Abstract
Genotoxic agents that cause double-strand breaks (DSBs) often generate damage at the break termini. Processing enzymes, including nucleases and polymerases, must remove damaged bases and/or add new bases before completion of repair. Artemis is a nuclease involved in mammalian nonhomologous end joining (NHEJ), but in Saccharomyces cerevisiae the nucleases and polymerases involved in NHEJ pathways are poorly understood. Only Pol4 has been shown to fill the gap that may form by imprecise pairing of overhanging 3′ DNA ends. We previously developed a chromosomal DSB assay in yeast to study factors involved in NHEJ. Here, we use this system to examine DNA polymerases required for NHEJ in yeast. We demonstrate that Pol2 is another major DNA polymerase involved in imprecise end joining. Pol1 modulates both imprecise end joining and more complex chromosomal rearrangements, and Pol3 is primarily involved in NHEJ-mediated chromosomal rearrangements. While Pol4 is the major polymerase to fill the gap that may form by imprecise pairing of overhanging 3′ DNA ends, Pol2 is important for the recession of 3′ flaps that can form during imprecise pairing. Indeed, a mutation in the 3′-5′ exonuclease domain of Pol2 dramatically reduces the frequency of end joins formed with initial 3′ flaps. Thus, Pol2 performs a key 3′ end-processing step in NHEJ.
Author Summary
Chromosomal DSBs caused by replication fork disruption, environmental factors, or endogenous nucleases are common yet potentially dangerous DNA lesions in all organisms. If they are repaired by homologous recombination (HR), the integrity of the genome is usually maintained. However, if the broken ends undergo NHEJ, sequences at the junction may be added, deleted, or substituted, and large segments of chromosomes can be rearranged. Partially overlapping sets of proteins are required for repair by either the HR or NHEJ pathway. Furthermore, different proteins may be used to process broken DNA ends, depending on the particular terminal structures. Since DNA synthesis occurs during HR in yeast and all three essential replicative polymerases are utilized, we asked how different polymerases might be involved in DSB repair by NHEJ. We find that Pol2, and particularly the enzyme's 3′ to 5′ nuclease activity, contributes to the removal of 3′ single strand flaps that can form during the initial joining of broken ends. We find that Pol1 and Pol3 modulate complex chromosomal rearrangements, and we confirm that Pol4 fills the gap that can form by imprecise pairing of overhanging 3′ DNA ends. Our work demonstrates that multiple DNA polymerases play important roles in NHEJ.
Introduction
DNA DSBs result from disruption of the phosphodiester backbone on both strands of a DNA double helix. They are induced by ionizing radiation and chemicals, including anticancer drugs, or can arise spontaneously during DNA replication [1]–[3]. Furthermore, DSBs occur normally as intermediates in V(D)J recombination, the process that helps to generate the vast range of antigen-binding sites of antibody and T-cell receptor proteins during lymphoid-cell development [4]. DNA DSBs are critical lesions that, if unrepaired or misrepaired, may be lethal for a cell or help in its malignant transformation.
DSBs can be repaired either by homologous recombination (HR) or by NHEJ [1], [3], [5]–[8]. While the former process is generally error-free, the latter process is potentially error-prone. DSB repair by HR requires extensive regions of sequence homology between donor and recipient DNA strands. In NHEJ the DNA ends are joined with little or no base pairing at the junction [9]. These repair mechanisms are evolutionarily conserved, but contribute unequally to overall DSB repair in different organisms. In mammals, DSBs are primarily repaired by NHEJ, while in yeast HR dominates. The budding yeast S. cerevisiae is the most intensely studied model system for DSB DNA repair. This organism has a classical NHEJ pathway that depends on Ku and DNA ligase IV, as well as Rad50, Mre11 and Xrs2, three proteins that have endo- and exonuclease activities [10],[11].
Agents that cause DSBs often create damaged or non-complementary bases at the break termini [12],[13]. In these circumstances, simple religation cannot occur, and additional factors must be used to process the DNA breaks to create suitable 5′ and 3′ ends for ligation. A nuclease, termed Artemis, has been shown to be important for mammalian NHEJ [14]. Purified Artemis protein possesses single-strand-specific 5′ to 3′ exonuclease activity. In conjunction with the DNA-dependent protein kinase (DNA-PK), Artemis has both 5′ to 3′ and 3′ to 5′ exonuclease activities [14]. S. cerevisiae lacks both Artemis and the catalytic component of DNA-PK, and the nuclease(s) and polymerase(s) involved in the yeast NHEJ pathway are not well understood. The Pol X family of DNA polymerases has been implicated in NHEJ, since Pol4, the only Pol X family member in yeast, is required for gap filling in some end configurations [15]. Pol X polymerases appear to be required for NHEJ only when gaps must be filled, indicating that they are not part of the core NHEJ complex [15],[16].
At least six nuclear DNA polymerases have been described in eukaryotic cells that participate in DNA replication and/or repair [17]. Pol1 (designated Pol α or CDC17 in yeast), Pol2 (Pol ɛ), and Pol3 (Pol δ or CDC2) together catalyze the essential functions of DNA replication. Pol2 and Pol3 are also involved in certain DNA repair events, notably nucleotide excision repair [18]. Rev3 (Pol ξ) and Rad30 (Pol η) mediate translesion bypass synthesis in yeast [19]. Pol β is a monomeric polymerase in vertebrates that mediates base excision repair [20],[21]. Moreover, Pol1, Pol2 and Pol3 are required for HR 22–24. Studies in mammals and yeast have provided evidence that Pol α [25], Pol ɛ [26], Pol4 [15] and Pol μ [27] play roles in NHEJ. However, much of these data were obtained through in vitro experiments, measuring religation of linear plasmids by cell extracts. Thus, it is not clear which DNA polymerases participate in NHEJ in vivo and under what circumstances. For example, DSBs on plasmids with 5′-overhangs do not depend on Pol4 for their repair [16]. Conversely, chromosomal breaks due to HO endonuclease, which generates breaks with 3′ overhangs, do rely on Pol4 for their repair [16].
In this paper we examine the roles of five DNA polymerases (Pol1, Pol2, Pol3, Pol4 and Rev3) in both imprecise end joining and in NHEJ-mediated chromosomal rearrangements in S. cerevisiae after a defined HO-induced DSB. These are all members of DNA polymerase families that are conserved from yeast to mammal [17]. Consistent with a previous report [15], we also observed that Pol4 is required for filling in gaps imprecise end joining. On the contrary, Pol2 is involved in deleting bases during imprecise end joining. Both Pol1 and Pol3 are required for chromosomal rearrangements, but Pol3 is not involved imprecise end joining. Our results suggest that most DNA polymerases are involved in NHEJ and each plays a distinct role in the detailed mechanisms of NHEJ.
