Abstract
Cold-shock proteins (CSPs) bind to single-stranded nucleic acids, thereby acting as a “RNA chaperone.” To gain deeper insights into the rather unspecific nature of ssDNA/RNA binding, we characterized the binding interface of CspB from Bacillus subtilis to a 25-mer of ssDNA (Y-Box25) using heteronuclear 2D NMR spectroscopy. Seventeen residues, including eight out of nine aromatic amino acids, are directly involved in the Y-Box25 interaction and were identified by extreme line broadening of their cross-peaks. Eight residues belong to the earlier proposed RNP binding motifs. A second set of seven backbone amides becomes evident by major chemical shift perturbations reporting remote conformational rearrangements upon binding. These residues are located in loop β3–β4 and loopβ4–β5, and include Ile18. The individual contributions of the so-identified residues were examined by fluorescence titration experiments of 15 CspB variants. Phenylalanine substitutions in- and outside the RNP motifs significantly reduce the binding affinity. Unrestricted possible backbone conformations of loop β3–β4 also markedly contribute to binding. Stopped-flow fluorescence kinetics revealed that the different binding affinities of CspB variants are determined by the dissociation rate, whereas the association rate remains unchanged. This might be of importance for the “RNA chaperone” activity of CspB.
Keywords: Cold-shock protein, CspB, single-stranded DNA, Y-Box, protein/ssDNA complex, NMR spectroscopy
Rapid adaptation to changes in the environment is essential for the survival of bacteria under extreme conditions such as chemical stress and heat or cold shock. A reduction in growth temperature elicits a cold-shock response, in the course of which a number of distinct essential proteins are induced (Jones and Inouye 1994), in particular members of the family of small cold-shock proteins (CSPs). CSPs are ubiquitous proteins found in psychrotrophic, mesophilic, thermophilic, and hyperthermophilic bacteria (Jones et al. 1987; Hebraud and Potier 1999; Phadtare et al. 1999). The induction originates from an increase in transcription of their genes (Jiang et al. 1993) and from the stabilization of their mRNA (Brandi et al. 1996; Goldenberg et al. 1996). The basal concentrations and levels of induction vary strongly among the CSPs due to their different functions during cell division or during the stationary phase (Yamanaka et al. 1998; Brandi et al. 1999). CSPs stimulate the transcription of cold-shock inducible genes (Jones et al. 1992) and facilitate the initiation of translation by destabilizing nonproductive secondary structures in mRNA at low temperature (Jiang et al. 1997) without apparent sequence specificity (Lopez et al. 1999, 2001; Lopez and Makhatadze 2000) and are therefore regarded as “RNA chaperones.”
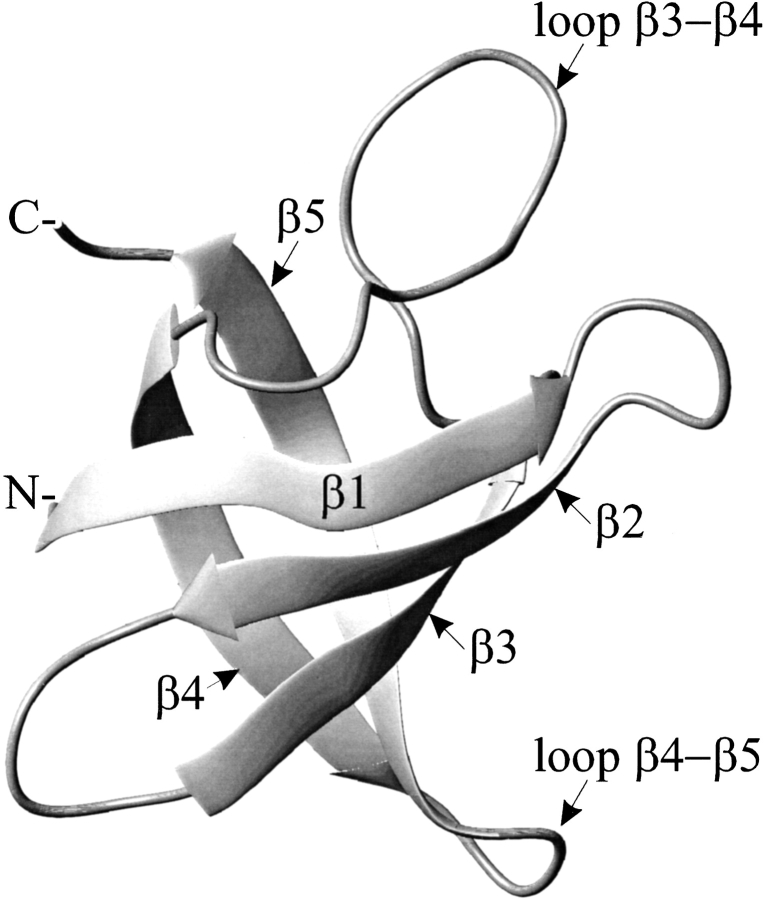
B. subtilis contains three homologous CSPs (CspB, CspC, CspD), which can complement each other in vivo (Graumann et al. 1997). The structure of the small acidic protein CspB (67 residues, Fig. 1 ▶) has been solved both in crystal and in solution, and revealed a very similar protein backbone conformation to CspA from Escherichia coli (Schindelin et al. 1993, 1994; Schnuchel et al. 1993; Newkirk et al. 1994; Feng et al. 1998). CspB comprises a five-stranded antiparallel β-barrel with perpendicularly arranged strands of subdomain I (β-strands 1, 2, and 3) and subdomain II (β-strands 4 and 5). It belongs to the OB-fold superfamily (Murzin 1993), which also includes ribosomal proteins S1 and S17, and translational initiation factor IF1 of E. coli (Sette et al. 1997; Draper and Reynaldo 1999). CSPs recognize DNA and RNA with the two conserved binding motifs RNP1 and RNP2, which comprise amino acids K13-V20 and V26-H29 in CspB, respectively. Investigations by mutagenesis and gel retardation experiments revealed that several aromatic and basic residues within the RNP motifs constitute the putative nucleic acid binding surface (Schröder et al. 1995). Until now, no structure of a cold-shock protein in complex with a single-stranded nucleic acid has been reported. CspB shows low sequence specificity with a preference for thymidine and uridine rich stretches (Schröder et al. 1995; Lopez et al. 1999, 2001; Phadtare and Inouye 1999). U-rich regions can be found upstream of the promotor in the unusually long 5′-untranslated region (5′-UTR) of the CspB mRNA, and T-rich stretches can be located at factor-independent transcription termination signals (Richardson 1993; Henkin 1996; Phadtare and Inouye 1999).
