Abstract
Autosomal recessive polycystic kidney disease is a hereditary fibrocystic disease that involves the kidneys and the biliary tract. Mutations in the PKHD1 gene are responsible for typical forms of autosomal recessive polycystic kidney disease. We have generated a mouse model with targeted mutation of Pkhd1 by disrupting exon 4, resulting in a mutant transcript with deletion of 66 codons and expression at ∼30% of wild-type levels. Pkhd1del4/del4 mice develop intrahepatic bile duct proliferation with progressive cyst formation and associated periportal fibrosis. In addition, these mice exhibit extrahepatic manifestations, including pancreatic cysts, splenomegaly, and common bile duct dilation. The kidneys are unaffected both histologically and functionally. Fibrocystin is expressed in the apical membranes and cilia of bile ducts and distal nephron segments but is absent from the proximal tubule. This pattern is unchanged in orthologous models of autosomal dominant polycystic kidney disease due to mutation in Pkd1 or Pkd2. Mutant fibrocystin in Pkhd1del4/del4 mice also retains this expression pattern. The hypomorphic Pkhd1del4/del4 mouse model provides evidence that reduced functional levels of fibrocystin are sufficient for cystogenesis and fibrosis in the liver and pancreas, but not the kidney, and supports the hypothesis of species-dependent differences in susceptibility of tissues to Pkhd1 mutations.
Autosomal recessive polycystic kidney disease (ARPKD) is a hereditary cystic disease involving the kidneys and the biliary tract. ARPKD is a significant cause of pediatric morbidity and mortality with an estimated incidence of 1 in 20,000 live births.1 The clinical spectrum is variable with ∼30 to 50% of affected neonates dying shortly after birth and other patients surviving to adulthood.2,3,4 The most severely affected fetuses have enlarged echogenic kidneys in utero with associated oligohydramnios indicative of intrauterine renal failure. Neonatal demise in this setting is primarily attributable to respiratory failure from associated pulmonary hypoplasia. The majority of affected individuals, however, do not have these severe manifestations in utero and survive the neonatal period. These individuals present later with a spectrum of associated morbidities that include systemic and portal hypertension, congenital hepatic fibrosis, and progressive renal insufficiency.3,5 Primary organ system involvement in the latter presentation of human ARPKD is restricted to the liver and kidneys, and the most common pathological lesions seen are biliary dysgenesis accompanied by portal tract fibrosis in the liver and fusiform dilatations of the renal collecting ducts radiating from the medulla to the cortex of the kidney.
Mutations in a single gene, PKHD1 (polycystic kidney and hepatic disease 1), have been found to underlie all presentations of ARPKD in humans.6,7 PKHD1 extends more than 469 kb on chromosome 6p21.1-p12. The longest predicted transcript consists of 67 exons and encodes a 4074-amino acid protein, called either fibrocystin or polyductin (heretofore referred to as fibrocystin for simplicity). This novel protein is predicted to have a single transmembrane-spanning domain near its carboxyl terminus. The extracellular domain is predicted to be heavily glycosylated and contains multiple iterations of immunoglobulin-like, plexin, transcription factor (IPT) domains and parallel β-helix 1 (PbH1) repeats. A single paralogous gene, PKHDL1, has been identified in mammalian species.8 Invertebrate model organisms including Drosophila melanogaster and Caenorhabditis elegans do not appear to have homologs of either gene. It has been proposed that fibrocystin functions as a cell surface receptor, a co-receptor, or a cell surface ligand,6,7 and it has recently been shown to undergo notch-like proteolytic cleavage resulting in regulated release from the apical surface of cells.9
To date, more than 300 different mutations have been detected in PKHD1 (http://www.humgen.rwth-aachen.de), with the majority predicted to be amino acid substitution mutations. Mutations reside almost exclusively in the large extracellular domain of the protein, with only one truncation mutation detected in the C-terminal cytoplasmic tail.10 Support for the hypothesis that ARPKD is a loss-of-function disease comes from several studies on patients in whom both mutations were predicted to be chain terminating. Patients with two predicted truncation mutations invariably present with the more severe perinatal lethal phenotype, whereas patients with later-onset disease have at least one missense mutation.11,12,13,14
The fibrocystin transcript is strongly expressed in the developing kidney, where it is primarily localized to the ureteric bud. In the adult kidney it appears in the collecting ducts and loops of Henle.15 Other sites of expression include the ductal plates and bile ducts of the liver, pancreatic ducts, as well as the heart, large vessels, testis, trachea, and sympathetic ganglia.15 In keeping with the transcript expression pattern, immunohistochemical studies of fibrocystin protein expression in kidney tissue have shown ureteric bud expression in the developing kidney and strong collecting duct and medullary thick ascending limb expression in adult kidney.16,17,18 Immunocytochemistry has consistently shown that fibrocystin is localized in primary cilia of cultured cells16,17,18,19,20,21 and immunoelectron microscopy in kidney tissues has confirmed the cilia location.17,20 Fibrocystin has been proposed to play a role in ciliogenesis because structural abnormalities were found in the cilia of biliary epithelia in the pck rat, a rodent model bearing a germline mutation in Pkhd1.19 Additional evidence that fibrocystin plays a role in ciliogenesis comes from Pkhd1 siRNA knockdown studies in cholangiocytes19 and renal epithelial cells,22 whereby the cells lacking fibrocystin do not form cilia.
Orthologous rodent models of mutation in Pkhd1 have been reported in both rat and mouse. The pck rat has a mutation in Pkhd1 that results in skipping of exon 367 but still forms a nearly full-length protein product.19 The pck rat develops renal cysts in thick ascending loops of Henle, distal tubules, and collecting ducts, and male animals are more severely affected than females.23 These animals also develop bile duct dilatation accompanied by mild portal fibrosis.23 Pancreatic lesions were not reported in the pck rat. An engineered mouse mutation with disruption of exon 40 still produces a modified transcript because of exon skipping.24 This mouse model shows bile duct proliferation with associated portal tract fibrosis and portal hypertension but does not have discernible abnormalities in the kidney.24 More recently renal phenotypes have been described in two mouse models harboring hypomorphic mutations in Pkhd1.25,26 The Pkhd1del2/del2 mouse develops renal cysts in the S3 segment of the proximal tubule but not in the collecting duct system.25 On the other hand, the Pkhd1del3−4/del3–4 model develops cysts in the collecting duct and thick ascending limbs of Henle’s loop.26
In the current study, we sought to investigate the role of fibrocystin in human disease by targeting the 5′ region of the murine Pkhd1. Partial deletion of exon 4 resulted in a hypomorphic allele with persistent protein expression because of cryptic splice site activation and exon skipping. The Pkhd1del4 allele mimics a phenotype of ARPKD that results in predominant bile duct dysgenesis accompanied by periportal fibrosis, dilated extrahepatic bile ducts, and splenomegaly. The mice develop significant pancreatic duct cysts that have not been described in human disease but do not develop kidney cysts. Wild-type and mutant fibrocystin are expressed on the apical and ciliary membranes of bile duct cells and of the distal nephron. This expression pattern of fibrocystin is not altered in kidney or bile duct cysts of mice bearing mutations in either of the dominant polycystic kidney disease genes, Pkd1 or Pkd2. In aggregate, the data show that rodent Pkhd1 is prone to alternative splicing, that hypomorphic alleles are sufficient to cause biliary fibrocystic disease and pancreatic duct cysts, and that the expression pattern of fibrocystin is independent of either polycystin-1 (PC1) or polycystin-2 (PC2).
Materials and Methods
Animal Care
All experiments were conducted in accordance with Yale University Institutional Animal Care and Use Committee guidelines and procedures. The Pkhd1 del4 mice described in this article are on a C57BL6/129 mixed background, unless stated otherwise. Mice of either gender were used in this study.
Construction of the Pkhd1 Knockout Mouse
A BAC clone, RPCI-22-534J18 (CHORI, Oakland, CA), previously identified as containing the first 32 exons of murine Pkhd1 gene, was digested with XbaI to isolate a 7.8-kb fragment containing exons 2 to 5. This fragment was subcloned into the XbaI site of pBluescript II KS vector containing a diphtheria toxin-negative selection cassette. A 0.8-kb fragment, containing the terminal 46 nucleotide residues of exon 4 and intronic sequence between exons 4 and 5, was replaced by insertion of a 1.8-kb PGK-Neo cassette into the AatII site. The Pkhd1 targeting vector was linearized at a unique NotI site in the pBluescript II KS vector and electroporated into 129Sv/E mouse embryonic stem cells. G418-resistant clones were picked and screened both 5′ and 3′ to the target region by long-range polymerase chain reaction (PCR), using rTth XL DNA polymerase (Applied Biosystems, Foster City, CA) according to the manufacturer’s recommendations, thereby generating 3.5- and 4.5-kb PCR products, respectively. The 5′-region was amplified using a forward primer designed outside the targeting vector upstream of exon 2 (5′-GGTCCCCATGTACTTTCCT-3′) and a reverse primer in the PGK promoter sequence of the Neo cassette (5′-ACATTCCACATCCACCGGTA-3′). The 3′ region was amplified with primers in the Neo cassette (5′-CGTTGGCTACCCGTGATATT-3′) and in the diphtheria toxin selection cassette downstream of the targeting vector (5′-TGCCCCTTCAGTATCCAAAC-3′). Two correctly targeted embryonic stem cell lines, 19 and 206, were expanded and injected into blastocysts to produce chimeric mice. Germline transmission was obtained, and mice on a mixed C57BL6/129sv background and heterozygous Pkhd1del4/+ mice were subsequently intercrossed to generate homozygous progeny.