Results
The In Vivo NHEJ Assay System
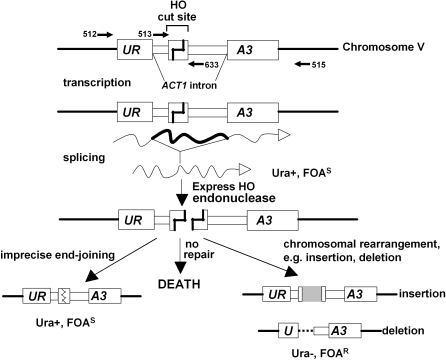
For our in vivo NHEJ assay, a copy of the ACT1 intron was placed within the URA3 gene on chromosome V and an HO endonuclease cut site was engineered in the middle of the intron (Figure 1) [28]. A galactose inducible copy of the HO-endonuclease gene is present at the ADE3 locus on chromosome VII. Other endogenous HO cut sites, as well as the endogenous ACT1 intron, were deleted. Therefore, upon growth on galactose, the HO endonuclease creates a unique DSB in the middle of URA3 that must be repaired for cells to survive (Figure 1). Because there are no other MAT related sequences in this haploid strain, the DSB cannot be repaired by the HR machinery, and most cells die. Survival is dependent on inefficient NHEJ repair pathways. Precise religation recreates the cut site, which can then be recut by the induced HO endonuclease. The most commonly observed stable repair event is imprecise NHEJ [29]. The imprecision of the end joining eliminates the endonuclease recognition sequence, but these sequence changes do not disrupt splicing because the repair occurs within a nonessential region of the intron. Repair events which interfere with the expression of URA3, however, can be identified by selecting for survivors that have become uracil auxotrophs (i.e., resistant to the drug 5-fluoro orotic acid or 5-FOA). Such repair events can include large insertions, deletions extending past the intron sequence, or chromosomal rearrangements such as translocations or inversions that separate the two halves of URA3 [28],[30],[31]. PCR amplification of the URA3::ACT1 intron::HO cut site allele, Southern blot analysis and/or sequencing of FOAR survivors allow us to distinguish such rearrangements (Figure S1).
Figure 1. The experimental system.
Structure of the URA3 allele on S. cerevisiae chromosome V used for these experiments. The position of various oligonucleotide primers (numbers and half arrows) used for PCR and sequencing are shown. The ACT1 intron placed into URA3 is normally spliced, resulting in uracil prototrophy (Ura+) and sensitivity to the drug FOAS. After creating a DSB within the engineered ACT1 intron with HO endonuclease, cells either die, are repaired in a way allowing normal splicing, or are repaired in a way that prevents splicing. The latter situation leads to a phenotype of uracil auxotrophy (Ura−) and resistance to 5-FOA (FOAR).
Addition of Bases during Imprecise End Joining Depends On Pol4
To determine which DNA polymerases participate in NHEJ in S. cerevisiae, we focused on five different types of DNA polymerases, including two non-essential DNA polymerases (Pol4 and Rev3) and three essential DNA polymerases (Pol1, Pol2 and Pol3). For the nonessential genes, wild-type and deletion mutants were initially grown in rich, galactose-containing, medium (YPGal) to induce HO endonuclease expression. The frequency of imprecise end joining was calculated as the ratio of colonies growing on YPGal compared to colonies growing on rich, glucose-containing medium (YPD) as described in Materials and Methods. The frequency of chromosomal rearrangements (and/or potential URA3 point-mutations) was estimated as the ratio of colonies growing on FOA-containing media compared to colonies growing on YPD, because insertion or deletion of large fragments results in disruption of splicing and consequent FOA resistance. Additionally, chromosomal translocations and inversions can also disrupt the integrity of the URA3 gene and cause FOA resistance [31]. It should be noted that the level of analysis carried out in this work (Figure S1) does not definitively distinguish translocations and inversions from insertions.
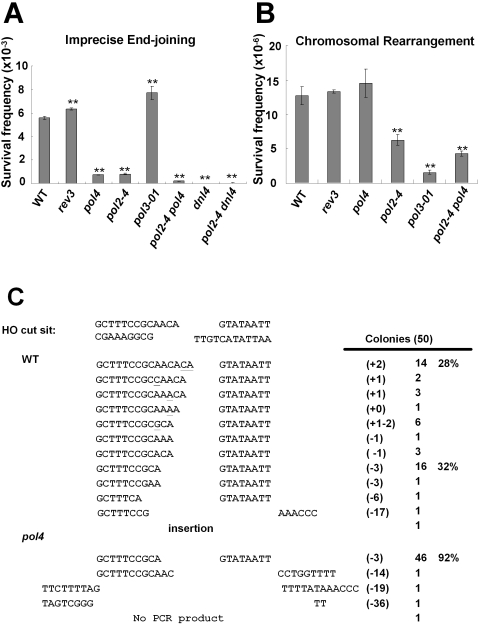
We first compared cell survival after a DSB in strains with or without disruption of nonessential polymerases. The wild-type strain, AGY673, had a frequency of imprecise end joining and chromosomal rearrangement events of 5.58×10−3 and 1.27×10−5 per plated cell, respectively (Figure 2A, 2B, and Table S1). Elimination of Rev3 (rev3) did not alter the frequency of imprecise end joining (6.33×10−3) or chromosomal rearrangement events (1.33×10−5), suggesting that Rev3 is not required for the NHEJ pathway. In contrast, elimination of Pol4 resulted in an 8.6-fold decrease in the frequency of imprecise end joining events, although the frequency of chromosomal rearrangement events was not significantly affected (Figure 2). Therefore, in agreement with previous findings, Pol4 appears to participate in imprecise end joining repair of NHEJ [15].
Figure 2. Measurements of NHEJ ability of non-essential DNA polymerases.
The efficiency of NHEJ of wild-type and polymerase defective mutants, rev3, pol4 and 3′ to 5′ exonuclease defective mutants, pol2-4, pol3-01, pol2-4 pol4, dnl4, and pol2-4 dnl4 were measured. (A) Frequencies of imprecise end joining. The survival frequency was evaluated by the ratio of YPGal/YPD. (B) Frequency of chromosomal rearrangement was evaluated by the ratio of FOAR/YPD. Each experiment was collected from at least four independent clones. *: P<0.05; **: P<0.01. (C) The sequence analysis of the imprecise end joining events from the WT and pol4 mutants. The structures of various imprecise end joining repairs were identified at the DSB site. Sequences of the HO cut site are shown, with the resulting 3′ overhanging terminal AACA shown on the cut site. Samples of independent joints were PCR amplified and sequenced. A total of 50 independent survivors on YPGal plates from WT and pol4 mutant were examined. Bases underlined indicate insertion or mutation. The numbers of inserted (+) and deleted (−) bases were denoted in brackets. N indicates the events of each repair pattern.