Figure 1.
Ribbon drawing of the cold-shock protein B from B. subtilis. The three-dimensional structure consists of two perpendicularly arranged subdomains (I: β-strands β1–β3, II: β-strands β4 and β5), which form a five-stranded β-barrel. The MOLMOL (Koradi et al. 1996) representation is based on the coordinates from the protein data bank file 1csp.pdb (Schindelin et al. 1993).
Domains homologous to the prokaryotic CSPs are the CSDs of eukaryotic Y-box transcription factors and nucleic acid-binding proteins, such as the telomere end binding protein (Graumann and Marahiel 1998; Horvath et al. 1998). CSDs are also involved in RNA binding of ribosomal proteins (Draper and Reynaldo 1999; Nakagawa et al. 1999) or of mitochondrial Y-box proteins (Pelletier et al. 2000).
In the present article we characterized the structural basis of CspB binding to a 25-mer ssDNA fragment termed Y-Box25, containing the Y-box core motif ATTGG, which is a cis-element in the promoter region of mammalian MHC II genes (Didier et al. 1988). Oligodeoxynucleotides containing Y-Box25 are the standard substrates of CspB used in previous studies (Schnuchel et al. 1993; Graumann and Marahiel 1994; Schröder et al. 1995; Schindler et al. 1999). Seventeen CspB residues involved in ssDNA binding were identified by line broadening of their backbone amides in a titration experiment followed by heteronuclear 2D NMR. They include eight residues of the proposed RNP motifs. Interestingly, seven amino acid residues in two loop regions that are not part of the classical RNP motifs show substantial changes in the chemical shifts of their backbone resonances upon binding. They report on remote conformational rearrangements in the nucleoprotein complex. The contributions of residues, identified by NMR, to the Gibbs free energy of binding were quantitatively characterized by equilibrium and kinetic fluorescence quenching experiments of 15 protein variants containing single amino acid substitutions. All examined aromatic and positively charged residues are necessary for a tight binding of Y-Box25. In addition, conformational restriction in loop β3–β4 decreases the binding affinity significantly. Reduced binding correlates with increased dissociation, rather than association, rate constants compared to the wild-type protein. This might be of importance for the “RNA chaperone” function of CspB.
Results
Identification of residues involved in the ssDNA binding of CspB by NMR spectroscopy
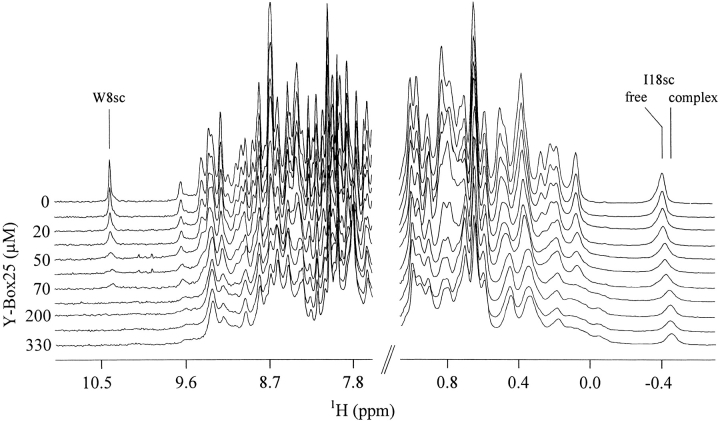
Cold-shock proteins bind to both ssDNA and RNA. To gain insights into contributions of individual residues, we used NMR spectroscopy. NMR signals of the backbone amides provide two probes to monitor complex formation: extreme resonance line broadening and chemical shift perturbation. Therefore, we performed a titration experiment by successively adding aliquots of a highly concentrated solution of Y-Box25 (Graumann and Marahiel 1994) to a solution of uniformly 15N-enriched CspB. After each titration step a 1D 1H and a 2D 1H/15N-HSQC spectrum was recorded. Figure 2 ▶ shows a stacked plot of 1D spectra acquired during the titration. In the absence of Y-Box25 both a good dispersion and sharp lines of the NMR signals are prevalent (Fig. 2 ▶, top spectrum). As expected, in the presence of high concentrations of Y-Box25 all resonances are broadened (Fig. 2 ▶, lower spectra) because of the increased molecular mass of the complex leading to decreased Brownian motion. Some resonances, for example, Trp8 Hɛ1, are extremely broadened and further completely lost at high Y-Box25 concentrations. The signal of Ile18 Hδ1 shifts to higher field upon ssDNA binding.