Southern Hybridization and Genotypic Analysis
Genomic DNA analysis was performed by digestion of DNA from liver with NcoI, which was then transferred to a nitrocellulose membrane and hybridized with an intron 5-containing 32P-labeled probe. The wild-type allele is 7.6 kb in size, and the mutant allele is 6.0 kb. Genomic DNA extracted from mouse tail biopsies was routinely genotyped by PCR using the following primers to recognize the wild-type allele: forward primer, 5′-TTAGGGAAGAATGGCTCTC-3′, and the reverse primer, 5′-TTCAGAGGGAGGAAAAGCAA-3′, to produce a 580-bp fragment. The Pkhd1del4 allele was amplified with forward primer 5′-TTAGGGAAGAATGGCTCTC-3′ and a reverse primer in the PGK-Neo cassette, 5′-GCCAGAGGCCACTTGTGTAG-3′, amplifying a 171-bp product.
Reverse Transcriptase (RT)-PCR Analysis
Total RNA was prepared from kidneys and livers of Pkhd1+/+ and Pkhd1del4/del4 mice using TRIzol reagent (Invitrogen, Carlsbad, CA) and was reverse-transcribed using a cDNA synthesis kit from Stratagene (La Jolla, CA). The 5′ region of the Pkhd1 gene was amplified from kidney and liver cDNA of Pkhd1+/+ and Pkhd1del4/del4 animals using the following primers: exon 1 forward primer 5′-CTGGCCTGTCACCGAATAGT-3′ and the reverse primer designed against exon 6, 5′-GATCAGACACCTGTTTTATATC-3′. The PCR products were run on a 1% agarose gel, extracted, and sequenced.
Quantitative PCR Analysis
An aliquot of 2 μg of total RNA was reverse-transcribed from wild-type and Pkhd1del4/del4 kidney tissues, using Omniscript reverse transcriptase (Qiagen, Valencia, CA) and random hexamers. Quantitative PCR of Pkhd1 transcripts was performed using TaqMan technology (Applied Biosystems). The TaqMan assay designed for mouse Pkhd1 exon 3/4 junction (specific for the exon 4-containing transcripts) consists of primers mPkhd1Ex4F 5′-TCACAGTTGTATTTGACGGTTTGGA-3′, mPkhd1Ex4R 5′-TCTCAGCTGCAGATAGACCTGT-3′, and the TaqMan probe, 5′-AAGTATTCTTTACCCCAACAATG-3′ in exon 4. The TaqMan assay for mouse Pkhd1 exon 3/5 junction (specific for exon 3/5-containing transcripts) consists of primers mPkhd1Ex3F 5′-CAGATTGAACCCGCAGAAGGTA-3′, mPkhd1Ex5R 5′-ATCAGAAGCAGATGCAGGGC- 3′, and the TaqMan probe 5′-TGTATTTGACGGTCTCTT-3′. The housekeeping gene GAPDH was used as a control for gene expression. The PCR conditions were 50°C for 2 minutes, 94°C for 10 minutes, followed by 40 cycles of 94°C for 15 seconds and 60°C for 30 seconds. The standard curves were generated from amplified cDNA products, and all reactions were performed in triplicate. The samples were normalized to GAPDH. Quantification was performed according to the manufacturer’s recommendations (Applied Biosystems).
Histochemistry
Tissues were fixed in 4% paraformaldehyde and embedded in paraffin. Five-μm sections of kidney, liver, and pancreas were stained with hematoxylin and eosin, periodic acid-Schiff, and Mallory trichrome.
Generation of Fibrocystin Polyclonal Antibodies
We raised polyclonal antibodies against different portions of the longest open reading frame product using fusion protein and synthetic peptide-based strategies. Two fusion proteins and two synthetic peptides were included in the immunization protocol. One fusion protein, pep7/4b, containing part of the murine fibrocystin extracellular region (amino acids 2694 to 2894) was coupled to GST at the N terminus. The corresponding cDNA fragment was amplified from mouse kidney cDNA using the following primers: forward 5′-AATTCCCGGTCACCTACCTGGTTTCAGGT-3′ and reverse 5′-GACACAATTGTCCTCAGCGACTCGAGCGGC-3′. This product was cloned into pGEX4T-3 and was sequence-verified. The GST-fibrocystin fusion protein was generated in BL21(DE3) pLysS E. coli and purified with glutathione-coated Sepharose beads (Amersham Biosciences, Piscataway, NJ) and eluted with glutathione-containing buffer (10 mmol/L glutathione, 50 mmol/L Tris-HCl, pH 8.0). Two mg of purified protein was used to immunize rabbits for polyclonal antibody production (Zymed, Carlsbad, CA) and affinity purified. The anti-P5P6 antiserum was raised against the amino acid sequences 3252 to 3266 (P5) and 3774 to 3785 (P6) in human fibrocystin and has been described previously.20
Generation of an Epitope-Tagged Human PKHD1 Construct
To construct an epitope-tagged full-length human PKHD1 cDNA, five overlapping fragments were generated by PCR using rTth XL DNA polymerase (Applied Biosystems). A sixth fragment bearing a C-terminal triple hemagluttinin (HA) tag was generated by polymerase chain reaction. Each fragment was cloned into pGEM-T-Easy vector (Promega, Madison, WI) and verified by restriction digestion and sequencing from both the 5′ and 3′ directions. The full-length human PKHD1 cDNA was subcloned into pcDNA3.1 vector via NotI and AseI restriction sites. Site-directed mutagenesis was used to insert a unique ClaI restriction site downstream of the leader sequence in the amino terminus of human PKHD1. A triple FLAG epitope tag was inserted at this unique site and sequence-verified.
Membrane Preparation and Western Blot Analysis
Membrane proteins were prepared from whole organs (kidneys, brain, pancreas, liver, lung, and heart) of adult mice. Fresh tissues were added to 4 vol of homogenization buffer (25 mmol/L Tris-HCl, pH 7.4, 20 mmol/L sucrose, 5 mmol/L ethylenediaminetetraacetic acid, and protease inhibitors) then homogenized with a motor-driven Dounce homogenizer. The homogenized tissue samples were first centrifuged at 500 × g, for 15 minutes at 4°C. The supernatant was transferred to a fresh tube and centrifuged for 15 minutes at 10,000 × g at 4°C. After the second centrifugation step, the supernatant was centrifuged for 60 minutes at 100,000 × g at 4°C. The resulting pellet represented the membrane fraction and was resuspended in homogenization buffer containing 1% Triton X-100, incubated on ice for 15 minutes, and centrifuged at 13,000 rpm for 15 minutes at 4°C. The protein concentration of supernatant containing the membrane fraction was determined using a protein assay kit from Bio-Rad (Hercules, CA) based on the Bradford assay. Membrane protein preparations were combined with a reducing sodium dodecyl sulfate sample buffer and boiled for 10 minutes. After brief centrifugation the supernatant was separated on a gradient 3 to 8% Novex polyacrylamide gel (Invitrogen) and transferred to a polyvinylidene difluoride membrane (Immobilon; Millipore, Billerica, MA) in 1× Novex transfer buffer (Invitrogen) containing 10% methanol and 0.1% sodium dodecyl sulfate. Prestained molecular weight markers were used (New England Biolabs, Ipswich, MA). The membrane was blocked for 1 hour at room temperature in 5% low-fat dry milk powder/0.1% Tween 20/1× phosphate-buffered saline (PBS) and then incubated with primary antiserum (pep7/4b, diluted 1:200 in blocking solution; P5/P6, diluted to 1:1000 in blocking solution) for 2 hours at room temperature. The membranes were washed with 0.1% Tween 20/1× PBS, five times each for 15 minutes and then incubated with horseradish peroxidase-linked anti-rabbit IgG secondary antibody (diluted 1:5000 in blocking solution; Jackson ImmunoResearch Laboratories, West Grove, PA) for 1 hour at room temperature. The membranes were washed for a further five times, 15 minutes each in 1× PBS/0.1%Tween-20. Antigen-antibody complexes were visualized using a chemiluminescence detection kit (ECL chemiluminescence, Amersham Biosciences).
