to detect insertions of interest.
We have developed a variation of enhancer-trap insertional mutagenesis that increases by several orders of magnitude the number of flies tested for patterns of expression. In place of the histological marker, lacZ, we have used a phenotypic marker, bride of sevenless (boss) (6, 7). Activation of the boss transgene, during a brief window of early eye development, leads to rescue of the boss mutant phenotype (8). Thus, expression of the boss transgene in the developing eye can be detected directly in live flies. Moreover, flies with boss expression restricted to the R8 neuron can be identified by simply assessing adult eye morphology under a dissecting microscope. In this paper, we describe the results of the screen and demonstrate that (i) it allows for the efficient isolation of R8-specific enhancer-trap lines, and (ii) it increases by several fold the number of mutants of interest isolated compared with traditional chemical or P-element-based mutageneses. With the appropriate phenotypic markers, this method can be used to screen for genes affecting the development of any adult structure.
MATERIALS AND METHODS
Fly Strains.
The following mutant alleles were used in the mutagenesis: boss1, sevX1, ry506, pn2, K-pn1, and w1118. The boss1 allele contains a 23-bp deletion that cause a frameshift predicted to lead to the production of a truncated protein product of 69 amino acids (7). The HSA1 stock contains a second chromosome with three P elements carrying the boss cDNA under the control of the hsp70 promoter and the rosy+ marker, P[boss; ry+] (8). Because all experiments are carried out without heat-shock treatment, the hsp70 promoter serves as a basal promoter. The Hobo on P (HOP) transposase source was provided by B. Calvi and M. Sanicola (W. Gelbart Lab., Harvard University) and consists of a hobo element carrying the Δ2-3 transposase gene and the mini-white+ marker. We mapped it to 1B cytologically and distal to pn by recombination. For complementation testing, we used the following mutant alleles: Gap1BJ61, gl60J, marbCD4, and fasIIeb112. All gene abbreviations are as listed by Lindsley and Zimm (9).
Dysgenic Crosses and Behavioral Test.
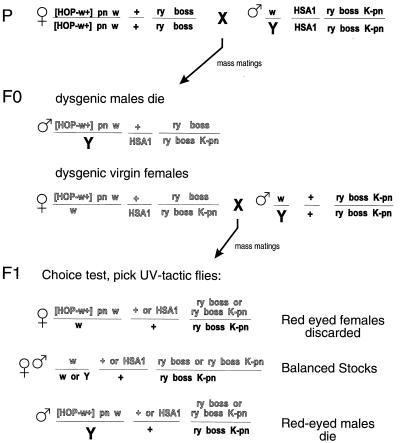
See Fig. 1 for an explanation of the genetic crosses. The phototactic choice test was carried out using a T-tube apparatus (11). Batches of ≈150 flies were given the choice of moving toward UV (350 nm) or visible (550 nm) light. Flies that moved toward the UV source were tested four more times; only flies that chose UV light all five times were scored as UV-tactic. This test takes only a few minutes to perform; the testing of 10,000 flies generally was completed within 4–5 hr. Adult eye morphology was assessed under a dissecting microscope. To balance the P element carrying chromosomes in a sev background, we generated stocks carrying a sev mutant X chromosome and the second or third chromosome balancers: CyO, P[sev+;ry+], or TM6B, boss1, P[sev+; ry+].
Figure 1.
Schematic diagram of genetic screen. The interaction between prune and Killer of prune eliminates male dysgenic progeny in the F0 and F1 generations (10). [HOP; w+], Hobo-element carrying the Δ2-3 transposase gene and the mini-white+ marker; [HSA1], second chromosome carrying three P[boss; ry+] elements (see Materials and Methods). Chromosomes from dysgenic F0 females shown in gray.
Histology.
Third-instar eye discs were stained as previously described using mAb anti-Boss1 (8). In situ hybridization to polytene chromosomes was carried out as described (12) with a digoxygenin labeled boss cDNA probe.
PCR and Genomic Southern Blot Analyses.