Figure 2.
Sections of 1D 1H NMR spectra recorded during the titration of CspB with Y-Box25. Spectra are shown for ssDNA concentrations between 0 and 330 μM in 20 mM sodium cacodylate/HCl pH 7.0 at 15°C and a fixed CspB concentration of 0.6 mM. The 1H resonance of Hɛ1 of the Trp8 side chain (W8sc) undergoing extreme line broadening and the shifting signal of Ile18 Hδ1 (I18sc) to higher field upon Y-Box25 binding have been specified.
Major rearrangements of the CspB secondary structure as a cause for changes in the NMR spectrum upon complex formation could be excluded by far UV-CD spectroscopy. The spectrum of the CspB/Y-Box25 complex is a linear combination of the spectra of free CspB and free Y-Box25 (data not shown).
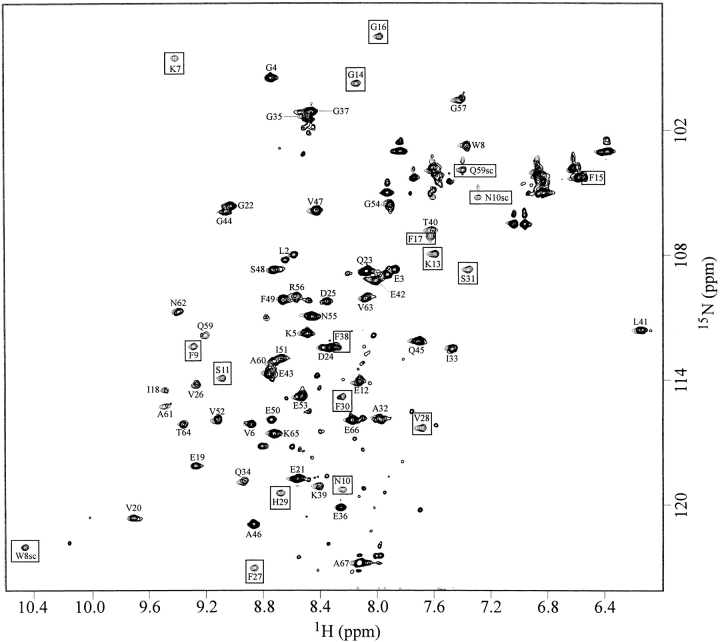
Complex formation was investigated in more detail by 2D 1H/15N-HSQC NMR spectroscopy. The cross-peaks of all 65 backbone amides in the 2D spectrum of 0.6 mM uncomplexed CspB (fine contours in Fig. 3 ▶) were assigned according to earlier studies (Schnuchel et al. 1993). Therefore, changes in line widths and chemical shifts upon ssDNA binding could be followed in a residue-specific fashion. Cross-peaks of backbone amides located in the hydrophobic core (e.g., V6, F49, I51) show nearly unmodified resonance frequencies and sharp lines. Therefore, we can conclude that the hydrophobic packing in the interior of the protein is not affected by the complex formation. This holds also for the backbone amides of all residues indicated in blue in Figure 4 ▶ and the side-chain amides of Q23, Q45, N55, and N62.
Figure 3.
2D 1H/15N-HSQC spectrum of 0.6 mM CspB in the absence and in the presence of 70 μM Y-Box25 at pH 7.0 and 15°C. The assigned cross-peaks of the amide backbone are labeled using the one-letter amino acid code and the sequence position. The indole 1H/15N cross-peak of the W8 side chain and the side-chain amides of N10 and Q59 are also labeled. Fine lines represent the spectrum in the absence of Y-Box25, and solid lines, the spectrum at 70 μM Y-Box25. The boxes indicate resonance signals, which vanished completely at a Y-Box25 concentration of 70 μM due to extreme line broadening. Nonlabeled cross-peaks with low signal intensities from impurities of probably unfolded peptide chains become visible at the lower contour level of the spectrum at 70 μM Y-Box25. They show no obvious dependence on the ssDNA concentration.
Figure 4.