A previous study showed that the predominant imprecise NHEJ repair products after an HO-induced DSB at the MAT a locus were either addition of two bases (+2 or +CA) or deletion of three bases (−3 or −ACA) [29]. These products are most plausibly caused by a 3′-terminal mismatch (HO (+2)) and by a 3 base flap mismatch (HO (−3)), respectively (see below for detailed description). Thus, we further analyzed the repair in wild-type and pol4 mutant strains. For wild-type survivors, ∼50% had added bases at the junction (including 28% with the +2 repair pattern) and 46% had deleted bases (including 32% with the −3 pattern) (Figure 2C and Table 1). For pol4 cells, however, 98% of survivors had deletion of bases at the junction (including 92% with the −3 pattern), while added bases were not observed in 40 independent colonies examined. This pattern confirms that Pol4 is required for addition of bases during imprecise end joining of NHEJ in vivo [15]. Interestingly, the absolute frequency of repair involving deletion of bases was also decreased more than 3 fold, from 2.57×10−3 in wild-type cells to 7.23×10−4 in pol4 cells (Table S1). These results suggest that Pol4 plays a role in imprecise end joining events in vivo where 3′flaps are generated.
Table 1. Repair patterns after a DSB at the URA3::ACT1 intron::HO cut site locus of non-essential DNA polymerase mutants.
| Strain | Imprecise end joininga(%) | Chromosomal rearrangementsb(%) | ||||||||
| +CA | −ACA | +base | −base | Others | Nc | insertions | deletions | mutation | Nc | |
| WT | 28 | 32 | 50 | 46 | 4 | 50 | 28 | 73 | 0 | 40 |
| rev3 | 35 | 35 | 55 | 45 | 0 | 20 | 40 | 60 | 0 | 40 |
| pol4 | 0 | 92 | 0 | 98 | 2 | 50 | 40 | 60 | 0 | 40 |
| pol2-4 | 69 | 2.2 | 76 | 22 | 2.2 | 45 | 45 | 55 | 0 | 40 |
| pol3-01 | 25 | 40 | 40 | 60 | 0 | 20 | 7.5 | 5 | 88 | 40 |
| pol2-4 pol4 | 0 | 78 | 0 | 100 | 0 | 50 | 70 | 15 | 15 | 40 |
| WT (α)d | 66 | 6.3 | 66 | 34 | 0 | 32 | ndd | nd | nd | nd |
| pol2-4 (α) | 76 | 0 | 87 | 14 | 0 | 37 | nd | nd | nd | nd |
| WT (NZ)d | 40 | 7.9 | 50 | 47 | 2.6 | 38 | nd | nd | nd | nd |
| pol2-4 (NZ) | 74 | 2.9 | 85 | 5.9 | 8.8 | 34 | nd | nd | nd | nd |
Analysis of survivors grown on YPGal plates.
Analysis of survivors grown on 5-FOA plates.
Total events examined.
nd, not determined; α, α factor arrested; NZ, nocodazole arrested.
Both Imprecise End Joining and Chromosomal Rearrangement Are Influenced by Essential Polymerases
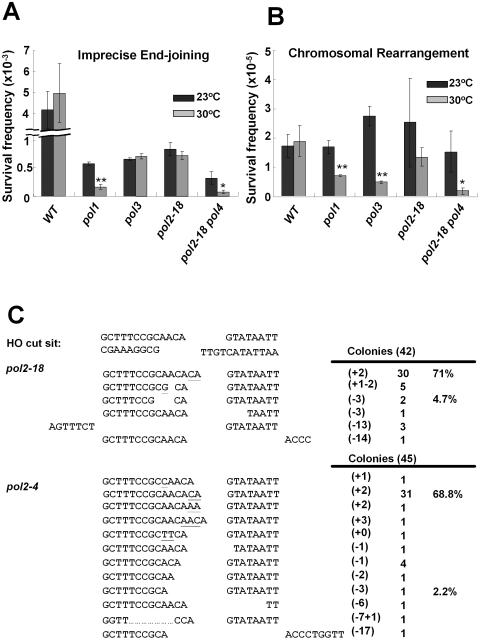
Three essential DNA polymerases (Pol1, Pol2, and Pol3) are required for DNA replication and HR. To investigate whether they also contribute to NHEJ, temperature-sensitive (ts) mutants with defects in polymerase activity were created in the AGY673 background. As shown in Figure 3A, the presence of any of the ts mutations resulted in a >10-fold drop in survival after a DSB, even at the permissive temperature. Despite this, the frequency of survival and pattern of end joining could be compared for each mutant at the permissive and semi-permissive temperatures, and the patterns of end joining could be compared to the WT strain at the equivalent temperature. As a control we analyzed survival of the ts mutants in the absence of a cut site at URA3. We found that strain with no cut sites (STY1553, STY1552 and STY1554) survived equally well at both permissive and semi-permissive temperatures in the absence or presence of HO-endonuclease (Table S2). These results indicate that the change in survival that we observed in the ts mutants after a DSB, was associated with the DSB and its repair, rather than a general effect of the ts mutations, the presence of HO endonuclease, or the growth in galactose versus glucose. Pol1 participates in DNA replication initiation and lagging strand DNA synthesis, as well as HR in yeast [22]–[24],[32]. For Pol1, we found that the frequencies of both imprecise end joining (YPGal/YPD) and chromosomal rearrangements (FOA/YPD) were reduced more than 50% compared with those cultured at permissive temperature (23°C), a decrease that was significant to a P<0.01 (Figure 3A, 3B, and Table S2). However, there was no significant difference in the pattern of imprecise end joining events or in the percentages of insertions or deletions (Table 2). These results suggest that the effect of Pol1 on NHEJ is indirect.
Figure 3. Measurements of NHEJ ability of essential DNA polymerases.
The NHEJ assay was performed for pol1, pol3, pol2-18 and pol2-18 pol4 at permissive (23°C) and semi-permissive (30°C) temperatures. (A) Frequencies of imprecise end joining. The survival frequency was evaluated by the ratio of YPGal/YPD. (B) Frequency of chromosomal rearrangement was evaluated by the ratio of FOAR/YPD. Each experiment was collected from at least five independent clones. Dark gray bars indicate 23°C incubation and light gray bars indicate 30°C incubation. *: P<0.05; **: P<0.01. (C) Survivors of pol2-18 or pol2-4 mutants from YPGal plates at 30°C were examined. Bases underlined indicate insertion or mutation. The numbers of inserted (+) and deleted (−) bases were denoted in brackets. N indicates the events of each repair pattern.