Immunocytochemistry
COS7 cells were transiently transfected with epitope-tagged fibrocystin construct and cultured on glass coverslips. The cells were washed with 1× PBS/100 mmol/L CaCl2/1 mmol/L MgCl2, fixed in Bouin’s reagent (15% saturated picric acid, 3.7% formaldehyde, 0.1 mol/L sodium phosphate, pH 7.4) for 20 minutes, washed thoroughly with PBS, and permeabilized in 0.2% Triton X-100/1× PBS/2% bovine serum albumin for 30 minutes before incubation with anti-FLAG (1:200; Sigma, St. Louis, MO), rat anti-HA (1:100, Sigma) and anti-fibrocystin pep7/4b (1:300) antibodies for 2 hours at room temperature in 2% bovine serum albumin/1× PBS. The coverslips were washed in high-salt PBS, two times for 5 minutes each and then 1× PBS buffer for a further 5 minutes before adding the fluorescence-conjugated secondary antibodies. The cells were washed three times for 5 minutes each in 1× PBS before being mounted in Vectashield DAPI-containing mounting medium (Vector Laboratories, Burlingame, CA). Analysis of the immunofluorescence staining was performed using a Axiophot confocal microscope (Carl Zeiss, Thornwood, NY) and an Eclipse Te2000U fluorescence microscope (Nikon, Toyo, Japan).
Immunohistochemistry
Mice were anesthetized by intraperitoneal injection of sodium pentobarbital. The tissues were cleared with PBS and fixed with 4% paraformaldehyde by cardiac perfusion. The kidneys and liver were extracted and prepared by paraffin embedding or cryosectioning. For the latter, tissues were equilibrated in 30% sucrose overnight and then frozen in liquid nitrogen. Five-μm sections were cut and used for immunohistochemical analysis. Sections were washed in 1× Tris-buffered saline (TBS) for 15 minutes and incubated in 0.1% sodium borohydride for 30 minutes. The sections were rinsed in 1× TBS then treated with 1% sodium dodecyl sulfate for 5 minutes. After washing, the sections were blocked for 1 hour in 0.2% bovine serum albumin/1× TBS/10% goat serum. The primary antibodies were incubated overnight in blocking solution at 4°C. The following day the sections were rinsed in 1× TBS, three times for 5 minutes, then incubated with secondary antibodies conjugated with CY3 or fluorescein isothiocyanate. Sections were mounted in Mowiol and analyzed using a Zeiss Axiophot confocal microscope.
Immunogold Labeling
Tissues were prepared as for immunohistochemistry and then cut into 30-μm sections and incubated in 0.1% bovine serum albumin/1× TBS/10% goat serum for at least 30 minutes. The sections were incubated overnight with primary anti-fibrocystin antibody (1:50) diluted in blocking solution. The sections were washed in high-salt TBS before adding the gold-conjugated secondary antibody (1:20) and incubated overnight at 4°C. Sections were washed six times in 1× TBS throughout a period of 1 hour and then fixed in 1.5% gluteraldehyde/100 mmol/L cacodylate buffer/5% sucrose for 1 hour. The samples were washed again in 1× TBS, three times for 5 minutes. Palade’s osmium tetroxide solution (veronal acetate-buffered 1% osmium tetroxide) was added for 1 hour on ice in a dark chamber, washed in distilled water and then immersed in veronal acetate buffer for 1 hour. The sections were washed in 1× TBS, three times for 5 minutes then dehydrated and embedded. Ultra-thin 80-nm sections were cut and stained with uranyl acetate and lead citrate and examined with a Zeiss EM910 electron microscope.
Serum Biochemical Analysis
Adult mice were anesthetized by an intraperitoneal injection of pentobarbital sodium and weighed. Blood samples were obtained by cardiac puncture, and serum was prepared by centrifugation at 6000 rpm for 10 minutes at 4°C and frozen. All serum biochemical measurements were performed in the National Institute of Diabetes and Digestive and Kidney Disease-funded Yale Mouse Phenotyping Core (Yale University, New Haven, CT).
Statistical Analysis
The Student’s t-test was used to determine the statistical differences between various experimental and control groups. The difference was considered statistically significant when *P < 0.05 or **P < 0.01.
Results
Generation of Pkhd1del4 Mice
A targeting construct was designed to inactivate Pkhd1 in the mouse by disrupting exon 4 of the gene (Supplementary Figure 1, see http://ajp.amjpathol.org). This mutation resulted in a deletion of the splice donor site of exon 4; skipping of exon 4 would produce an out-of-frame splice product from exon 3 to exon 5. Mice homozygous for the mutant Pkhd1del4 alleles were born on a mixed C57BL6/129Sv background with expected Mendelian ratio (104 of 441; 23.5%). Pkhd1del4/del4 mice grew and developed normally and lived to at least 16 months of age.
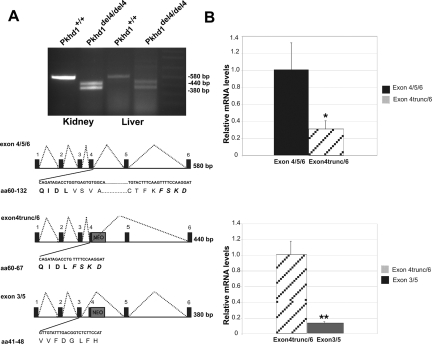
mRNA transcripts were analyzed from kidneys and livers of both control and Pkhd1del4/del4 animals in the region encompassing exons 1 to 6 using RT-PCR. The expected 580-bp product from amplification of the wild-type allele was observed in control mice; however, amplification of the mutant allele from tissues of Pkhd1del4/del4 mice yielded the 380-bp product expected from exon 4 skipping, as well as a 440-bp product (Figure 1A). Sequencing confirmed that the 380-bp product represents the exon 3 to 5 out-of-frame transcript (exon 3/5) leading to premature termination. The 440-bp band was the result of activation of a cryptic donor splice site in exon 4 and represented an in-frame transcript from within exon 4 to the consensus acceptor splice site of exon 6. Sequence analysis shows that the predicted protein product contains a 66-amino acid deletion (exon 4trunc/6) (Figure 1A). Quantitative real-time PCR showed that the expression of the mutant exon 4trunc/6 in-frame product in Pkhd1del4/del4 kidneys was ∼30% of the exon 4/5/6 product in wild type kidneys (Figure 1B, top). Furthermore, expression of the mutant exon 4trunc/6 in-frame product was 10-fold higher than that of the smaller out-of-frame exon 3/5 product in mutant mouse kidneys (Figure 1B, bottom). The latter finding may result from preferential degradation of the out-of-frame product by nonsense-mediated mRNA decay.
Figure 1.
Analysis of the Pkhd1 transcript in Pkhd1del4/del4 kidney and liver tissues. A: Detection of an alternatively spliced transcript in the Pkhd1del4/del4 mice. RT-PCR products were amplified using primers designed against exon 1 (forward primer) and reverse primer in exon 6. Schematic diagram of the products generated in the Pkhd1del4/del4 mice compared to the wild-type mice. The insertion of the Neo cassette in exon 4 is represented by the gray box. The 580-bp product represents the wild-type transcript exon 4/5/6 encompassing amino acids 60 to 132. The bold amino acids represent the coding region from exon 4, and the bold italicized amino acids represent the coding region from exon 6. The 440-bp product represents the exon 4trunc/6 transcript lacking exon 5, deleting 66 amino acids. The smaller 380-bp PCR fragment producing the exon 3/5 transcript resulting in a frameshift product. B: Quantitative RT-PCR analysis of kidney samples from wild-type and Pkhd1del4/del4 mice. The top panel uses an exon 4-specific TaqMan probe (Supplementary Table 1, see http://ajp.amjpathol.org) to amplify the exon 4-containing products from both wild-type (exon 4/5/6) and mutant mice (exon 4trunc/6) to show ∼70% reduction of the mutant in-frame transcript compared to wild-type. The bottom panel indicates that the level of expression of the out-of-frame exon 3/5 transcript is ∼10% of the level of the in-frame exon 4trunc/6 transcript in Pkhd1del4/del4 kidneys. Differences were significant P < 0.05 (*) and P < 0.005 (**) by paired Student’s t-test. For each data point, n = 4.