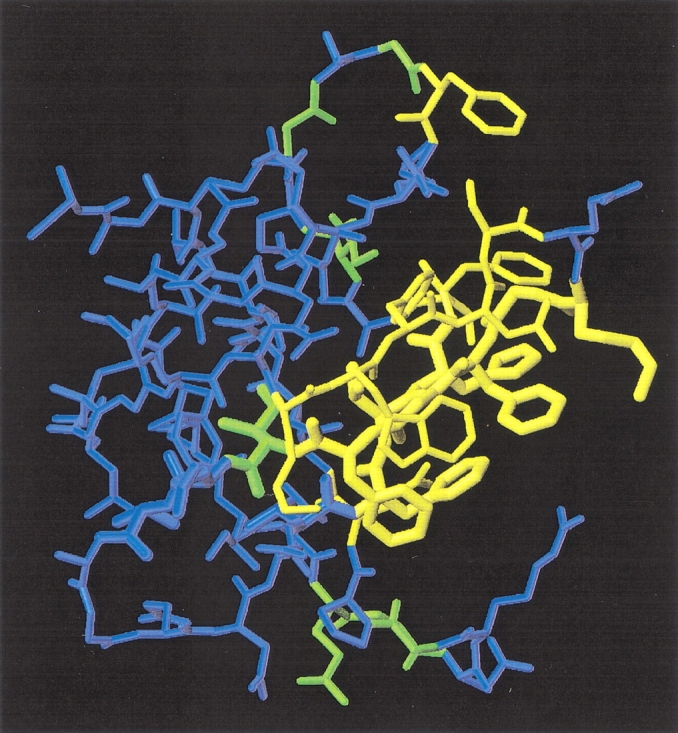
Identification of residues involved in ssDNA binding of CspB. Amino acid residues with cross-peaks undergoing extreme line broadening upon ssDNA binding at 70 μM Y-Box25 are represented in yellow (K7, W8, F9, N10, S11, K13, G14, F15, G16, F17, F27, V28, H29, F30, S31, F38, Q59). Residues with cross-peaks of backbone amide protons, which exhibit a substantial chemical shift perturbation during the NMR titration experiment, are indicated in green (I18, I33, G35, G37, E53, G54, G57). Residues of the RNP motifs are depicted with an increased bond radius. This representation was drawn in the same orientation as Figure 1 ▶ using the program MOLMOL (Koradi et al. 1996).
The extend of line broadening depends on the exchange rate constants as well as on the difference in chemical shift and the population of the respective nuclei within exchanging conformations (Sandström 1982). Line broadening becomes evident when the lifetimes of exchange between two conformations A and B are in the range of 2π/(νA − νB), with (νA − νB) being the difference in resonance frequency of the nucleus in the respective conformations. The concentration-dependent association rate and the populations of protein and ssDNA change during the titration experiment. Additionally, each nucleus exhibits an individual chemical shift difference between the exchanging conformations. In the case of CspB, binding of three CspB molecules at different sites of Y-Box25 may cause a heterogeneity in the ensemble of bound protein, with a possible exchange between adjacent sites without complete dissociation. It turned out that during the NMR titration there is only a small range of ssDNA concentrations, where CspB residues showing extreme line broadening can be clearly distinguished from residues showing distinct chemical shift changes within one spectrum.
This concentration range was found around a 10-fold excess of CspB over the Y-Box25 oligodeoxynucleotide. The 2D 1H/15N-HSQC NMR spectrum of CspB at an ssDNA concentration of 70 μM is plotted with solid contours in Figure 3 ▶. The boxes in Figure 3 ▶ indicate cross-peaks of residues with signal intensities broadened beyond the detection level in the presence of 70 μM Y-Box25. They include backbone amide resonances of K7, F9, N10, S11, K13, G14, F15, G16, F17, F27, V28, H29, F30, S31, F38, and the side-chain resonances of W8, N10, and Q59. These 17 residues are indicated in yellow in the three-dimensional structure representation of CspB in Figure 4 ▶. All of them are located at one side of the CspB molecule, with eight of them belonging to the classical RNA binding motifs RNP1 (K13–V20) and RNP2 (V26–H29). The backbone amides of seven out of the nine aromatic residues of CspB are extremely broadened (F9, F15, F17, F27, H29, F30, F38) as well as residues with positively charged side chains (K7, K13).
Chemical shift perturbations report about alterations in the chemical environment of protein nuclei upon binding to the nucleic acid. Changes in chemical shifts could be followed only for residues lacking the above-mentioned extreme line broadening. We found a gradual shift for some CspB residues with an increasing Y-Box25 concentration without extreme line broadening. The most likely reason for this observation is a small difference in the resonance frequencies between exchanging protein conformations.
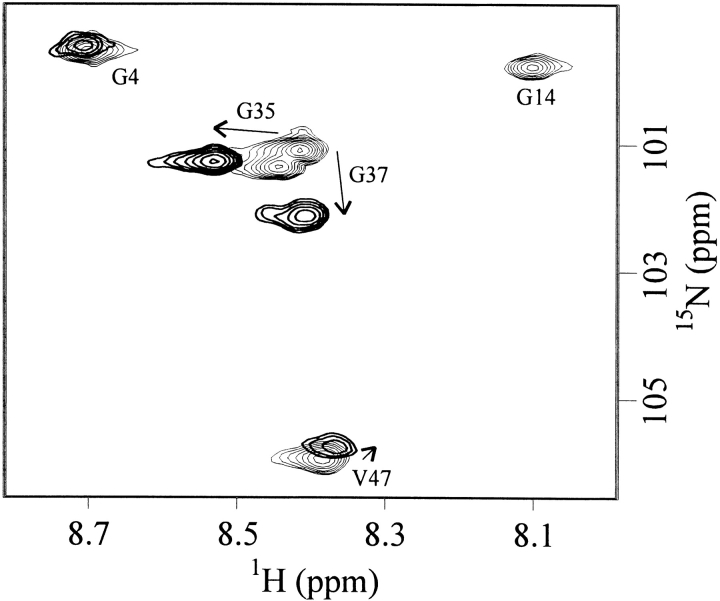
A section of a HSQC spectrum in both the absence and the presence of an equimolar concentration of Y-Box25 is shown in Figure 5 ▶. All possible responses of the protein resonances to complex formation are evident here: minor (G4), medium (V47), and large (G35, G37) chemical shift perturbations as well as extreme line broadening (G14). Most resonances detectable during the entire titration indeed exhibit very small chemical shift perturbations (Δδ[1H] < 0.05 ppm, Δδ[15N] < 0.25 ppm), suggesting minor changes in the structural environment of the respective nuclei. In contrast, cross-peaks of Ile 18 at the C-terminal end of β-strands β2 and amino acids forming the loop between the two subdomains (I33, G35, G37) and the loop connecting the β-strands β4 and β5 (E53, G54, G57) display larger chemical shift changes (Δδ[1H] > 0.05 ppm, Δδ[15N] > 0.5 ppm) in their resonance frequency. These residues are highlighted in green in Figure 4 ▶.