Table 2. Repair patterns after a DSB at the URA3::ACT1 intron::HO cut site locus of essential DNA polymerase mutants.
| Strain | Tm(°C) | Imprecise end joininga(%) | Chromosomal rearrangementsb(%) | ||||||||
| +CA | −ACA | +base | −base | Others | Nc | insertions | deletions | mutation | Nc | ||
| WT | 23 | 15 | 25 | 35 | 65 | 0 | 20 | 75 | 25 | 0 | 20 |
| 30 | 40 | 25 | 40 | 50 | 10 | 20 | 50 | 50 | 0 | 20 | |
| pol1 | 23 | 35 | 35 | 50 | 50 | 0 | 20 | 55 | 45 | 0 | 40 |
| 30 | 40 | 35 | 60 | 40 | 0 | 20 | 60 | 40 | 0 | 40 | |
| pol3 | 23 | 25 | 40 | 45 | 55 | 0 | 20 | 80 | 20 | 0 | 40 |
| 30 | 30 | 35 | 40 | 60 | 0 | 20 | 80 | 20 | 0 | 40 | |
| pol2-18 | 23 | 42 | 28 | 64 | 36 | 0 | 50 | 75 | 7 | 18 | 40 |
| 30 | 71 | 4.7 | 83 | 17 | 0 | 42 | 88 | 7 | 5 | 40 | |
| pol2-18 | 23 | 0 | 80 | 0 | 100 | 0 | 50 | 60 | 7 | 33 | 40 |
| pol4 | 30 | 0 | 74 | 0 | 100 | 0 | 50 | 53 | 17 | 30 | 40 |
Analysis of survivors grown on YPGal plates.
Analysis of survivors grown on 5-FOA plates.
Total events examined.
Pol3 is essential for both leading strand and lagging strand DNA synthesis [32]. Interestingly, we found that while the frequency of imprecise end joining did not change, the frequency of chromosomal rearrangements was significantly reduced in the pol3 mutant at the semi-permissive temperature (Figure 3A, 3B, and Table S2). The repair pattern of the pol3 ts mutant showed no obvious difference compared to WT for either end joining or chromosomal rearrangements (Table 3), implying that the major consequence of the pol3 ts mutation at the semi-permissive temperature was a defect in the ability to generate complex chromosomal rearrangements.
Table 3. Yeast strains used in this study.
| Strain | Genotype | Source |
| AGY628 | ho,hml::ADE1,mata::hisG, hmr::ADE, ade1, lys5, trp1::hisG, ade3::GAL-HO, ura3-52, LEU2, intronless ACT1 | [44] |
| AGY673 | AGY628 URA3::ai::HO cut site | [44] |
| YHA322 | MATα pol2-3::LEU2 leu2-3,112 ura3-52 trp1-289 his4 [YCppol2-18] | [46] |
| STY891 | AGY673 pol4::TRP1 | This study |
| STY890 | AGY673 rev3::TRP1 | This study |
| STY969 | AGY673 pol1 (cdc17-1) | This study |
| STY970 | AGY673 pol3 (cdc2-2) | This study |
| STY966a | AGY673 pol2::LEU2 [YCppol2-18] | This study |
| STY1358 | AGY673 pol2-4 | This study |
| STY1359 | AGY673 pol3-01 | This study |
| STY1356b | AGY673 pol2::LEU2 [YCppol2-18] pol4::TRP1 | This study |
| STY1360c | AGY673 pol2-4 pol4::TRP1 | This study |
| STY1544 | AGY673 dnl4::KanMX4 | This study |
| STY1546 | AGY673 pol2-4 dnl4::KanMX4 | This study |
| STY1549 | AGY673 pol3 (cdc2-2) ura3Δ | This study |
| STY1550 | AGY673 pol1 (cdc17-1) ura3Δ | This study |
| STY1551 | AGY673 pol2::LEU2 [YCppol2-18] ura3Δ | This study |
| STY1552 | AGY673 pol3 (cdc2-2) URA3::ai | This study |
| STY1553 | AGY673 pol1 (cdc17-1) URA3::ai | This study |
| STY1554 | AGY673 pol2::LEU2 [YCppol2-18] URA3::ai | This study |
From three backcrosses of YHA322 with AGY673.
pol4 of STY966.
pol4 of STY1358.
Pol2 Contributes to Imprecise End Joining through Its 3′ to 5′ Exonuclease Activity
Pol2 interacts with PCNA in S phase and is required for the S-phase checkpoint, for assembly of replication complexes at origins, and for leading strand replication [33],[34]. A pol2 polymerase mutant (pol2-18) at the semi-permissive temperature showed no obvious difference in the frequency of imprecise end joining or chromosomal rearrangements compared with the same strain at 23°C (Figure 3A and 3B). Sequence analysis of the imprecise end joining repair pattern, however, revealed that the proportion of −3 events was reduced more than 5 fold at 30°C compared to 23°C, while the proportion of +2 events was nearly doubled (Table 3). These data suggest that Pol2 is particularly involved in generating the −3 product during imprecise end joining repair.
A likely step in the creation of the −3 end joining product is deletion of the 3 base 3′ flaps from both broken ends. Pol2 might directly participle in this step through its associated exonuclease activity. To examine this possibility, we constructed pol2-4 and pol3-01 mutant strains which are defective in the 3′ to 5′ exonuclease activity but do not affect the DNA polymerase activity of Pol2 and Pol3, respectively [35],[36]. These are both non-lethal mutant strains and both have been shown to display mutator phenotypes [35],[37]. Interestingly, pol2-4 but not pol3-01 affects the frequency of imprecise end joining (Figure 2A), and a reduction of the −3 pathway was observed in the pol2-4 strain, but not in the pol3-01 strain (Figure 3C and Table 1). The absolute frequency of base loss during repair was decreased more than 10 fold, from 2.57×10−3 in wild-type cells to 1.83×10−4 in pol2-4 cells (Table S1). In particular the absolute frequency of the −3 product was decreased over 98-fold, from 1.79×10−3 in wild-type cells to 1.82×10−5 in pol2-4 cells (Table S1). These results support our hypothesis that the 3′ to 5′ exonuclease activity of Pol2 is involved in the processing step that degrades the 3′ flaps to create the −3 end joining product. Further, we confirmed that the events we observed in the pol2-4 strain were in fact due to NHEJ. We compared survival in isogenic dnl4 and pol2-4 dnl4 yeast cells relative to the wild type and pol2-4 strains (Figure 2 and Table S1), and found an ∼100 fold drop in the absence of dnl4, a key component of the NHEJ pathway. Of note, we also observed that 87.5% of FOA resistant survivors in pol3-01 cells showed the parental size product at the DSB site (Table 1). Sequencing results of the HO cut site junctions in twenty independent survivors revealed that the junctions of each survivor contain different imprecise end joining sequences, suggesting that they are not siblings from a single event (data not shown). Since these changes should not affect splicing of the ACT1 intron, the observed FOA resistance likely resulted from mutations in the URA3 coding region. These data support the previously described mutator phenotype of pol3-01 [35],[36].