Pkhd1del4/del4 Mice Develop Liver Cysts Accompanied by Portal Tract Fibrosis
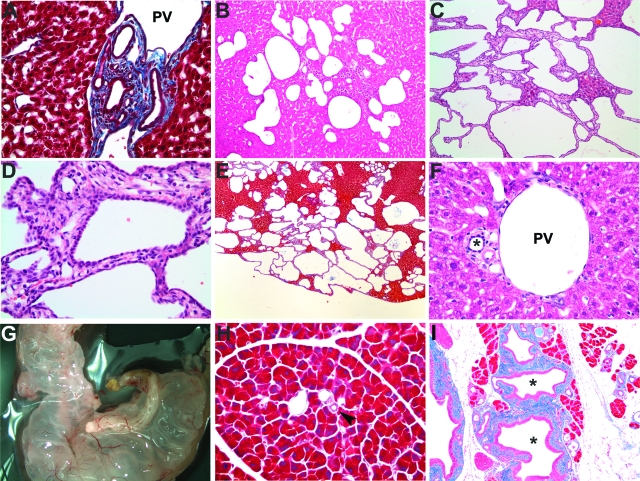
Cohorts of Pkhd1del4/del4 and control mice were studied serially at 2 weeks and at 3, 6, 9, and 12 months. Intrahepatic bile duct proliferation, with features reminiscent of von Meyenburg complexes and ductal plate malformation with bile ducts surrounding the portal vein, was present as early as 2 weeks (Figure 2A). After 3 months, the Pkhd1del4/del4 mice exhibited progressive liver cyst formation, accompanied by periportal fibrosis (Figure 2, B–E). Synthetic and biliary clearance functions of the liver, as measured by total and direct bilirubin, albumin, alanine aminotransferase, and alkaline phosphatase, were not significantly different between wild-type and Pkhd1del4/del4 mice (Supplementary Table 1, see http://ajp.amjpathol.org). Hence, despite the progressive bile duct-derived cystic liver disease, normal hepatic functions were maintained for up to 12 months in Pkhd1del4/del4 mice.
Figure 2.
Histological analysis of Pkhd1del4/del4 livers and pancreas. A: Mallory trichrome staining of 2-week-old Pkhd1del4/del4 liver. B–D: H&E staining of Pkhd1del4/del4 liver at 3 months (B) and at 12 months (C, D). E: Mallory trichrome staining of Pkhd1del4/del4 liver at 12 months. F: H&E staining of 12-month-old wild-type bile ducts indicated by the asterisk. G: Grossly cystic pancreas from a 3-month-old Pkhd1del4/del4 animal. H and I: Mallory trichrome staining of the pancreas from 6-month-old wild-type (H) and Pkhd1del4/del4 (I) mice showing pancreatic ductal dilatation in mutant mice. The pancreatic ducts are highlighted by an arrowhead in H and by an asterisk in I. PV, portal vein. Original magnifications: ×400 (A, D, F); ×100 (B, C, H, I); ×40 (E).
Extrahepatic Manifestations of the Pkhd1del4 Mutation
Additionally, prominent extrahepatic manifestations involving the pancreas, common bile duct, and spleen were observed in Pkhd1del4/del4 mice. The majority of Pkhd1del4/del4 mice showed dilatation of the pancreatic ducts, and 10% (9 of 89) developed large cysts in the pancreas beginning as early as 1 month of age (Figure 2G). The dilated pancreatic ducts were lined with tall columnar mucinous epithelia (stained positive for mucin, data not shown) and showed significant periductal fibrosis (Figure 2I). Grossly cystic common bile ducts were present in 27% (24 of 89) of the mice by 9 months of age. The frequency of this manifestation was age-dependent with 38% (21 of 55) of mice aged 9 months or older having cystic common bile ducts. Splenomegaly was present in ∼50% of the Pkhd1del4/del4 cohorts and began as early as 3 months, perhaps indicating evolving portal hypertension (Table 1).
Table 1.
Absolute Organ Weights (g) of Age-Matched Pkhd1del4/del4 Mice and Their Littermate Controls
| Organ | 3 months
|
6 months
|
12 months
|
|||
|---|---|---|---|---|---|---|
| Control (n) | Pkhd1del4/del4 (n) | Control (n) | Pkhd1del4/del4 (n) | Control (n) | Pkhd1del4/del4 (n) | |
| Liver | 1.0 ± 0.0 (12) | 1.5 ± 0.1 (13)** | 1.2 ± 0.1 (8) | 1.3 ± 0.1 (16) | 1.4 ± 0.1 (18) | 3.5 ± 0.3 (37)** |
| Spleen | 0.108 ± 0.01 (9) | 0.164 ± 0.028 (10) | 0.084 ± 0.006 (7) | 0.222 ± 0.045 (16)* | 0.115 ± 0.012 (18) | 0.358 ± 0.049 (36)** |
| Kidney | 0.271 ± 0.039 (12) | 0.289 ± 0.016 (12) | 0.353 ± 0.026 (8) | 0.33 ± 0.018 (16) | 0.35 ± 0.02 (14) | 0.5 ± 0.022 (36)* |
Mean values are accompanied by the standard error; the number of mice per study is noted in parentheses.
P < 0.05,
P < 0.005.
Histological examination of kidneys revealed normal glomerular and tubular structures up to 12 months of age, with no apparent renal cysts. No significant elevation in blood urea nitrogen (BUN) was detected in 12-month-old homozygous Pkhd1del4 mice, indicating preserved renal function (Supplementary Table 1, see http://ajp.amjpathol. org). Pkhd1del4/del4 mice backcrossed onto a congenic 129/REJ background developed similar phenotypes in the liver and pancreas as those seen on the mixed background. Likewise, the kidneys of 129/REJ mice appeared histologically normal up to 12 months of age. Therefore, at least on these genetic backgrounds, we observed no effects of genetic background either on the development of liver and pancreatic abnormalities or on the absence of a kidney phenotype.
Fibrocystin Is Found at the Apical Membrane and Cilia
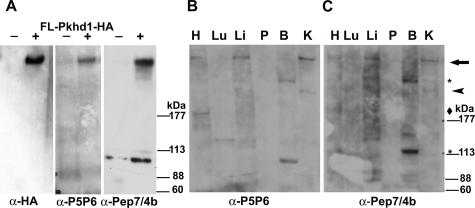
Given the presence of an in-frame cryptic splice variant of Pkhd1 mRNA resulting from the introduced mutation, the expression pattern of fibrocystin was investigated in tissues from Pkhd1del4/del4 mice. In addition to the previously described P5/P6 antibody directed against residues 3252 to 3266 (P5) and 3774 to 3785 (P6) of human fibrocystin,16 the polyclonal antibody pep7/4b, directed against amino acids 2694 to 2894 in the extracellular domain of mouse fibrocystin, was generated and characterized. The specificity of the pep7/4b antibody was evaluated by immunostaining COS7 cells transiently transfected with the dual epitope-tagged fibrocystin under permeabilized conditions (Supplementary Figure 2, see http://ajp.amjpathol.org). In addition, using P5P6 and pep7/4b antisera, the same >400-kDa band was recognized on immunoblots of cells transfected with a full-length human fibrocystin with a C-terminal HA epitope tag (Figure 3A).
Figure 3.
Characterization of polyclonal fibrocystin antisera by immunoblot analysis. A: Immunoblot of COS7 cells transiently transfected with the full-length epitope-tagged Pkhd1 cDNA (+, Fl-Pkhd1-HA) or vector only (−). The blot was initially probed with polyclonal anti-fibrocystin P5P6 and then reprobed, after stripping, with polyclonal anti-HA. The same differentially expressed transfect protein migrating at >400-kDa band was detected by anti-HA, P5P6, and pep7/4b antisera in the transfected sample lane. B and C: Immunoblot with polyclonal anti-P5P6 (B) and a parallel immunoblot with the polyclonal anti-Pep7/4b (C). The slowest migrating band (arrow) detected by both Pep7/4b and P5P6 antisera in kidney, liver, and heart ran at a migration that it is in the >400-kDa range (B, C). Smaller bands were likewise detected by both antisera independently in the various tissues, suggesting that these are true fibrocystin variant species. They are indicated by the arrowhead for the ∼250-kDa band detected in kidney, diamond for the ∼200-kDa band in the heart, and asterisk for the two bands, ∼280 kDa and ∼120 kDa, seen in the brain. H, heart; Lu, lung; Li, liver; P, pancreas; B, brain; K, kidney.
Next, the pattern of expression of fibrocystin protein products in multiple mouse tissues was defined using both P5P6 and the novel pep7/4b antibodies. The novel pep7/4b antibody detected the same >400-kDa band as the P5P6 antisera in membrane protein fractions of mouse kidney, liver, and heart extracts (Figure 3, B and C). Migration of this full-length fibrocystin in tissue was similar to that of full-length fibrocystin in transfected cell lines (Figure 3A). Additional faster-migrating bands were detected using both the P5P6 and pep7/4b antisera in a tissue-specific manner using independent immunoblots (Figure 3, B and C). Pep7/4b and P5P6 antisera recognized an ∼250-kDa band in the kidney, as well as an ∼200-kDa band in the heart. Two additional bands of ∼280 kDa and 120 kDa were detected by both antisera in mouse brain. The finding that independently derived polyclonal antisera recognize the same pattern of fragments in these tissues strongly supports the presence of tissue-specific variants of fibrocystin.