Figure 5.
Chemical shift perturbation upon ssDNA binding. Expansions of HSQC spectra in the absence (fine contours) and in the presence of an equimolar Y-Box25 concentration (strong contours) are overlaid. Alterations of the resonance frequency are small (G4), medium (V47), or large (G35, G37). The respective cross-peak of G14 in the complexed protein is broadened beyond detection.
ssDNA binding of CspB monitored by Trp fluorescence quenching
The NMR titration experiment revealed that most of the aromatic and some of the basic residues of CspB are involved in ssDNA binding. Trp8 dominates the fluorescence emission spectrum of CspB and binding of ssDNA quenches its fluorescence (Lopez et al. 1999, 2001). Therefore, we could quantify the Y-Box25 binding of CspB wild-type protein and variants, which were selected according to the results from the NMR titration experiment.
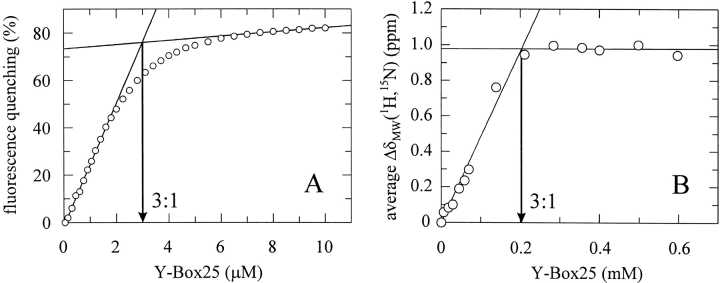
To determine the number of CspB molecules bound to one Y-Box25 molecule, we performed a titration experiment at 10 μM CspB (Fig. 6A ▶). It yields an almost linear change in fluorescence up to 2 μM Y-Box25, indicating stoichiometric binding conditions. The intersection of the two lines in the titration profile at 3 μM Y-Box25 allows estimation of the ratio between CspB and Y-Box25 as 3:1, implying that one Y-Box25 molecule is able to bind three CspB molecules. A 3:1 stoichiometry was confirmed in the titration experiment monitored by NMR by analyzing changes in the chemical shifts of 11 residues at various Y-Box25 concentrations up to 0.6 mM (Fig. 6B ▶). The coincidence indicates that the stoichiometry and mechanism of complex formation remains the same over a wide range of experimental conditions (0.25 to 10 μM CspB for fluorescence quenching or 0.6 mM for NMR spectroscopy).
Figure 6.
Stoichiometric titration of CspB with Y-Box25 probed by (A) fluorescence quenching and (B) NMR chemical shifts. An intersection at (A) 3 μM Y-Box25 (10 μM CspB) and (B) 0.2 mM Y-Box25 (0.6 mM CspB) implies that one Y-Box25 molecule binds three CspB molecules. (B) The mean weighted difference in the chemical shifts for 11 residues of β-strands β4 and β5 (E43, G44, A46, V47, S48, I51, G54, N55, G57, N62, K65) were calculated according to ΔδMW(1H,15N) = [(Δδ(1H)2 + 1/25•Δδ(15N)2)/2]0.5 (Grzesiek et al. 1996).
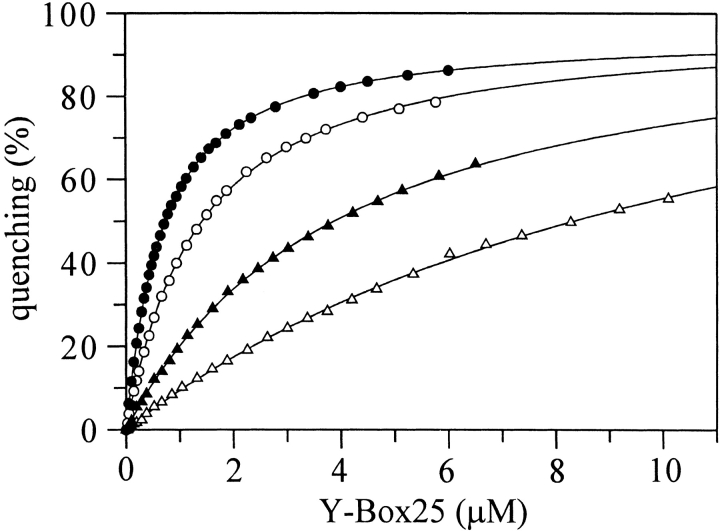
Next, we determined contributions to the binding affinity of individual CspB residues (identified by NMR) with fluorescence quenching titrations. To this end 15 protein variants with single amino acid substitutions (Table 1) were examined. Figure 7 ▶ depicts representative titrations with Y-Box25 of wild-type CspB and the variants G35A, F38A, and G54P.