Genomic fly DNA isolation, Southern blot analysis, and PCR analysis were performed following standard protocols (13, 14). PCR primers used to detect boss yielded distinguishable products for the boss gene, the boss1 allele, and the boss cDNA. The insert at the scabrous locus was mapped by PCR amplification of the DNA between the 5′ end of the P element and the 5′ sequence of the scabrous transcription unit. A boss cDNA probe was used in the genomic Southern blot analyses to determine the number of P elements present in 29 lines. One line contained two new inserts. Of the 28 with one new insert, 7 contained two or three of the original HSA1 P elements.
RESULTS AND DISCUSSION
Boss as an Enhancer-Trap Marker in Live Flies.
Pattern formation in the eye anlage, the eye imaginal disc, occurs as a wave in the third and final stage of larval development (reviewed in ref. 1). The leading edge of this wave, referred to as the morphogenetic furrow (MF), sweeps across the eye disc. Ahead of the MF, cells are undifferentiated; immediately posterior to the MF, cells aggregate into proto-ommatidial clusters and begin to differentiate into R cells. This is followed by the differentiation of the cone cells and then the pigment cells surrounding each cluster of developing R cells (1).
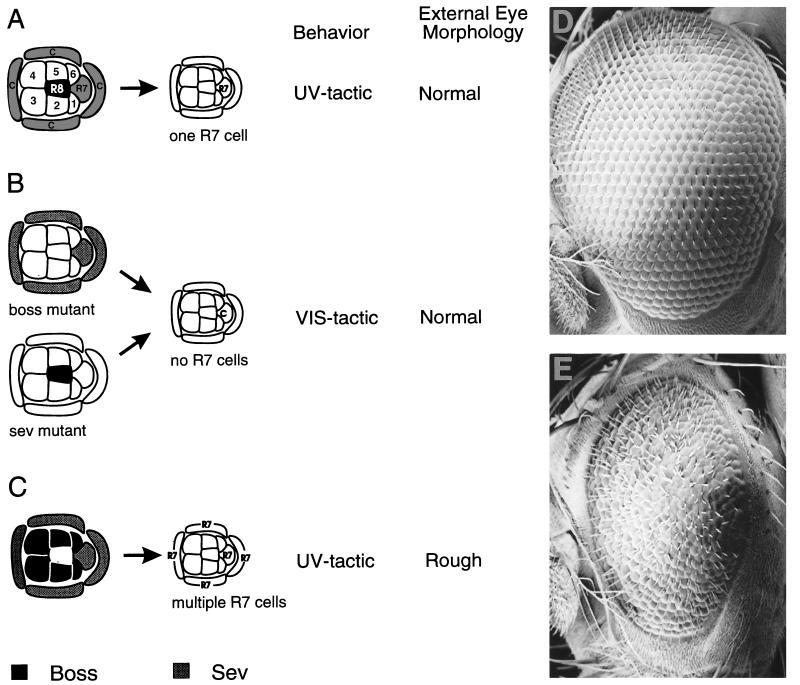
The development of the R7 photoreceptor neuron has been studied in considerable detail (reviewed in ref. 2). The R7 precursor cell is induced to become R7 by a neighboring cell, R8. The inductive signal is a membrane glycoprotein encoded by the boss gene and expressed only in R8. The receptor, Sev, is a receptor tyrosine kinase and it is expressed in many cells in the developing ommatidium (i.e., R1, R3, R4, R6, R7, and the cone cell precursors). Among the Sev-expressing cells, only R7 and the cone cell precursors are competent to respond to the inductive signal. During normal development, however, activation of the Sev pathway is restricted to the R7 precursor cell (Fig. 2A). The cone cell precursors do not assume an R7 fate because they are physically isolated from the surface of the Boss-ligand-expressing R8 cell.
Figure 2.