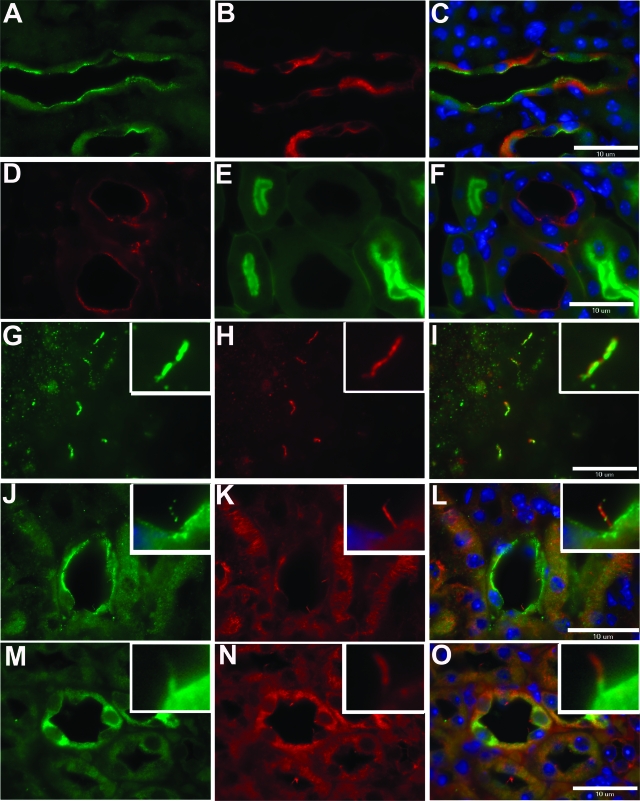
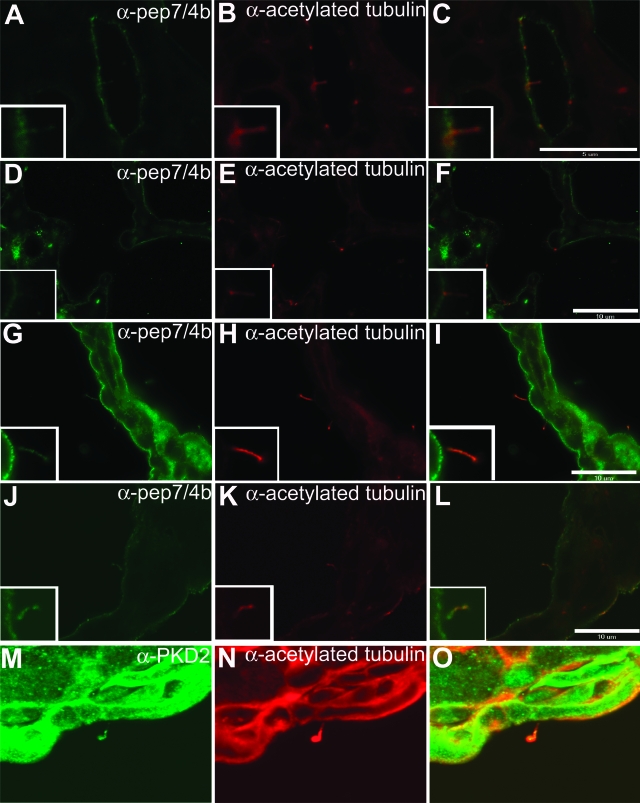
Immunohistochemical labeling of adult mouse kidney using pep7/4b showed that fibrocystin is expressed at the apical membranes of renal collecting ducts as indicated by co-labeling with Dolichos biflorus agglutinin (Figure 4, A–C). Fibrocystin is not expressed in proximal tubules (Figure 4, D–F). Fibrocystin expression is also detectable in Tamm-Horsfall-positive cells of the thick ascending limb of Henle’s loop (data not shown). In addition to the apical membrane expression, native fibrocystin immunoreactivity co-localizes with the acetylated α-tubulin signal in the primary cilia of polarized IMCD3 cells grown in culture (Figure 4, G–I) and of normal kidney tubule cells in kidney sections (Figure 4, J–L). Mutant fibrocystin in the Pkhd1del4/del4 kidney showed qualitatively reduced apical membrane and cilia expression, but relatively increased cytosolic expression (Figure 4, M–O).
Figure 4.
Fibrocystin is expressed at the apical plasma membrane and in cilia in kidney tissue. Endogenous fibrocystin is located at the apical plasma membrane in wild-type renal collecting ducts (A) co-localizing with Dolichos biflorous agglutinin (B, C). Fibrocystin (D) is not detected in proximal tubule segments stained with Lotus tetragonolobus lectin (E, F). Endogenous fibrocystin (G, J) co-localizes with acetylated α-tubulin in IMCD3 cells (H, I) and in collecting ducts (K, L). Mutant fibrocystin is detected in Pkhd1del4/del4 kidneys, with a predominant cytoplasmic location and some protein trafficking to the apical and ciliary membrane (M–O). A, D, G, J, M: Anti-fibrocystin pep7/4b; C, F, I, L, O: merged images. Scale bars = 10 μm.
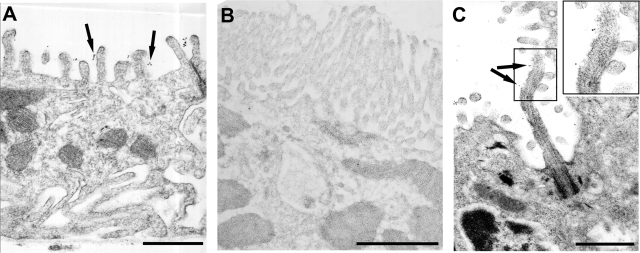
Immunogold electron microscope labeling using the pep7/4b directed toward predicted extracellular domain of fibrocystin showed gold particles on the luminal side of the apical plasma membrane in microvilli of intercalated cells (Figure 5A) and principal cells (data not shown). Proximal tubule cells found in the same section and distinguished by the presence of a brush border, were completely devoid of gold particles (Figure 5B). In aggregate, these findings confirm the exclusive expression of fibrocystin in distal nephron segments and collecting ducts, the localization of fibrocystin in the apical plasma membrane, and the topology of the putative extracellular domain. Immunogold electron microscopy in the liver showed fibrocystin-positive gold grains along the extracellular aspect of the ciliary shaft and microvilli of normal bile duct epithelial cells (Figure 5C), demonstrating fibrocystin also traffics to primary cilia.
Figure 5.
Immunogold-labeled fibrocystin is found at the apical membranes of renal collecting ducts and alongside cholangiocyte cilia. A: Immunogold-labeled anti-pep7/4b particles in the intercalated cell appear on the extracellular aspect of the apical microvillus border (arrows). B: Proximal tubule from the same electron microscope grid shown with brush border completely devoid of anti-fibrocystin immunogold particles. C: Anti-fibrocystin immunogold particles alongside the extracellular aspect of a cholangiocyte cilium as well as along the microvilli (arrows). Scale bars =1 μm.
Expression of Fibrocystin in Bile Ducts of Mouse Models of Polycystic Liver Diseases
Fibrocystin expression was analyzed in the bile duct cells of Pkhd1del4/del4 mice, as well as in mouse models of polycystic liver disease because of mutations in Pkd1 and Pkd2. Mutant fibrocystin is detected in the apical membranes and cilia of distal nephron segments in Pkhd1del4/del4 mice (Figure 4, M–O). Immunostaining against fibrocystin revealed that the respective wild-type and mutant fibrocystin proteins are detectable in the apical membranes and cilia of biliary epithelia of wild-type and Pkhd1del4/del4 cystic livers (Figure 6, A–F). However, the ciliary expression of fibrocystin in cyst-lining bile duct epithelia of the liver was significantly reduced, with fibrocystin labeling noted in 134 of 161 acetylated α-tubulin-positive cilia of wild-type cholangiocytes and in 66 of 180 of mutant cholangiocytes (χ2 = 72.98, P < 0.001). These data are consistent with the quantitative reduction in stable in-frame fibrocystin transcript and qualitative reduction in protein expression in kidney tubules noted earlier.
Figure 6.
Trafficking of fibrocystin and PC2 in cyst-lining bile duct epithelia of ADPKD and Pkhd1del4/del4 mouse models. A-C: Fibrocystin is expressed at the apical membranes and cilia of wild-type hepatic bile ducts. D–F: In Pkhd1del4/del4 mice, mutant fibrocystin can still be detected in the apical membranes and cilia of cyst-lining epithelia in the liver. G–L: Immunohistochemical analysis of wild-type fibrocystin expression in cyst-lining epithelia of Pkd1 cystic kidney (G–I) and Pkd2 cystic liver (J–L) reveals normal-appearing expression of fibrocystin in apical membranes and cilia in these cystic tissues. Cilia were identified by co-staining with acetylated α-tubulin (B, E, H, K). PC2 is expressed normally in cilia of cyst-lining biliary epithelia of Pkhd1del4/del4 mice. Original magnification, ×240 (M–O).
Finally, we investigated whether fibrocystin expression was altered in cystic epithelia resulting from loss of the Pkd1 (Figure 6, G–I) or Pkd2 (Figure 6, J–L). Using the pep7/4b antibody, we found that fibrocystin remained expressed in the apical membranes and the cilia of bile duct cyst-lining cells in both mouse models. Conversely, we investigated expression of the Pkd2 gene product polycystin-2 (PC2) in the cystic bile duct epithelia of Pkhd1del4/del4 livers and found that the expression of PC2 in cilia was not altered (Figure 6, M–O). These data indicate that, at least at the level of resolution provided by tissue immunofluorescence, expression and trafficking of fibrocystin is independent of PC1 and PC2. Furthermore, PC2 is normally expressed in liver tissue despite the severe biliary dysgenesis resulting from mutant fibrocystin.