Table 1.
Dissociation constants of CspB/Y-Box25 complexes and thermodynamic stability of the respective CspB variant
| Protein | KD•10−6 (M) | Tm (°C) | ΔGU (15°C) (kJ/mole) |
| Wild type | 3.9 ± 0.1 | 55.7 ± 0.1 | 13.1 ± 0.4 |
| F9Aa | 15.1 ± 0.6 | 34.0 ± 0.2 | 2.9 ± 0.1 |
| K13Qa | 21.6 ± 0.2 | 55.3 ± 0.2 | 12.7 ± 0.7 |
| F15Aa | 27.2 ± 0.8 | 36.2 ± 0.1 | 4.7 ± 0.1 |
| F15Ya,c | 1.9 ± 0.1 | 55.5 ± 0.1 | 14.0 ± 0.6 |
| F17Aa | 48.4 ± 1.6 | 41.2 ± 0.3 | 6.7 ± 0.4 |
| F27Aa | 17.0 ± 0.3 | 47.9 ± 0.6 | 8.2 ± 1.1 |
| H29Qa | 27.5 ± 1.0 | 48.7 ± 0.3 | 9.1 ± 0.8 |
| F30Aa | 40.8 ± 0.6 | 54.5 ± 0.1 | 13.4 ± 0.4 |
| F30Wa,c | 1.4 ± 0.1 | 57.8 ± 0.1 | 14.1 ± 0.2 |
| G35Ab | 1.8 ± 0.1 | 48.5 ± 0.1 | 9.1 ± 0.2 |
| G35Pb | 26.1 ± 0.8 | 44.2 ± 0.3 | 6.7 ± 0.5 |
| F38Aa | 35.4 ± 1.1 | 56.8 ± 0.1 | 13.4 ± 0.6 |
| G54Ab | 3.2 ± 0.1 | 46.2 ± 0.3 | 7.4 ± 0.6 |
| G54Pb | 12.1 ± 0.3 | 36.0 ± 1.1 | 2.5 ± 0.4 |
| P58Ad | 4.4 ± 0.1 | 49.6 ± 0.2 | 9.4 ± 0.4 |
a CspB revealed at this positions extreme line broadening in the NMR titration experiment.
b CspB revealed at this positions strong chemical shift changes in the NMR titration experiment.
c Substitution based on thermophilic cold-shock proteins from B. caldolyticus and T. maritima.
d Substitution to increase the number of possible backbone conformations of loop β4–β5.
Figure 7.
Quenching effect on the Trp8 fluorescence of CspB upon binding to Y-Box25. The hyperbolic binding isotherms of 0.25 μM wild-type CspB (open circles), G35A (filled circles), F38A (open triangles), and G54P (filled triangles) depict different Y-Box25 binding affinities in 50 mM sodium cacodylate/HCl, 100 mM KCl, pH 7.0 at 15°C. The resulting KD values from fits using Equation 1 (solid lines) are given in Table 1.
Dissociation constants (KD) were calculated from the experimental binding isotherms by assuming independent binding of three protein molecules to one Y-Box25 molecule (see equation 1). All binding curves are well represented by this simple model and the results are summarized in Table 1. Replacing one highly conserved phenylalanine in the binding motif RNP1 (F17) or RNP2 (F30) by alanine causes a 10-fold reduction in affinity. F15A and F27A lead to a moderate decrease in affinity (sevenfold and fourfold, respectively). Eliminating positively charged amino acids reduces the binding affinity as well: K13Q by 5.5-fold (RNP1) and H29Q by sevenfold (RNP2). [A fluorescence quenching titration of CspB wild type under high salt concentrations (1 M KCl) revealed an unchanged dissociation constant (M. Zeeb and J. Balbach, unpubl.) so that the overall electrostatic interactions should play a minor role.] For CspB variants with a very low binding affinity (e.g., F17A and F30A) the binding stoichiometry could not be determined with similar accuracy as for CspB variants with a high affinity (e.g., P58A), but there is no evidence for significant deviations from the 3:1 stoichiometry.
Aromatic residues located outside the RNP1 and RNP2 motif also contribute to the affinity for Y-Box25. The KD of F9A (in the C-terminal part of β-strand β1), F30A (in the C-terminal part of β-strand β3) and F38A (in the subdomain connecting loop between β3 and β4) are four- to tenfold increased compared to the wild-type protein. The importance of F9 and F38 has not been identified and analyzed in previous binding studies.
Cold-shock proteins from thermophilic bacteria such as Bacillus caldolyticus or Thermotoga maritima bind more tightly to Y-Box25 at 15°C with respective KD values of 0.7 and 0.01 μM (M. Zeeb and J. Balbach, unpubl.). Sequence comparisons revealed that both thermophilic proteins differ only at one or two positions (BcCsp or TmCsp) in the RNP1 and RNP2 motif from CspB. They contain Tyr15 and a Trp30 instead of Phe15 and Phe30. Therefore, we produced CspB variants F15Y and F30W. Both proteins revealed a twofold enhanced binding affinity to Y-Box25 compared to wild-type CspB, showing that the composition of aromatic residues involved in binding can still be improved.