Induction of R7. (A) Wild-type ommatidium. Expression of Boss in the R8 cell induces the R7 cell precursor to form an R7 neuron. (B) In a boss or sev mutant the R7 cell precursor becomes a nonneuronal cone cell. (C) Ectopic expression of Boss induces cone cell precursors to form R7 neurons. These three outcomes can be distinguished in live flies based on phototactic behavior and external eye morphology. (D) Scanning electron micrograph of a wild-type eye; an eye lacking R7 cells (sev or boss) is indistinguishable from wild type at this level of analysis. (E) Scanning electron micrograph of a “rough” eye due to ectopic expression of Boss from a line isolated in the screen. C, cone cells; 1–6, R1–R6 neurons.
Because R7 is the primary receptor for UV light, flies containing R7 can be easily separated from flies lacking R7 (boss or sev mutant flies; Fig. 2B) by a simple behavioral test, the phototactic choice test. When given a choice between UV and visible light, 90% of boss mutant flies move toward the visible light (VIS-tactic), whereas 90% of the wild-type flies move toward the UV light (UV-tactic). UV-tactic behavior can be conferred upon boss mutant flies by expressing boss in the eye disc during a period in which Sev-expressing precursor cells are competent to respond. Using boss in place of lacZ as an enhancer-trap marker enabled us to screen behaviorally for genes expressed in the eye imaginal disc within the first ≈30 h of ommatidial development (2, 15).
In addition, ectopic expression of Boss on cells in contact with cone cells drive them into an R7 fate (Fig. 2C) causing an abnormal or “rough” morphology of the adult eye (compare Fig. 2 D to E). Selective expression in R8, however, restores the normal R7 cell, conferring UV-tactic behavior without disrupting eye morphology (Fig. 2 A and D). As a result, one can readily enrich for flies in which Boss is expressed solely in the R8 cell by assessing the external eye morphology under the dissecting microscope.
Identification of UV-Tactic Flies Resulting from Ectopic Expression of boss.
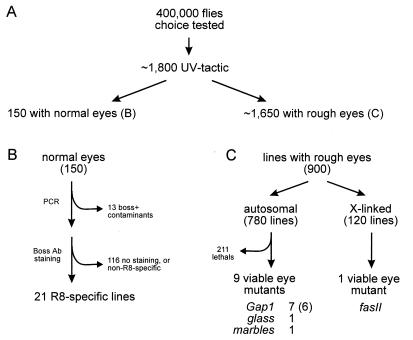
P elements carrying the boss cDNA fused to a basal promoter, P[boss; ry+], were mobilized from silent positions on a second chromosome to other sites in the genome of boss mutant flies in a hybrid dysgenic cross (Fig. 1). A second chromosome carrying three silent copies of P[boss; ry+] (HSA1) was used to increase the frequency of transposition; this did not result in a large number of lines with multiple insertions (<4%; see Materials and Methods). UV-tactic flies were selected directly using the phototactic choice test. From ≈400,000 F1 progeny screened, some 1800 UV-tactic F1 flies were isolated (Fig. 3A).
Figure 3.
Summary diagram of screen results. (A) Enhancer-trap selection. (B) Identification of R8-specific pattern. (C) Isolation of eye mutants.