Since both Pol2 and Pol4 affect the frequency and pattern of imprecise end joining after an HO-induced DSB, we next asked whether their effects were additive. To this end we tested both pol2-18 pol4 and pol2-4 pol4 double mutant strains with our assay (Figures 2 and 3, and Tables 2 and 3). Strikingly, both pol2-18 pol4 and pol2-4 pol4 double mutant strains showed further reduction of imprecise end joining events compared to those of the single mutations (Figures 2A and 3A). Examining the pattern of repair, we found that the −3 product predominated in the double mutants (Tables 2, 3, S1 and S2). These results suggest that the +2 product pathway is completely Pol4-dependent. The absolute frequency of repair with deletions decreased from 7.23×10−4 in pol4 cells to 2.58×10−4 in pol2-4 pol4 cells (Table S1). Therefore, in contrast to Pol4, Pol2 is not the exclusive nuclease for processing the −3 product pathway.
Our data also provide evidence for interplay between Pol4 and Pol2 in base deletion and addition during imprecise end joining repair. Although Pol4 has its major effect on base addition, the absolute frequency of base deletion products decreased 3.6 fold, from 2.57×10−3 in wild-type cells to 7.23×10−4 in pol4 cells. Conversely, whereas Pol2 is associated with base deletions, the absolute frequency of base addition products decreased >44 fold, from 2.79×10−3 in wild-type cells to 6.24×10−5 in pol2-4 cells (Table S1).
Lack of Cell Cycle Dependence of Pol2-Mediated 3′-End Processing of NHEJ
Moore and Haber [29] have shown that specific NHEJ repair products have a cell cycle dependence, with the proportion of −3 products increasing and the proportion of +2 products decreasing in G1. One possible explanation for our observed Pol2-mediated 3′-end processing defect is that the cell cycle in the pol2 mutants is altered, possibly even arrested at some stage. In that case, the decreased proportion of −3 products could be related to the cell cycle stage, rather than a direct consequence of Pol2's role in NHEJ. To test whether Pol2-mediated 3′-end processing of NHEJ varies by cell cycle stage, we induced HO cleavage after arresting wild-type or pol2 cells either at G1 or G2/M. As shown in Tables 2 and S1, pol2-4 cells exhibited a reduced frequency of repair via base deletion, regardless of whether the cells were non-synchronized, arrested in G1 or arrested in G2/M. These results indicate that reduction of base deletion during NHEJ in pol2 cells is not cell cycle dependent.
Discussion
From yeast to humans, NHEJ plays a role in the repair of DSBs. The range of proteins required for the various forms of NHEJ, however, have not yet been defined. DNA polymerases should be important contributors to DNA repair. Gaps often occur which require reconstruction and the nuclease activities associated with DNA polymerases can be utilized for processing repair intermediates. The essential DNA polymerases Pol1, Pol2 and Pol3 have previously been implicated as being required for HR-type DSB repair [22]–[24]. Pol4 was the first DNA polymerase found to participate in repairing 3′ overhangs during NHEJ in yeast [15]. Interestingly, two Pol4 homologs, Pol μ and Pol λ, have also been implicated in NHEJ in humans [27],[38],[39]. The fact that Pol4 directly interacts with DNA ligase IV implies that Pol4 might recruit other NHEJ factors to the DSB sites [11],[40],[41]. Indeed, we have observed, by chromatin immunoprecipitation, that Pol4 is present at DSB sites (Tseng and Teng, unpublished observation).
In this study, we aimed to clarify the in vivo roles of DNA polymerases in NHEJ-type DSB repair. Of the DNA polymerases we examined, only Rev3, showed no involvement in some aspect of NHEJ. This exception is interesting in that Rev3 has been shown, by chromatin immunoprecipitation, to localize to the site of HO-induced DSBs [42]. Pol4 is specifically required for adding bases during imprecise end joining; Pol3 is required for some unknown aspect of generating complex chromosomal rearrangements; both Pol1 and Pol2 play roles in both imprecise end joining and chromosomal rearrangements. Although Pol4has a primary rolein resynthesizing gaps and Pol2contributes to deleting the flaps at imprecise pairing sites at a DSB, we found that Pol4and Pol2can also influence, respectively, base deletion and addition during imprecise end joining repair. These results provide evidence for interplay between Pol4 and Pol2. We speculate that eliminating either one of these polymerases may influence the ability of the other to repair the DSB. This may also account for the observed overall reduction in survival in the presence of any of the ts mutations, even at the permissive temperature (Figure 3A). Even subtle changes in these essential proteins could tip the balance away from successful repair. Our study reflects the diversity, collaboration, and redundancy of multiple DNA polymerases, both essential and non-essential, in eukaryotic repair processes. DSBs generate different DNA end structures, which need to be recognized and repaired by complexes that likely include more than one DNA polymerase.
Our data demonstrates that Pol2, and more specifically, the 3′ to 5′ exonuclease activity of Pol2, plays a significant role in generating imprecise NHEJ joints that require removal of 3′ terminal flaps (Figure 4). However, unlike filling in gaps resulting from 3′ terminal mismatches, which seems to be an exclusive function of Pol4, flap removal can be carried out by nucleases other than Pol2, although at reduced efficiency. The frequencies of imprecise end joining in pol2-18 pol4 and pol2-4 pol4 double mutant strains were significantly decreased compared to those of wild-type or either single mutant, but in both double mutant strains DSB repair was predominantly through the −3 pathway of imprecise end joining. Since the pol4 mutant used in this study contains a complete truncation of the POL4 open reading frame, our data imply that other unidentified 3′ to 5′ exonucleases might be utilized in this NHEJ process. Further studies with additional mutant genes should help to delineate the roles and compartmentalization of particular repair factors in different aspects of NHEJ.
Figure 4. Proposed pathways for imprecise end joining of the HO endonuclease induced DSB at MATa.
The 4-bp 3′ overhanging sequence resulting from HO endonuclease cleavage is indicated. The two ends must be brought together into synapsis. Precise religation would lead to re-cutting. Possible end processing and subsequent alignment of complementary base pairs that would lead to insertions or deletions are shown. When 3′ overhangs mispair, Pol4 is required to fill in the gap [15],[16]. When 3′ flaps are generated by mis-alignment, the 3′ to 5′ exonuclease activity of Pol2 is the predominate exonuclease to remove the flaps.