Gly35 and Gly37 experience a very strong chemical shift perturbation of amide crosspeaks upon complex formation (Fig. 5 ▶). They are located in the loop between β-strand β3 and β4. To modulate the possible backbone conformations of this loop Gly35 was mutated to Ala or Pro. The G35A substitution restricts the expected backbone conformations to negative φ angles at this position but left the affinity for Y-Box25 virtually unchanged (Table 1). A further restriction by introducing a proline reduces the KD by a factor of 6.5 comparable to the extent of complex destabilization by mutations inside RNP1 (F15A, sevenfold) or RNP2 (H29Q, sevenfold). The same substitutions were performed for Gly54 in the loop between β-strands β4 and β5. The G54A mutation left the KD for Y-Box25 almost constant, and inserting a proline at position 54 leads only to a threefold increase of the dissociation constant. The affinity was also unchanged when the possible backbone conformations of this loop were enhanced by a P58A substitution. Together, these results indicate that conformational restrictions of the CspB backbone in the loop between β4 and β5 are of minor importance for the Y-Box25 binding affinity whereas conformational freedom in the loop between β3 and β4 is necessary for a tight binding.
Heat-induced unfolding transitions
To investigate whether variations in the ssDNA binding affinities of the examined CspB variants are caused by different thermodynamic stabilities, thermal transitions were measured for all variants. The transition midpoints and the extrapolated Gibbs free energies of unfolding (ΔGU) at 15°C are given in Table 1. All protein variants depict a positive ΔGU, and all variants have reached the native baseline in the thermal transitions at 15°C. Therefore, we can conclude that they all were fully native under the experimental conditions of the Y-Box25 binding studies. Additionally, we found no correlation between the thermodynamic stability and the binding affinity.
Association and dissociation kinetics of protein/ssDNA complexes
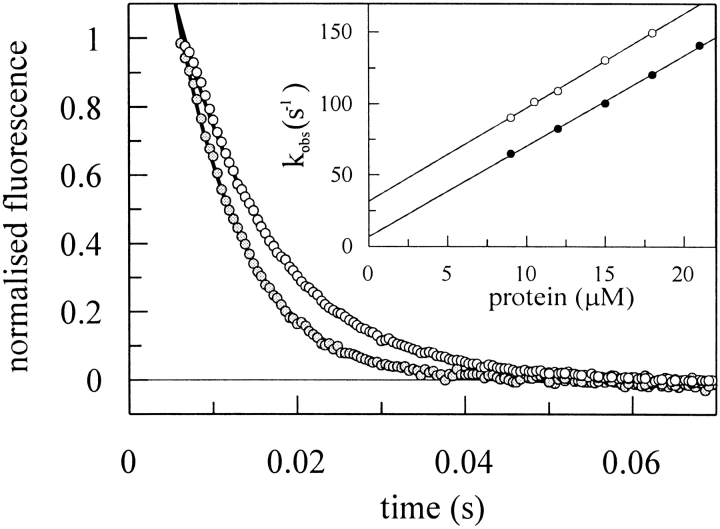
To elucidate whether the variations in binding affinity of the CspB variants originate from changes in the rate of association or of dissociation of the nucleoprotein complex, we measured the kinetics of complex formation by stopped-flow fluorescence. The association (kon) and dissociation (koff) rate constants were determined from the concentration dependence of the observed rate constant (kobs) under pseudo first-order conditions (Bernasconi 1976) as displayed in Figure 8 ▶. All kinetic curves are well described by a simple one-step mechanism P + D ⇋ PD, where P represents the protein, D the nucleic acid, and PD the nucleoprotein complex. Considering the 3:1 stoichiometry of the CspB/Y-Box25 complex it is assumed that the binding sites are equivalent and noninteracting. Analyzing the data of Figure 8 ▶ according to the equation kobs = kon•[P] + koff provides the association rate constant as the slope and the dissociation rate constant as the intercept with the ordinate (Fig. 8 ▶, inset). Because koff is measured in the kon•[P] >> koff regime, the extrapolation to the intersection with the ordinate is not very accurate, compared to the slope to determine kon (Bernasconi 1976). Even though the koff values from the intercept (e.g., 28.9 ± 3.3 sec−1 for wild-type CspB) were only marginally affected by extrapolation errors, we calculated the koff value as the product of kon and the equilibrium dissociation constant KD determined by the fluorescence quenching titration experiment (e.g., 26.9 ± 1.9 sec−1 for wild-type CspB). The association and dissociation rate constants of five protein variants are shown in Table 2.
Figure 8.
Association kinetics of CspB with Y-Box25. Final CspB concentrations in 50 mM sodium cacodylate/HCl, 100 mM KCl pH 7.0 at 15°C were 9 μM (open circles) and 15 μM (filled circles) in the presence of 1.5 μM Y-Box25. Continuous lines represent the best fits of a single exponential equation to the data. The inset shows the concentration dependence of the observed pseudo first-order rate constant of the wild-type protein (open circles) and the F15Y mutant (filled circles) including the linear fits (results are listed in Table 2).