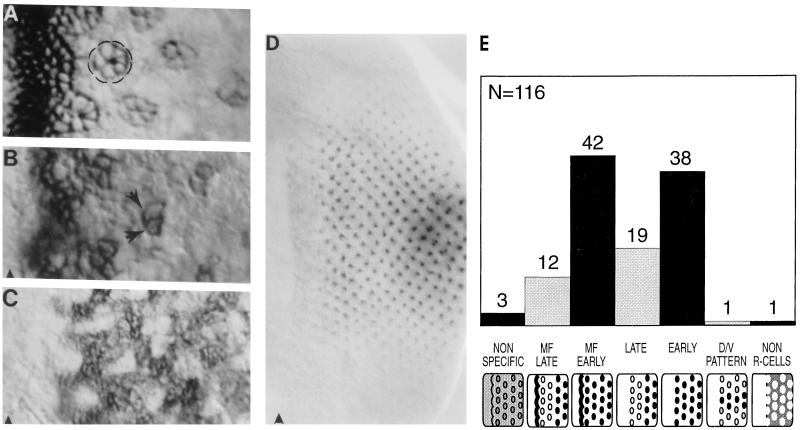
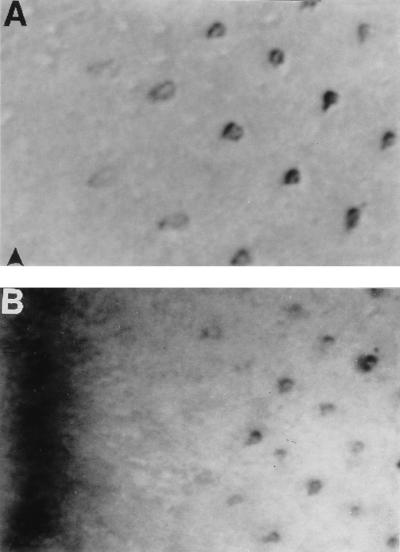
As a direct means of assessing enhancer-trap expression patterns, eye imaginal discs from a sample of 116 UV-tactic lines were stained with an anti-Boss mAb (8). Many different patterns of expression were observed (Fig. 4). None of the lines stained showed R8-specific expression characteristic of the endogenous pattern of Boss expression. Consistent with this observation, these randomly selected lines had flies with “rough” eyes reflecting the induction of multiple R7 neurons in many ommatidia (Fig. 2C).
Figure 4.
Enhancer-traps with ectopic Boss expression. In all panels the position of the MF is indicated by an arrowhead. (A and B) Expression in many cells within the MF becomes restricted to the developing neuronal clusters (circled in A) or a subset of neurons (R3 and R4 marked by arrows in B). (C) Many cells between clusters stain. (D) Several neurons per cluster express Boss, but staining is strongest in the center of the eye field and fades to undetectable levels toward the dorsal and ventral margins. (E) Distribution of enhancer-trap patterns with ectopic Boss expression. A total of 116 lines were stained with anti-Boss antibody. The number of lines displaying each pattern is indicated over the bar. The expression patterns are defined as: ubiquitous (NON SPECIFIC); in many cells within the MF and then reappearing in late clusters (MF LATE); in many cells within the MF and persisting in all or a subset of the R cells (MF EARLY) (see A and B); in late neuronal clusters many rows behind the MF (LATE); in all or a subset of the R cells from a few rows behind the MF (EARLY); differentially expressed along the dorso-ventral axis (D/V PATTERN) (see D); and posterior to the MF in many cells between clusters (NON R-CELLS; see C).
Enrichment of R8-Specific Enhancer-Traps by a Simple Morphology Screen in Live Flies.
Out of the ≈1800 UV-tactic fly lines obtained in the behavioral screen, we identified 150 with normal eye morphology (Fig. 3B). These were tested for the presence of the boss transgene, P[boss; ry+] and absence of the wild-type boss gene by PCR (see Materials and Methods). Thirteen lines were judged to be wild-type (i.e., boss+) contaminants. These contaminants are likely to have been introduced during the phototactic choice test in which the behavior of 400,000 flies was assessed. The remaining lines contained at least one P[boss; ry+] construct.
To directly assess the pattern of transgenic Boss expression, third-instar eye imaginal discs from the remaining 139 lines were stained with Boss antibody. In 9 lines, Boss expression was restricted to the R8 cell (Fig. 5A; type I pattern), while in another 14 lines, expression was seen in many cells at the MF and then specifically in R8 posterior to the MF (Fig. 5B; type II pattern). The remaining lines showed no staining or very weak staining in multiple cells, and were discarded. Presumably, in these lines, low level of Boss expression led to induction of only a small number of R7 cell. Indeed, a restoration of R7 to only some 15–20% of the ommatidia is sufficient to confer UV selectivity (S.L.Z., unpublished work). Using this approach, we isolated 21 lines (19 independent insertions) in which Boss was largely restricted to R8.
Figure 5.
Enhancer-traps with R8-specific patterns. In all panels the position of the MF is indicated by an arrowhead. (A) Expression in R8 only (type I pattern). (B) Expression in many cells within the MF resolves to R8 a few rows posterior to the MF (type II pattern).