Materials and Methods
Yeast Strain and Plasmid Constructions
General yeast manipulations were performed as described [43]. The S. cerevisiae strains used in this study are listed in Table 3. Yeast strains used in the study were derivatives of AGY628 or AGY673 [44]. The fragments of POL4 (coding sequence 286∼969) and REV3 (coding sequence 2652∼3863) were PCR-amplified from yeast genomic DNA and cloned into PvuII digested pRS304. pRS304pol4 and pRS304rev3 were linearized by AflII and HpaI, respectively. And these linearized YIPs were transformed into AGY673 using single crossover approach [43] to create pol4 (STY891) and rev3 (STY890) mutant strains. pSD218 [45] (kindly provided by Dr. Daniel E Gottschling) was used as previously described to create the pol1(cdc17-1) mutant in AGY673 (STY969). pST738 was constructed by ligating PCR-amplified pol3(cdc2-2) (coding sequence −600∼1950) from yeast strain UCC5898 (generously provided by Dr. Daniel E Gottschling) [45] to pRS306. BglII linearized pST738 was used to transform AGY628. pol3 of AGY628 was obtained by the two step pop in and pop out method, selecting first for Ura+ and then for Ura− by FOA and screened for temperature-sensitivity at 37°C for pol3 of AGY628. pol3 of AGY628 was then transformed with the PCR fragment of URA3::ai::HO to create the pol3 ts mutant of AGY673 (STY970). The pol2-18 mutant strain (STY966) was constructed by backcrossing YAH322 [46] (kindly provided by Dr. Akio Sugino) with AGY673 three times, each time selecting for spores with temperature sensitivity. YIpBI and YIpAM26 (kindly provided by Dr. Akio Sugino) were used to create pol2-4 (STY1358) and pol3-01 (STY1359) mutants respectively, as previously described by selecting Ura+ transformants using the single crossover replacement method, and then FOA selection for popouts removed the wild-type sequence [37],[43]. Genotype was confirmed by PCR and sequencing. STY1356 (pol2-18 pol4) and STY1360 (pol2-4 pol4) were constructed by disrupting POL4 in STY966 and STY1356 using pRS304pol4, respectively. A ura3 deletion fragment was PCR amplified from an FOA-resistant clone of AGY673. “No cut” site controls were created using the double crossover approach [43] by transforming the ura3 deletion fragment into STY969, STY973 and STY966 and selecting for intergrants on FOA plates to obtain STY1553, STY1552 and STY1554, respectively. The dnl4 strain was constructed by transformation of a PCR product into AGY673 and STY1358 using the genomic DNA from a BY4741 dnl4 strain (Invitrogen) as a template and oligonucleotides flanking the DNL4 gene as primers. Transformants with double crossover at DNL4 were selected for G418 resistance and insertion sites were confirmed by PCR. All primer sequences for PCR are available upon request.
Media and Growth Conditions
Yeast cells were grown in yeast extract-peptone-dextrose (YPD) or synthetic complete media (SC) with appropriate amino acids missing [47]. Yeast extract-peptone-galactose (YEP-galactose) and yeast extract-peptone-raffinose (YEP-raffinose) contain 2% galactose (w/v) and 2% raffinose (w/v), respectively, instead of dextrose (2%). 5- fluoro-orotic acid (5-FOA) plates are SC glucose plates supplemented with 1 mg/ml of 5-FOA [48],[49].
The NHEJ Assay
The NHEJ assay system was established in the Gabriel lab [28]. This system contains a positive selection for chromosomal changes, including insertions, deletions, translocations and inversions, associated with repair of a defined chromosomal DSB. As shown in Figure 1, a functional URA3 allele was created on chromosome V, which contains a copy of the ACT1 intron as well as the Y–Z junction from the MATa locus. The Y-Z junction includes the recognition sequence and cleavage site for the HO endonuclease, was created on chromosome V. All MAT related sequences had been deleted and this strain contains an integrated galactose-inducible HO endonuclease gene.
Induction of the HO Endonuclease, Measurement of DSB Repair Efficiency (Survival Frequency), and 5-FOA Resistance Frequency
For non-essential polymerase mutants, multiple independent colonies from each strain were grown at 30°C in YEP-raffinose liquid medium to a final concentration of OD595 ∼1. Appropriate dilutions of cells were then plated on YPD or YEP-galactose (YPGal) plates. Colonies were counted after 4 days of growth. Colonies on the YEP-galactose plates were replica plated onto synthetic complete 5-FOA-containing media to measure the frequency of 5-FOA resistance among survivors of HO endonuclease induction. For essential polymerase mutants, multiple independent colonies from each strain were grown at 23°C in YEP-raffinose liquid medium to a final concentration of OD595 ∼1. Appropriate dilutions of cells were then plated on YPD or YPGal plates and incubated at 23°C or 30°C. Colonies were counted after four days of growth. Alternatively, yeast cells were diluted to YPGal liquid medium to OD595 ∼0.5, incubated at 23°C or 30°C for 20 hours, and then serial diluted to plate on YPGal and FOA plates. The frequency for imprecise end joining is the ratio of the number of colonies growing on YPGal vs. YPD from per ml of culture. The frequency of chromosomal rearrangements is the number of colonies growing on 5-FOA vs. YPD from per ml of culture. It was notable that colony sizes were smaller and much more heterogeneous on YPGal and FOA plates from all ts mutants, at both the permissive and semi-restrictive temperatures, necessitating an arbitrary cutoff for tiny colonies that were hard to score.
All values are expressed as means (±) standard error. Differences between groups were tested using the student's t-test. For non-essential polymerase mutants, significance tests were compared against frequency of wild type. For essential polymerase mutants, significance tests were compared against frequencies at permissive temperature. Samples of survivors growing on YPGal were analyzed by PCR using primer RAG512 and RAG515 flanking the URA3 gene (Figure 1) [28], and PCR products were sequenced using internal primer RAG513 or RAG633 (Figure 1) [28]. Survivors growing on FOA were further analyzed by Southern blot analysis (Figure S1). For preparing probes, the URA3::ACT1 intron::HO cut site fragment was amplified by PCR using primer RAG512 and RAG515. The 799 (probe A) and 878 (probe B) base pairs XhoI-digested PCR fragments were used as probes for Southern hybridization. In most cases, 40 survivors from four to seven independent cultures were examined for each strain. Based on Southern blot analysis, a rearrangement was termed an insertion if we observed two or more hybridizing bands, a deletion if we observed only a single band, and a mutation if the single band was the same size as the parent strain. Note that potential translocations or inversions were read as insertions. Further, very small insertions could be mis-read as mutations because of minimal change in band size, although we did not see evidence of this after sequencing twenty same size products from the pol3-01 strain (data not shown).
Cell-Cycle Experiments
Multiple independent colonies from each strain were grown at 30°C overnight in YEP-raffinose liquid medium. Yeast cells were diluted to YEP-raffinose liquid medium to OD595 0.1 to refresh for three hours, incubated at 30°C for additional four hours in 100 µM α-factor or 20 µg/ml nocodazole. Cultures were split and half continued to grow in YEP-raffinose with α-factor or nocodazole while the other half received galactose to a final concentration of 2% for one hour. Yeast cells from YEP-raffinose and YPGal were then serial diluted on YPD and YPGal plates, respectively. The frequency of imprecise end joining was determined as the ratio of the number of colonies growing on YPGal vs. YPD from per ml of culture.
Supporting Information
Southern blot analysis of 5-FOA-resistant survivors after HO endonuclease induced.