Discussion
We have generated a mouse model that develops biliary dysgenesis accompanied by periportal fibrosis as a result of mutation in Pkhd1. Despite the progressive liver disease, these mice are viable at 12 months of age with no apparent decline in synthetic liver function, hence making it a good model to study the biliary dysgenesis associated with ARPKD. These mice also develop extrahepatic phenotypes involving the pancreas, extrahepatic bile ducts, and spleen, which occur in a more variable manner owing to either variable expressivity or possible genetic background effects.
Pancreatic ductal ectasia was noted in the majority of the Pkhd1del4/del4 mice studied; however, only 10% of the mice developed gross pancreatic cysts. Pancreatic abnormalities are not commonly associated with the human form of ARPKD, nor have they been detected in orthologous rodent models, including the pck rat and a previously reported mouse model targeting exon 40 of Pkhd1.23,24 However, as in Pkhd1del2,25 Pkhd1del3–4 26 and other cilia-related fibrocystic disease models including Pkd1,27 Pkd2,28 cpk,29 jcpk,30 orpk,31,32 and inv,33 pancreatic cysts were also detected in the Pkhd1del4 mutant model. It is possible that the pancreatic phenotype in mice reflects some species variation because this presentation differs from human ARPKD and the pck rat model, in which kidney and liver phenotypes predominate and the pancreas does not appear to be affected.23
The presence of splenomegaly in a large percentage of the mice may be secondary to portal hypertension,34,35 which is commonly associated with hepatic fibrosis and is a clinical manifestation of older patients with ARPKD.5 Extrahepatic bile duct dilatation was seen in more than 25% of the mice studied but increased in prevalence to ∼40% in mice 9 months of age and older. Intrahepatic ductal dilation is mostly associated with ARPKD and Caroli’s disease36; however, more recent clinical studies have shown that extrahepatic bile duct dilation can also be a prominent clinical feature in ARPKD patients.37 Fibrocystin is expressed in rat extrahepatic bile duct and it has been suggested that Pkhd1 plays a role in regulating the luminal diameter of the extrahepatic bile duct.37 The intra- and extrahepatic bile ducts and the pancreatic ducts descend from the same progenitor.38,39 The commonality of the fibrocystin-dependent phenotype we find in these tissues suggests that fibrocystin functions correspondingly in all three tissues of common embryonic origin.
Surprisingly, the kidneys, the organs predominantly affected in ARPKD, appear to be normal both at the functional and histological levels in Pkhd1del4 mice. Mutation analyses of ARPKD patients included individuals with predominant liver disease manifest as congenital hepatic fibrosis or Caroli’s disease and only mild renal involvement.6,7,10,12,13,14,40 In such families, the mutations invariably include at least one predicted amino acid substitution that may act in a hypomorphic manner. In the case of the Pkhd1del4 mouse model, the transcript bears a deletion of 66 amino acids that includes part of exon 4 and all of exon 5. The mutant protein is presumed to act in a hypomorphic manner because the mouse retains normal renal tubular structure and function yet has abnormalities in the liver and pancreas. Interestingly, another published Pkhd1 mouse model, generated by disrupting exon 40, also yielded an in-frame product with the loss of amino acids 2160 to 2223 and resulted in cystic liver disease and portal tract fibrosis but no pancreatic or kidney abnormalities.24 Two other murine models, Pkhd1del2 25 and Pkhd1del3–4 26 develop renal cysts but only in aged mice; none of the orthologous models recapitulate the severe neonatal phenotype seen in humans suggesting the possibility that species differences may play a role in the relative resistance of mouse kidneys to cyst formation because of mutation in Pkhd1. The location of the mutations in the genes may also affect the severity of the disease, hence the clinical lesion in mice, and this is borne out in human studies in which genotype-phenotype interactions have been reported.11,12,13,14 Human mutations in the region of the Pkhd1del4 mutation have resulted in severe loss of function. For example, one mutation, IVS5 + 1G→T, predicted to result in loss of the exon 5 splice donor site causing aberrant splicing of exon 5 was found in a child with the severe perinatal lethal presentation associated with presumed complete loss-of-function alleles.12
In our mouse model, targeting of the 5′region of the Pkhd1 gene generated the predicted mutant transcript and an unexpected in-frame splice variant. Alternative splice forms of the PKHD1 gene have been reported in both human6 and mouse15 tissues. In the mouse, it has been shown by Northern blot analysis and in situ hybridization that various-sized transcripts were recognized by different exon-containing probes in a tissue-specific manner. However, exon 5- and exon 41-containing transcripts were most abundant in the kidney although their expression differed in other tissues.15 Our data would suggest that exon 5 is not essential for fibrocystin function in the mouse kidney.
At the protein level, we analyzed fibrocystin expression using two independently generated polyclonal antisera directed against extracellular epitopes. These antisera gave similar patterns of expression in multiple tissues. Fibrocystin antibodies, pep7/4b and P5P6, recognized a band migrating at >400 kDa in the kidney, liver, and heart. In addition, we identified smaller products that were common to both antisera that appear in a tissue-dependent manner in the kidney, liver, heart, lung, and brain. These may represent protein products of tissue-specific alternate transcripts or posttranslational processing such as proteolytic cleavage. Because the level of fibrocystin protein expression may be developmentally regulated, Pkhd1del4 mice of various ages were evaluated ranging from postnatal day 3 up to 1 month of age. We failed to detect the mutant protein by immunoblot analysis with either of the fibrocystin-specific antibodies. Coupled with the reduced intensity of expression observed in immunohistochemistry in both kidney and liver, it is likely that mutant fibrocystin has reduced stability and steady-state expression because of improper folding and trafficking.
Using co-localization with nephron segment markers, we found fibrocystin expression is restricted to the distal nephron including the thick ascending limb of Henle’s loop and the collecting duct.16,17,21 In the liver, fibrocystin is expressed in intra- and extrahepatic bile ducts. This expression pattern is consistent with the histopathological phenotype of ARPKD. We found the subcellular location of fibrocystin is in the apical plasma membrane and ciliary membrane in kidney tubule and bile duct cells.16,17,19,21 In the pck rat, mutant fibrocystin is expressed in cystic epithelium. However, it does not reach the cilia, suggesting that improper trafficking of the protein may lead to cyst formation.19 In the liver cyst-lining epithelium of the Pkhd1del4/del4 mouse, fibrocystin was detected both at the apical membrane and in cilia. Protein dosage effects and the presence of a hypomorphic product may result in cystogenesis because of a possible functional threshold effect. It has been shown that whereas embryonic lethality occurs with complete Pkd1 inactivation in mice,27 the reduction in Pkd1 gene dosage is sufficient to initiate cystogenesis in the kidney.41,42 Reduced functional protein may be sufficient for the kidney to retain normal structure, but not the liver or pancreas. Either reduced levels below a certain threshold or reduced or lost function despite proper organellar location may underlie the biliary dysgenesis in our model.
Many of the cystic disease-related proteins have been localized to cilia or basal bodies. PC1 and PC2 have been shown to interact via their C termini43 and play a role in sensing fluid flow and transducing Ca2+-dependent signals from the cilium to the cell.44 Fibrocystin is localized to the cilium and has been recently found in the same protein complex as PC2.18,45 This suggests that fibrocystin and the autosomal dominant disease proteins may share a common pathway. We addressed the question whether lack of PC1 or PC2 might influence location or level of fibrocystin expression by examining fibrocystic expression in orthologous cystic models because of mutation in either Pkd1 or Pkd2. Fibrocystin traffics to the apical membrane and primary cilia in bile duct cystic tissue lacking PC1 or PC2. Conversely, at least for PC2, expression and trafficking is independent of fibrocystin mutation in cystic bile ducts. These data suggest that fibrocystin and polycystin functions are not interrelated at the level of protein trafficking and cilia location.
In aggregate, we have generated a mouse model that resembles the presentation of ARPKD predominated by congenital hepatic fibrosis. Our data suggest that the Pkhd1del4 allele results in insufficient functional protein, at least in bile duct and pancreatic duct epithelia, despite trafficking to the apical membrane and cilia. The Pkhd1del4 allele provides a useful model to study the pathogenesis of the biliary dysgenesis associated with ARPKD, and combination of this allele with other hypomorphic fibrocystin alleles may shed further light on the role of mouse fibrocystin in kidney cyst development.
Acknowledgments
We thank Sue Ann Mentone for her technical expertise with the electron microscope, Dayne Okuhara for statistical analysis, Howard Crawford for critically reading the manuscript, and Carlo Spirli and Luca Fabris (University of Padova, Padova, Italy) for helpful suggestions.
Footnotes
Address reprint requests to Stefan Somlo, Yale University School of Medicine, Section of Nephrology, P.O. Box 208029, 333 Cedar St., New Haven, CT 06520. E-mail: stefan.somlo@yale.edu.