To test whether these expression patterns reflected the expression of nearby genes, we looked for insertions mapping to known R8-specific loci. One gene with a type I pattern (boss) and two genes with type II patterns (scabrous and atonal) have been identified previously (7, 16, 17). The site of insertion for 14 of our lines were mapped by in situ hybridization and shown to localize to six different regions of the polytene chromosomes (Table 1). Among these, we identified two lines with independent insertions near the scabrous locus. In both lines, the inserts map to 49 C2–D3 and the enhancer-trap expression pattern is identical to that of previously isolated P[lacZ] insertions into scabrous (16). One of the inserts maps ≈300 bp upstream of the scabrous transcription unit (see Materials and Methods).
Table 1.
Map positions of enhancer-traps with expression in R8 only (R8) or in the MF and R8 (MF + R8)
| No. of Lines | Map position* | Expression pattern |
|---|---|---|
| 2 | 71C (III) | R8 |
| 1 | 83A (III) | R8 |
| 1 | 66A (III) | R8 |
| 7 (5)† | 70A (III) | MF + R8 |
| 2 | 49C2–D3 (II) | MF + R8 |
| 1 | 16A (X) | R8 |
Cytological position and chromosome linkage.
Number of independent insertions.
The other five cytological positions do not correspond to any known R8-specific loci and thus may identify new R8-specific genes. None of these lines displayed homozygous mutant phenotypes as assessed by lethality, adult eye morphology, pseudopupil test (18), or staining of the neuronal projection pattern with mAb 24B10 (19). The analysis of the potential role in eye development of the neighboring R8-specific genes requires the generation of mutations through imprecise excision or local hopping strategies.
Identification of Insertions Disrupting Genes Required Early in Eye Development.
To assess the mutagenic potential of the P-element insertions, 900 lines (780 autosomal and 120 X-chromosome-linked insertions) were tested for recessive loss-of-function phenotypes. The homozygous phenotypes were checked in a sev mutant background to suppress the dominant “rough” eye phenotype caused by ectopic boss expression (Fig. 2C). Approximately 70% of the autosomal lines mapped to the third chromosome, with the remaining 30% mapping to the second chromosome. Of the inserts on the third, 20% were homozygous lethal, whereas 40% of the second chromosome insertions resulted in lethality. A total of 211 lines were found to carry lethal mutations. Because the parental stock was isogenized for the third chromosome only, we suspect that background lethality, and/or lethality due to mobilization of one or more of the P[boss; ry+] inserts, may account for both the lower recovery of second chromosome inserts and the higher lethality observed on this chromosome. Of the 900 insertions tested, 569 autosomal and 120 X-chromosome-linked lines generated homozygous or hemizygous adults (Fig. 3C). Twelve autosomal and one X-chromosome-linked insertion led to mutant eye phenotypes.
To establish whether the mutant eye phenotypes were due to the P element, the cytological locations of the insertions were determined. Insertions mapped to seven cytological regions. Deficiency mapping demonstrated that for three lines, eye phenotypes were not associated with the insertions. One or more of these mutations may have already been present in the background or may have been the result of dysgenesis followed by loss of the P element. The nine remaining lines had eye phenotypes due to the P element (Fig. 3C). Seven lines mapped to 67D and were shown to be alleles of gap1 (20). Of these, six were independent insertions since they originated from different dysgenic parents. Insertions into 91A1/2, 61C1-6, and 4B1-5 were shown to be alleles of glass (gl), marbles (marb), and fasciclin II (fasII), respectively (21–23). All four genes are required early in development of the Drosophila eye (Table 2). Only P alleles of gap1 were previously identified (20), whereas large-scale P-element screens for gl and marb mutants (380,000 and 100,000 chromosomes, respectively) were unsuccessful (21, 22).
Table 2.