(0.55 MB DOC)
Quantitative analysis of repair events at the URA3::ACT1 intron::HO cut site locus of non-essential DNA polymerase mutants.
(0.05 MB DOC)
Quantitative analysis of repair events at the URA3::ACT1 intron::HO cut site locus of essential DNA polymerase mutants.
(0.06 MB DOC)
Acknowledgments
We thank Drs. Daniel E Gottschling and Akio Sugino for providing plasmids and strains. We also thank Mr. Hung Ting-Wei for his expert technical assistance.
Footnotes
The authors have declared that no competing interests exist.
This work was supported by grants from the National Science Council (NRPGM-95-3112-B-002-023) and National Health Research Institute of Taiwan (NHRI-EX95-9328SI).
References
- 1.Pastink A, Lohman PH. Repair and consequences of double-strand breaks in DNA. Mutat Res. 1999;428:141–156. doi: 10.1016/s1383-5742(99)00042-3. [DOI] [PubMed] [Google Scholar]
- 2.Hoeijmakers JH. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411:366–374. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- 3.Pastink A, Eeken JC, Lohman PH. Genomic integrity and the repair of double-strand DNA breaks. Mutat Res. 2001;480–481:37–50. doi: 10.1016/s0027-5107(01)00167-1. [DOI] [PubMed] [Google Scholar]
- 4.Lieber MR, Ma Y, Pannicke U, Schwarz K. The mechanism of vertebrate nonhomologous DNA end joining and its role in V(D)J recombination. DNA Repair (Amst) 2004;3:817–826. doi: 10.1016/j.dnarep.2004.03.015. [DOI] [PubMed] [Google Scholar]
- 5.van den Bosch M, Lohman PH, Pastink A. DNA double-strand break repair by homologous recombination. Biol Chem. 2002;383:873–892. doi: 10.1515/BC.2002.095. [DOI] [PubMed] [Google Scholar]
- 6.Valerie K, Povirk LF. Regulation and mechanisms of mammalian double-strand break repair. Oncogene. 2003;22:5792–5812. doi: 10.1038/sj.onc.1206679. [DOI] [PubMed] [Google Scholar]
- 7.West SC. Molecular views of recombination proteins and their control. Nat Rev Mol Cell Biol. 2003;4:435–445. doi: 10.1038/nrm1127. [DOI] [PubMed] [Google Scholar]
- 8.Dudas A, Chovanec M. DNA double-strand break repair by homologous recombination. Mutat Res. 2004;566:131–167. doi: 10.1016/j.mrrev.2003.07.001. [DOI] [PubMed] [Google Scholar]
- 9.Haber JE. Partners and pathwaysrepairing a double-strand break. Trends Genet. 2000;16:259–264. doi: 10.1016/s0168-9525(00)02022-9. [DOI] [PubMed] [Google Scholar]
- 10.Chen C, Kolodner RD. Gross chromosomal rearrangements in Saccharomyces cerevisiae replication and recombination defective mutants. Nat Genet. 1999;23:81–85. doi: 10.1038/12687. [DOI] [PubMed] [Google Scholar]
- 11.Daley JM, Palmbos PL, Wu D, Wilson TE. Nonhomologous end joining in yeast. Annu Rev Genet. 2005;39:431–451. doi: 10.1146/annurev.genet.39.073003.113340. [DOI] [PubMed] [Google Scholar]
- 12.Coquerelle T, Bopp A, Kessler B, Hagen U. Strand breaks and K' end-groups in DNA of irradiated thymocytes. Int J Radiat Biol Relat Stud Phys Chem Med. 1973;24:397–404. doi: 10.1080/09553007314551251. [DOI] [PubMed] [Google Scholar]
- 13.Pastwa E, Neumann RD, Mezhevaya K, Winters TA. Repair of radiation-induced DNA double-strand breaks is dependent upon radiation quality and the structural complexity of double-strand breaks. Radiat Res. 2003;159:251–261. doi: 10.1667/0033-7587(2003)159[0251:roridd]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 14.Ma Y, Pannicke U, Schwarz K, Lieber MR. Hairpin opening and overhang processing by an Artemis/DNA-dependent protein kinase complex in nonhomologous end joining and V(D)J recombination. Cell. 2002;108:781–794. doi: 10.1016/s0092-8674(02)00671-2. [DOI] [PubMed] [Google Scholar]
- 15.Wilson TE, Lieber MR. Efficient processing of DNA ends during yeast nonhomologous end joining. Evidence for a DNA polymerase beta (Pol4)-dependent pathway. J Biol Chem. 1999;274:23599–23609. doi: 10.1074/jbc.274.33.23599. [DOI] [PubMed] [Google Scholar]
- 16.Daley JM, Laan RL, Suresh A, Wilson TE. DNA joint dependence of pol X family polymerase action in nonhomologous end joining. J Biol Chem. 2005;280:29030–29037. doi: 10.1074/jbc.M505277200. [DOI] [PubMed] [Google Scholar]
- 17.Burgers PM. Eukaryotic DNA polymerases in DNA replication and DNA repair. Chromosoma. 1998;107:218–227. doi: 10.1007/s004120050300. [DOI] [PubMed] [Google Scholar]
- 18.Wu X, Guo D, Yuan F, Wang Z. Accessibility of DNA polymerases to repair synthesis during nucleotide excision repair in yeast cell-free extracts. Nucleic Acids Res. 2001;29:3123–3130. doi: 10.1093/nar/29.14.3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson RE, Prakash S, Prakash L. Efficient bypass of a thymine-thymine dimer by yeast DNA polymerase, Poleta. Science. 1999;283:1001–1004. doi: 10.1126/science.283.5404.1001. [DOI] [PubMed] [Google Scholar]
- 20.Sobol RW, Horton JK, Kuhn R, Gu H, Singhal RK, et al. Requirement of mammalian DNA polymerase-beta in base-excision repair. Nature. 1996;379:183–186. doi: 10.1038/379183a0. [DOI] [PubMed] [Google Scholar]
- 21.Wilson DM, 3rd, Thompson LH. Life without DNA repair. Proc Natl Acad Sci U S A. 1997;94:12754–12757. doi: 10.1073/pnas.94.24.12754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holmes AM, Haber JE. Double-strand break repair in yeast requires both leading and lagging strand DNA polymerases. Cell. 1999;96:415–424. doi: 10.1016/s0092-8674(00)80554-1. [DOI] [PubMed] [Google Scholar]
- 23.Tsai YL, Tseng SF, Chang SH, Lin CC, Teng SC. Involvement of replicative polymerases, Tel1p, Mec1p, Cdc13p, and the Ku complex in telomere-telomere recombination. Mol Cell Biol. 2002;22:5679–5687. doi: 10.1128/MCB.22.16.5679-5687.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lydeard JR, Jain S, Yamaguchi M, Haber JE. Break-induced replication and telomerase-independent telomere maintenance require Pol32. Nature. 2007;448:820–823. doi: 10.1038/nature06047. [DOI] [PubMed] [Google Scholar]
- 25.Pospiech H, Rytkonen AK, Syvaoja JE. The role of DNA polymerase activity in human non-homologous end joining. Nucleic Acids Res. 2001;29:3277–3288. doi: 10.1093/nar/29.15.3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gottlich B, Reichenberger S, Feldmann E, Pfeiffer P. Rejoining of DNA double-strand breaks in vitro by single-strand annealing. Eur J Biochem. 1998;258:387–395. doi: 10.1046/j.1432-1327.1998.2580387.x. [DOI] [PubMed] [Google Scholar]