Supported by the Polycystic Kidney Disease Foundation (grant 77aR2 to A.R.G.), the National Institutes of Health (training grant T32 DK007276 to E.L.E.), the Kidney and Urology Foundation of America (to A.R.G.), and the Joseph LeRoy and Ann C. Warner Fund (to S.S.).
A.-R.G. and E.L.E. contributed equally to this article.
Supplemental material for this article can be found on http://ajp. amjpathol.org.
Current address of E.L.E.: INSERM U574, Hopital Necker Enfants-Malades, Paris V University, Paris, France.
References
- Zerres K, Mucher G, Becker J, Steinkamm C, Rudnik-Schoneborn S, Heikkila P, Rapola J, Salonen R, Germino GG, Onuchic LF, Germino GG, Onuchic L, Somlo S, Avner ED, Harman LA, Stockwin JM, Guay-Woodford LM. Prenatal diagnosis of autosomal recessive polycystic kidney disease (ARPKD): molecular genetics, clinical experience, and fetal morphology. Am J Med Genet. 1998;76:137–144. [PubMed] [Google Scholar]
- Kaplan BS, Fay J, Shah V, Dillon MJ, Barratt TM. Autosomal recessive polycystic kidney-disease. Pediatr Nephrol. 1989;3:43–49. doi: 10.1007/BF00859625. [DOI] [PubMed] [Google Scholar]
- Roy S, Dillon MJ, Trompeter RS, Barratt M. Autosomal recessive polycystic kidney disease: long-term outcome of neonatal survivors. Pediatr Nephrol. 1997;11:302–306. doi: 10.1007/s004670050281. [DOI] [PubMed] [Google Scholar]
- Fonck C, Chauveau D, Gagnadoux MF, Pirson Y, Grunfeld JP. Autosomal recessive polycystic kidney disease in adulthood. Nephrol Dial Transplant. 2001;16:1648–1652. doi: 10.1093/ndt/16.8.1648. [DOI] [PubMed] [Google Scholar]
- Guay-Woodford LM, Muecher G, Hopkins SD, Avner ED, Germino GG, Guillot AP, Herrin J, Holleman R, Irons DA, Primack W. The severe perinatal form of autosomal recessive polycystic kidney disease maps to chromosome 6p21.1-p12: implications for genetic counseling. Am J Hum Genet. 1995;56:1101–1107. [PMC free article] [PubMed] [Google Scholar]
- Onuchic LF, Furu L, Nagasawa Y, Hou X, Eggermann T, Ren Z, Bergmann C, Senderek J, Esquivel E, Zeltner R, Rudnik-Schoneborn S, Mrug M, Sweeney W, Avner ED, Zerres K, Guay-Woodford LM, Somlo S, Germino GG. PKHD1, the polycystic kidney and hepatic disease 1 gene, encodes a novel large protein containing multiple immunoglobulin-like plexin-transcription-factor domains and parallel beta-helix 1 repeats. Am J Hum Genet. 2002;70:1305–1317. doi: 10.1086/340448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward CJ, Hogan MC, Rossetti S, Walker D, Sneddon T, Wang X, Kubly V, Cunningham JM, Bacallao R, Ishibashi M, Milliner DS, Torres VE, Harris PC. The gene mutated in autosomal recessive polycystic kidney disease encodes a large, receptor-like protein. Nat Genet. 2002;30:259–269. doi: 10.1038/ng833. [DOI] [PubMed] [Google Scholar]
- Hogan MC, Griffin MD, Rossetti S, Torres VE, Ward CJ, Harris PC. PKHDL1, a homolog of the autosomal recessive polycystic kidney disease gene, encodes a receptor with inducible T lymphocyte expression. Hum Mol Genet. 2003;12:685–698. [PubMed] [Google Scholar]
- Kaimori JY, Nagasawa Y, Menezes LF, Garcia-Gonzalez MA, Deng J, Imai E, Onuchic LF, Guay-Woodford LM, Germino GG. Polyductin undergoes notch-like processing and regulated release from primary cilia. Hum Mol Genet. 2007;16:942–956. doi: 10.1093/hmg/ddm039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann C, Senderek J, Kupper F, Schneider F, Dornia C, Windelen E, Eggermann T, Rudnik-Schoneborn S, Kirfel J, Furu L, Onuchic LF, Rossetti S, Harris PC, Somlo S, Guay-Woodford L, Germino GG, Moser M, Buttner R, Zerres K. PKHD1 mutations in autosomal recessive polycystic kidney disease (ARPKD). Hum Mutat. 2004;23:453–463. doi: 10.1002/humu.20029. [DOI] [PubMed] [Google Scholar]
- Bergmann C, Senderek J, Sedlacek B, Pegiazoglou I, Puglia P, Eggermann T, Rudnik-Schoneborn S, Furu L, Onuchic LF, De Baca M, Germino GG, Guay-Woodford L, Somlo S, Moser M, Buttner R, Zerres K. Spectrum of mutations in the gene for autosomal recessive polycystic kidney disease (ARPKD/PKHD1). J Am Soc Nephrol. 2003;14:76–89. doi: 10.1097/01.asn.0000039578.55705.6e. [DOI] [PubMed] [Google Scholar]
- Furu L, Onuchic LF, Gharavi A, Hou X, Esquivel EL, Nagasawa Y, Bergmann C, Senderek J, Avner E, Zerres K, Germino GG, Guay-Woodford LM, Somlo S. Milder presentation of recessive polycystic kidney disease requires presence of amino acid substitution mutations. J Am Soc Nephrol. 2003;14:2004–2014. doi: 10.1097/01.asn.0000078805.87038.05. [DOI] [PubMed] [Google Scholar]
- Rossetti S, Torra R, Coto E, Consugar M, Kubly V, Malaga S, Navarro M, El-Youssef M, Torres VE, Harris PC. A complete mutation screen of PKHD1 in autosomal-recessive polycystic kidney disease (ARPKD) pedigrees. Kidney Int. 2003;64:391–403. doi: 10.1046/j.1523-1755.2003.00111.x. [DOI] [PubMed] [Google Scholar]
- Sharp AM, Messiaen LM, Page G, Antignac C, Gubler MC, Onuchic LF, Germino GG, Guay-Woodford LM. Comprehensive genomic analysis of PKHD1 mutations in ARPKD cohorts. J Med Genet. 2005;42:336–349. doi: 10.1136/jmg.2004.024489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagasawa Y, Matthiesen S, Onuchic LF, Hou X, Bergmann C, Esquivel E, Senderek J, Ren Z, Zeltner R, Furu L, Avner E, Moser M, Somlo S, Guay-Woodford L, Buttner R, Zerres K, Germino GG. Identification and characterization of Pkhd1, the mouse orthologue of the human ARPKD gene. J Am Soc Nephrol. 2002;13:2246–2258. doi: 10.1097/01.asn.0000030392.19694.9d. [DOI] [PubMed] [Google Scholar]
- Ward CJ, Yuan D, Masyuk TV, Wang X, Punyashthiti R, Whelan S, Bacallao R, Torra R, Larusso NF, Torres VE, Harris PC. Cellular and subcellular localization of the ARPKD protein; fibrocystin is expressed on primary cilia. Hum Mol Genet. 2003;12:2703–2710. doi: 10.1093/hmg/ddg274. [DOI] [PubMed] [Google Scholar]
- Menezes LF, Cai Y, Nagasawa Y, Silva AM, Watkins ML, Da Silva AM, Somlo S, Guay-Woodford LM, Germino GG, Onuchic LF. Polyductin, the PKHD1 gene product, comprises isoforms expressed in plasma membrane, primary cilium, and cytoplasm. Kidney Int. 2004;66:1345–1355. doi: 10.1111/j.1523-1755.2004.00844.x. [DOI] [PubMed] [Google Scholar]
- Wang S, Zhang J, Nauli SM, Li X, Starremans PG, Luo Y, Roberts KA, Zhou J. Fibrocystin/polyductin, found in the same protein complex with polycystin-2, regulates calcium responses in kidney epithelia. Mol Cell Biol. 2007;8:3241–3252. doi: 10.1128/MCB.00072-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masyuk TV, Huang BQ, Ward CJ, Masyuk AI, Yuan D, Splinter PL, Punyashthiti R, Ritman EL, Torres VE, Harris PC, Larusso NF. Defects in cholangiocyte fibrocystin expression and ciliary structure in the PCK rat. Gastroenterology. 2003;125:1303–1310. doi: 10.1016/j.gastro.2003.09.001. [DOI] [PubMed] [Google Scholar]
- Wang S, Luo Y, Wilson PD, Witman GB, Zhou J. The autosomal recessive polycystic kidney disease protein is localized to primary cilia, with concentration in the basal body area. J Am Soc Nephrol. 2004;15:592–602. doi: 10.1097/01.asn.0000113793.12558.1d. [DOI] [PubMed] [Google Scholar]