Summary of eye mutants isolated in the screen
| Mutant | Lines | Map position* | Disc phenotype | Ref. |
|---|---|---|---|---|
| Gap1 | 7 (6)† | 67D (III) | The cone cells are transformed into R7 neurons | 20 |
| gl | 1 | 91A1-2 (III) | Differentiation into photoreceptor-type neurons is blocked at an early stage | 21 |
| marb | 1 | 61C1-6 (III) | Nuclear migration in developing R-cells is impaired | 22 |
| fasII | 1 | 4B1-5 (X) | Expression of atonal in the MF is reduced; abnormal development and cell death are observed in eye discs mutant for this P-element allele | 23 |
Cytological position and chromosome-linkage.
Number of independent insertions.
In summary, of the 900 mutagenized chromosomes screened for recessive eye phenotypes, mutations in four genes were identified. All four genes function at early stages of eye development, consistent with the use of boss as the phenotypic marker for the enhancer-trap selection scheme.
CONCLUSIONS
We have designed a modified enhancer-trap method that provides a selection for insertions expressed in a limited spectrum of spatiotemporal patterns during development. The method relies on the use of a phenotypic marker as the enhancer reporter. The reporter determines the time and tissue specificity of the gene pool sampled. As shown for the R8-specific enhancer-traps, the identification of genes with very restricted expression patterns is possible with the appropriate rescue marker. This method can be adapted to a wide variety of other tissues and developmental times by selecting appropriate rescue markers. Indeed, while this manuscript was in preparation, a similar approach to enhancer detection in live flies was described by Calleja et al. (24). Using the cuticle pigmentation marker, yellow, body-specific enhancers driving expression in third instar larvae or pupae were identified.
The use of a phenotypic marker, rather than a reporter requiring a histological stain, also increases by many fold the number of enhancer-trap insertions with developmentally restricted expression patterns that can be assessed for homozygous mutant phenotypes. As a result, the mutation frequency obtained represents an improvement over “single-P-element insertional mutagenesis” (25, 26) or a previously described chemical-mutagen-based screen for mutations affecting early stages of compound eye development (27). In the P-element screen by Cooley et al. (25), out of 1,317 lines established, five viable mutants with adult phenotypes were isolated and 139 lines were homozygous lethal. Of the viable mutants, none had eye phenotypes. In contrast, using the phenotypic enhancer-trap, we were able to obtain a comparable mutation frequency specifically for genes required in the early stages of eye development. These results also represent an improvement of several fold in the frequency of mutations over the ethylmethane sulfonate mutagenesis for eye development mutants described by Baker et al. (27). In this screen, from 17,846 mutagenized chromosomes, of which 4,526 were homozygous viable, 33 visible eye mutants mapping to 23 complementation groups were isolated. Only two mutations, however, affected genes required in the early stages of eye development. In conclusions, the approach to insertional mutagenesis we have described achieves mutation frequencies comparable to chemical mutagenesis coupled with the ease of cloning and molecular characterization provided by the P element.
Acknowledgments
We thank J. Merriam, T. Chouard, P. Garrity, and K. Zavitz for critically reading the manuscript; the Bloomington Stock Center, B. Calvi and M. Sanicola, K. Moses, J. Fischer-Vize, C. Goodman, and U. Banerjee for fly stocks; and P. C. Guillinta for assistance on the project. This work was supported by a grant from the National Institutes of Health (S.L.Z.). F.P. is an associate of the Howard Hughes Medical Institute. S.L.Z. is an investigator of the Howard Hughes Medical Institute.