- 27.Mahajan KN, Nick McElhinny SA, Mitchell BS, Ramsden DA. Association of DNA polymerase mu (pol mu) with Ku and ligase IV: role for pol mu in end-joining double-strand break repair. Mol Cell Biol. 2002;22:5194–5202. doi: 10.1128/MCB.22.14.5194-5202.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu X, Gabriel A. Patching broken chromosomes with extranuclear cellular DNA. Mol Cell. 1999;4:873–881. doi: 10.1016/s1097-2765(00)80397-4. [DOI] [PubMed] [Google Scholar]
- 29.Moore JK, Haber JE. Cell cycle and genetic requirements of two pathways of nonhomologous end-joining repair of double-strand breaks in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:2164–2173. doi: 10.1128/mcb.16.5.2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu X, Gabriel A. Ku-dependent and Ku-independent end-joining pathways lead to chromosomal rearrangements during double-strand break repair in Saccharomyces cerevisiae. Genetics. 2003;163:843–856. doi: 10.1093/genetics/163.3.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu X, Gabriel A. Reciprocal translocations in Saccharomyces cerevisiae formed by nonhomologous end joining. Genetics. 2004;166:741–751. doi: 10.1093/genetics/166.2.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hubscher U, Maga G, Spadari S. Eukaryotic DNA polymerases. Annu Rev Biochem. 2002;71:133–163. doi: 10.1146/annurev.biochem.71.090501.150041. [DOI] [PubMed] [Google Scholar]
- 33.Garg P, Burgers PM. DNA polymerases that propagate the eukaryotic DNA replication fork. Crit Rev Biochem Mol Biol. 2005;40:115–128. doi: 10.1080/10409230590935433. [DOI] [PubMed] [Google Scholar]
- 34.Pursell ZF, Isoz I, Lundstrom EB, Johansson E, Kunkel TA. Yeast DNA polymerase epsilon participates in leading-strand DNA replication. Science. 2007;317:127–130. doi: 10.1126/science.1144067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morrison A, Sugino A. The 3′–>5′ exonucleases of both DNA polymerases delta and epsilon participate in correcting errors of DNA replication in Saccharomyces cerevisiae. Mol Gen Genet. 1994;242:289–296. doi: 10.1007/BF00280418. [DOI] [PubMed] [Google Scholar]
- 36.Shcherbakova PV, Pavlov YI. 3′–>5′ exonucleases of DNA polymerases epsilon and delta correct base analog induced DNA replication errors on opposite DNA strands in Saccharomyces cerevisiae. Genetics. 1996;142:717–726. doi: 10.1093/genetics/142.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morrison A, Johnson AL, Johnston LH, Sugino A. Pathway correcting DNA replication errors in Saccharomyces cerevisiae. Embo J. 1993;12:1467–1473. doi: 10.1002/j.1460-2075.1993.tb05790.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nick McElhinny SA, Havener JM, Garcia-Diaz M, Juarez R, Bebenek K, et al. A gradient of template dependence defines distinct biological roles for family X polymerases in nonhomologous end joining. Mol Cell. 2005;19:357–366. doi: 10.1016/j.molcel.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 39.Capp JP, Boudsocq F, Bertrand P, Laroche-Clary A, Pourquier P, et al. The DNA polymerase lambda is required for the repair of non-compatible DNA double strand breaks by NHEJ in mammalian cells. Nucleic Acids Res. 2006;34:2998–3007. doi: 10.1093/nar/gkl380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tseng HM, Tomkinson AE. A physical and functional interaction between yeast Pol4 and Dnl4-Lif1 links DNA synthesis and ligation in nonhomologous end joining. J Biol Chem. 2002;277:45630–45637. doi: 10.1074/jbc.M206861200. [DOI] [PubMed] [Google Scholar]
- 41.Dudasova Z, Dudas A, Chovanec M. Non-homologous end-joining factors of Saccharomyces cerevisiae. FEMS Microbiol Rev. 2004;28:581–601. doi: 10.1016/j.femsre.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 42.Hirano Y, Sugimoto K. ATR homolog Mec1 controls association of DNA polymerase zeta-Rev1 complex with regions near a double-strand break. Curr Biol. 2006;16:586–590. doi: 10.1016/j.cub.2006.01.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rose M, Winston F, Hieter P. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1990. Methods in Yeast Genetics. [Google Scholar]
- 44.Haviv-Chesner A, Kobayashi Y, Gabriel A, Kupiec M. Capture of linear fragments at a double-strand break in yeast. Nucleic Acids Res. 2007;35:5192–5202. doi: 10.1093/nar/gkm521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Diede SJ, Gottschling DE. Telomerase-mediated telomere addition in vivo requires DNA primase and DNA polymerases alpha and delta. Cell. 1999;99:723–733. doi: 10.1016/s0092-8674(00)81670-0. [DOI] [PubMed] [Google Scholar]
- 46.Araki H, Leem SH, Phongdara A, Sugino A. Dpb11, which interacts with DNA polymerase II(epsilon) in Saccharomyces cerevisiae, has a dual role in S-phase progression and at a cell cycle checkpoint. Proc Natl Acad Sci U S A. 1995;92:11791–11795. doi: 10.1073/pnas.92.25.11791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sherman F, Fink GR, Hicks JB. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1986. Methods in YeasGenetics: A Laboratory Manual. [Google Scholar]
- 48.Boeke JD, LaCroute F, Fink GR. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol Gen Genet. 1984;197:345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- 49.Sikorski RS, Boeke JD. In vitro mutagenesis and plasmid shuffling: from cloned gene to mutant yeast. Methods Enzymol. 1991;194:302–318. doi: 10.1016/0076-6879(91)94023-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Southern blot analysis of 5-FOA-resistant survivors after HO endonuclease induced.
(0.55 MB DOC)
Quantitative analysis of repair events at the URA3::ACT1 intron::HO cut site locus of non-essential DNA polymerase mutants.
(0.05 MB DOC)
Quantitative analysis of repair events at the URA3::ACT1 intron::HO cut site locus of essential DNA polymerase mutants.
(0.06 MB DOC)