- Zhang MZ, Mai W, Li C, Cho SY, Hao C, Moeckel G, Zhao R, Kim I, Wang J, Xiong H, Wang H, Sato Y, Wu Y, Nakanuma Y, Lilova M, Pei Y, Harris RC, Li S, Coffey RJ, Sun L, Wu D, Chen XZ, Breyer MD, Zhao ZJ, McKanna JA, Wu G. PKHD1 protein encoded by the gene for autosomal recessive polycystic kidney disease associates with basal bodies and primary cilia in renal epithelial cells. Proc Natl Acad Sci USA. 2004;101:2311–2316. doi: 10.1073/pnas.0400073101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mai WY, Chen D, Ding TB, Kim IY, Park SJ, Cho SY, Chu JSF, Liang D, Wang N, Wu DQ, Li S, Zhao P, Zent R, Wu GQ. Inhibition of Pkhd1 impairs tubulomorphogenesis of cultured IMCD cells. Mol Biol Cell. 2005;16:4398–4409. doi: 10.1091/mbc.E04-11-1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lager DJ, Qian Q, Bengal RJ, Ishibashi M, Torres VE. The pck rat: a new model that resembles human autosomal dominant polycystic kidney and liver disease. Kidney Int. 2001;59:126–136. doi: 10.1046/j.1523-1755.2001.00473.x. [DOI] [PubMed] [Google Scholar]
- Moser M, Matthiesen S, Kirfel J, Schorle H, Bergmann C, Senderek J, Rudnik-Schoneborn S, Zerres K, Buettner R. A mouse model for cystic biliary dysgenesis in autosomal recessive polycystic kidney disease (ARPKD). Hepatology. 2005;41:1113–1121. doi: 10.1002/hep.20655. [DOI] [PubMed] [Google Scholar]
- Woollard JR, Punyashtiti R, Richardson S, Masyuk TV, Whelan S, Huang BQ, Lager DJ, vanDeursen J, Torres VE, Gattone VH, LaRusso NF, Harris PC, Ward CJ. A mouse model of autosomal recessive polycystic kidney disease with biliary duct and proximal tubule dilatation. Kidney Int. 2007;72:328–336. doi: 10.1038/sj.ki.5002294. [DOI] [PubMed] [Google Scholar]
- Garcia-Gonzalez MA, Menezes LF, Piontek KB, Kaimori J, Huso DL, Watnick T, Onuchic LF, Guay-Woodford LM, Germino GG. Genetic interaction studies link autosomal dominant and recessive polycystic kidney disease in a common pathway. Hum Mol Genet. 2007;16:1940–1950. doi: 10.1093/hmg/ddm141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W, Peissel B, Babakhanlou H, Pavlova A, Geng L, Fan X, Larson C, Brent G, Zhou J. Perinatal lethality with kidney and pancreas defects in mice with a targeted Pkd1 mutation. Nat Genet. 1997;17:179–181. doi: 10.1038/ng1097-179. [DOI] [PubMed] [Google Scholar]
- Wu GQ, Markowitz GS, Li L, D’Agati VD, Factor SM, Geng L, Tibara S, Tuchman J, Cai YQ, Park JH, van Adelsberg J, Hou H, Kucherlapati R, Edelmann W, Somlo S. Cardiac defects and renal failure in mice with targeted mutations in Pkd2. Nat Genet. 2000;24:75–78. doi: 10.1038/71724. [DOI] [PubMed] [Google Scholar]
- Gattone VH, MacNaughton KA, Kraybill AL. Murine autosomal recessive polycystic kidney disease with multiorgan involvement induced by the cpk gene. Anat Rec. 1996;245:488–499. doi: 10.1002/(SICI)1097-0185(199607)245:3<488::AID-AR5>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Flaherty L, Bryda EC, Collins D, Rudofsky U, Montogomery JC. New mouse model for polycystic kidney disease with both recessive and dominant gene effects. Kidney Int. 1995;47:552–558. doi: 10.1038/ki.1995.69. [DOI] [PubMed] [Google Scholar]
- Moyer JH, Lee-Tischler MJ, Kwon HY, Schrick JJ, Avner ED, Sweeney WE, Godfrey VL, Cacheiro NL, Wilkinson JE, Woychik RP. Candidate gene associated with a mutation causing recessive polycystic kidney disease in mice. Science. 1994;264:1329–1333. doi: 10.1126/science.8191288. [DOI] [PubMed] [Google Scholar]
- Cano DA, Murcia NS, Pazour GJ, Hebrok M. Orpk mouse model of polycystic kidney disease reveals essential role of primary cilia in pancreatic tissue organization. Development. 2004;131:3457–3467. doi: 10.1242/dev.01189. [DOI] [PubMed] [Google Scholar]
- Morgan D, Turnpenny L, Goodship J, Dai W, Majumder K, Matthews L, Gardner A, Schuster G, Vien L, Harrison W, Elder FF, Penman-Splitt M, Overbeek P, Strachan T. Inversin, a novel gene in the vertebrate left-right axis pathway, is partially deleted in the inv mouse. Nat Genet. 1998;20:149–156. doi: 10.1038/2450. [DOI] [PubMed] [Google Scholar]
- Farrell GC, Zaluzny L. Portal vein ligation selectively lowers hepatic cytochrome P450 levels in rats. Gastroenterology. 1983;85:275–282. [PubMed] [Google Scholar]
- Dieguez B, Aller MA, Nava MP, Palma MD, Arias JL, Lopez L, Arias J. Chronic portal hypertension in the rat by triple portal stenosing ligation. J Invest Surg. 2002;15:329–336. doi: 10.1080/08941930290086146. [DOI] [PubMed] [Google Scholar]
- Caroli J. Diseases of the intrahepatic biliary tree. Clin Gastroenterol. 1973;2:147–161. [PubMed] [Google Scholar]
- Goilav B, Norton KI, Satlin LM, Guay-Woodford L, Chen F, Magid MS, Emre S, Shneider BL. Predominant extrahepatic biliary disease in autosomal recessive polycystic kidney disease: a new association. Pediatr Transplant. 2006;10:294–298. doi: 10.1111/j.1399-3046.2005.00456.x. [DOI] [PubMed] [Google Scholar]
- Crawford JM. Development of the intrahepatic biliary tree. Semin Liver Dis. 2002;22:213–226. doi: 10.1055/s-2002-34508. [DOI] [PubMed] [Google Scholar]
- Mortelé KJ, Rocha TC, Streeter JL, Taylor AJ. Multimodality imaging of pancreatic and biliary congenital anomalies. Radiographics. 2006;26:715–731. doi: 10.1148/rg.263055164. [DOI] [PubMed] [Google Scholar]
- Adeva M, El-Youssef M, Rossetti S, Kamath PS, Kubly V, Consugar MB, Milliner DM, King BF, Torres VE, Harris PC. Clinical and molecular characterization defines a broadened spectrum of autosomal recessive polycystic kidney disease (ARPKD) 8. Medicine (Baltimore) 2006;85:1–21. doi: 10.1097/01.md.0000200165.90373.9a. [DOI] [PubMed] [Google Scholar]
- Lantinga-van Leeuwen I, Dauwerse JG, Baelde HJ, Leonhard WN, van de Wal AM, Ward CJ, Verbeek S, Deruiter MC, Breuning MH, de Heer E, Peters DJ. Lowering of Pkd1 expression is sufficient to cause polycystic kidney disease. Hum Mol Genet. 2004;13:3069–3077. doi: 10.1093/hmg/ddh336. [DOI] [PubMed] [Google Scholar]
- Jiang ST, Chiou YY, Wang E, Lin HK, Lin YT, Chi YC, Wang CK, Tang MJ, Li H. Defining a link with autosomal-dominant polycystic kidney disease in mice with congenitally low expression of Pkd1. Am J Pathol. 2006;168:205–220. doi: 10.2353/ajpath.2006.050342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsiokas L, Kim E, Arnould T, Sukhatme VP, Walz G. Homo- and heterodimeric interactions between the gene products of PKD1 and PKD2. Proc Natl Acad Sci USA. 1997;94:6965–6970. doi: 10.1073/pnas.94.13.6965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nauli SM, Alenghat FJ, Luo Y, Williams E, Vassilev P, Li X, Elia AE, Lu W, Brown EM, Quinn SJ, Ingber DE, Zhou J. Polycystins 1 and 2 mediate mechanosensation in the primary cilium of kidney cells. Nat Genet. 2003;33:129–137. doi: 10.1038/ng1076. [DOI] [PubMed] [Google Scholar]
- Wu Y, Dai XQ, Li L, Chen CX, Mai M, Hussain Z, Long W, Montalbetti N, Li L, Glynne R, Wang S, Cantiello HF, Wu G, Chen XZ. Kinesin-2 mediates physical and functional interactions between polycystin-2 and fibrocystin. Hum Mol Genet. 2006;15:3280–3292. doi: 10.1093/hmg/ddl404. [DOI] [PubMed] [Google Scholar]