ABBREVIATION
- MF
morphogenetic furrow
References
- 1.Wolff T, Ready D F. In: The Development of Drosophila melanogaster. Bates M, Martinez Arias A, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1993. pp. 1277–1325. [Google Scholar]
- 2.Dickson B, Hafen E. In: The Development of Drosophila melanogaster. Bates M, Martinez Arias A, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1993. pp. 1327–1362. [Google Scholar]
- 3.Ashburner M. Drosophila, A Laboratory Handbook. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. pp. 299–403. [Google Scholar]
- 4.Engels W R. In: Transposable Elements. Saedler H, Gierl A, editors. Berlin: Springer; 1996. pp. 103–123. [Google Scholar]
- 5.O’Kane C J, Gehring W J. Proc Natl Acad Sci USA. 1987;84:9123–9127. doi: 10.1073/pnas.84.24.9123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reinke R, Zipursky S L. Cell. 1988;55:321–330. doi: 10.1016/0092-8674(88)90055-4. [DOI] [PubMed] [Google Scholar]
- 7.Hart A C, Krämer H, Van Vactor D L, Jr, Paidhungat M, Zipursky S L. Genes Dev. 1990;4:1835–1847. doi: 10.1101/gad.4.11.1835. [DOI] [PubMed] [Google Scholar]
- 8.Van Vactor D L, Jr, Cagan R L, Krämer H, Zipursky S L. Cell. 1991;67:1145–1155. doi: 10.1016/0092-8674(91)90291-6. [DOI] [PubMed] [Google Scholar]
- 9.Lindsley D L, Zimm G G. The Genome of Drosophila melanogaster. San Diego: Academic; 1992. [Google Scholar]
- 10.Sturtevant A H. Genetics. 1956;41:118–123. doi: 10.1093/genetics/41.1.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heisenberg M, Buchner E. J Comp Physiol. 1977;117:127–162. [Google Scholar]
- 12.Ashburner M. Drosophila: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 13.Maniatis T, Fritsch E F, Sambrook J. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1982. [Google Scholar]
- 14.Saiki R. In: PCR Protocols: A Guide to Methods and Applications. Innis M A, Gelfand D H, Sninsky J J, White T J, editors. San Diego: Academic; 1990. pp. 13–20. [Google Scholar]
- 15.Cagan R L, Thomas B J, Zipursky S L. In: Molecular Basis of Morphogenesis. Bernfield M, editor. New York: Wiley–Liss; 1993. pp. 109–133. [Google Scholar]
- 16.Mlodzik M, Baker N E, Rubin G M. Genes Dev. 1990;4:1848–1861. doi: 10.1101/gad.4.11.1848. [DOI] [PubMed] [Google Scholar]
- 17.Jarman A P, Grell E H, Ackerman L, Jan L Y, Yan Y N. Nature (London) 1994;369:398–400. doi: 10.1038/369398a0. [DOI] [PubMed] [Google Scholar]
- 18.Franceschini N, Kirschfeld K. Kybernetik. 1971;9:159–182. doi: 10.1007/BF02215177. [DOI] [PubMed] [Google Scholar]
- 19.Zipursky S L, Venkatesh T R, Teplow D B, Benzer S. Cell. 1984;36:15–26. doi: 10.1016/0092-8674(84)90069-2. [DOI] [PubMed] [Google Scholar]
- 20.Gaul U, Mardon G, Rubin G M. Cell. 1992;68:1007–1019. doi: 10.1016/0092-8674(92)90073-l. [DOI] [PubMed] [Google Scholar]
- 21.Moses K, Ellis M C, Rubin G M. Nature (London) 1989;340:531–536. doi: 10.1038/340531a0. [DOI] [PubMed] [Google Scholar]
- 22.Fischer-Vize J A, Mosley K L. Development (Cambridge, UK) 1994;120:2609–2618. doi: 10.1242/dev.120.9.2609. [DOI] [PubMed] [Google Scholar]
- 23.Garcia-Alonso L, VanBerkum M F A, Grenningloh G, Schuster C, Goodman C S. Proc Natl Acad Sci USA. 1995;92:10501–10505. doi: 10.1073/pnas.92.23.10501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Calleja M, Moreno E, Pelaz S, Morata G. Science. 1996;274:252–255. doi: 10.1126/science.274.5285.252. [DOI] [PubMed] [Google Scholar]
- 25.Cooley L, Kelley R, Spradling A. Science. 1988;239:1121–1128. doi: 10.1126/science.2830671. [DOI] [PubMed] [Google Scholar]
- 26.Török T, Tick G, Alvarado M, Kiss I. Genetics. 1993;135:71–80. doi: 10.1093/genetics/135.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baker N E, Moses K, Nakahara D, Ellis M C, Carthew R W, Rubin G M. J Neurogenet. 1992;8:85–100. doi: 10.3109/01677069209084154. [DOI] [PubMed] [Google Scholar]