Table 2.
Association and dissociation rate constants of CspB wild type and variant proteins for the binding to Y-Box25
| Protein | kon•106 (M−1•s−1) | koff (s−1)e |
| Wild type | 6.9 ± 0.3 | 26.9 ± 1.9 |
| F15Ya,c | 6.3 ± 0.1 | 11.4 ± 0.2 |
| F30Wa,c | 6.9 ± 0.2 | 9.7 ± 0.3 |
| G35Ab | 6.3 ± 0.3 | 12.9 ± 0.6 |
| G54Ab | 6.2 ± 0.3 | 19.8 ± 1.6 |
| P58Ad | 6.5 ± 0.3 | 28.6 ± 1.9 |
a–d See legend of Table 1.
e Calculated as product of kon and the equilibrium dissociation constant (see Table 1).
We found uniform association rate constants for all examined protein variants (between 6.2•106 M−1•sec−1 and 6.9•106 M−1•sec−1) so that the variation in the Y-Box25 affinity originates from differences in the dissociation rate constants (9.7 sec−1 to 28.6 sec−1). The rate constants could not be detected for protein variants with a dissociation constant higher than 10 μM because the apparent rates were too fast to be determined by the stopped-flow technique.
Discussion
We identified by an NMR titration experiment those residues of the cold-shock protein CspB, which are involved in binding of the single-stranded DNA fragment Y-Box25. The individual contributions of these residues to the binding affinity have been elucidated by various single residue substitutions.
The 3:1 stoichiometry of the CspB/Y-Box25 complex was found by two independent probes: quenching of the fluorescence of Trp 8, and NMR resonances of backbone amides. Both datasets are well explained by assuming three independent binding sites for CspB at Y-Box25. The apparently single exponential binding kinetics supports this simple model and virtually identical KD values are derived from the ratio of the measured kon and koff rates and from the equilibrium titration. There is no evidence for more complex behavior, but of course, protein–protein interactions found for example in the CopG operator complex (Gomis-Rüth et al. 1998) and mechanisms such as “sliding” (Berg et al. 1981) found essential for the DNA replisome (Hingorani and O’Donnell 2000) or for RecB (Phillips et al. 1997) cannot be excluded rigorously.
Extremely broadened NMR resonances were observed for 17 contiguous residues at the surface of CspB (yellow residues in Fig. 4 ▶). They include backbone amides of seven out of the nine aromatic residues (F9, F15, F17, F27, H29, F30, F38), two residues with positively charged side chains (K7, K13), and also the side chains of W8, N10, and Q59. Only eight of these residues belong to the earlier proposed RNP motifs derived from an alignment of different CSPs and CSDs. The RNP motifs are drawn by an increased bond radius in Figure 4 ▶, indicating the importance of the NMR titration data to map the entire binding interface. Schröder et al. classified the aromatic residues W8, F15, F17, F27, H29, F30, and basic residues K7 and K13 (all belong mainly to the RNP1 and RNP2 motifs) as essential for tight ssDNA binding (Schröder et al. 1995). They could not detect complex formation for CspB variants with F → A substitutions (F15, F17, F27) and for H29Q in gel retardation experiments. Here we could show that these mutations lead to a 4–10-fold increase in the respective dissociation constant. Newkirk et al. probed the DNA binding epitope of CspA from E. coli in complex with a 24-mer corresponding to the 5′ leader region of cspA mRNA (Newkirk et al. 1994). They used one single high ssDNA concentration and found strong perturbations of chemical shifts and/or line broadening for 26 residues, including six residues from the RNP motifs. Corresponding residues 7, 11, 13, 14, 16, 27, 29, 33, 35, 38, and 57 (CspB numbering) show perturbed NMR resonances in both CspA and CspB upon ssDNA binding. This comparison suggests that different residues close to the conserved RNP motifs facilitate the substrate specificity of the cold-shock proteins for different nucleic acid fragments.
Our NMR titration experiment is restricted to analyses of the backbone and side-chain amides, and well-resolved side-chain resonances of W8 and I18. Therefore, we complemented the NMR study by a mutational analysis of residues that are supposedly involved in Y-Box25 binding. Substitutions in the nucleic acid binding motifs RNP1 or RNP2 reduced the binding affinity. Phe → Ala mutations of the highly conserved phenylalanines lead to a loss of key interactions for complex stabilization. Outside the RNP motifs, F9 and F38 are important for binding.
Undetectably broad resonance signals might originate from an intermediate exchange between free and bound forms of CspB on the NMR chemical shift time scale. Broadened interface resonances have indeed been detected for entropically advantageous fluctuations in the network of protein–DNA hydrogen bonds (Billeter et al. 1993; Foster et al. 1997). Changes in the dynamics of aromatic rings also caused line broadening of proximal nuclei (Iwahara et al. 2001), which might apply for the CspB/Y-Box25 interaction dominated by aromatic residues.
The cross-peaks of several amino acid residues shift gradually with increasing Y-Box25 concentration. This suggests that different CspB conformations are in fast exchange on the NMR time scale with rates above about 300 sec−1. An extrapolation of the observed binding rates to NMR concentrations (Fig. 8 ▶, inset) supports this assumption.
The residues with perturbed backbone chemical shifts in the Y-Box25 complex (highlighted in green in Fig. 4 ▶) are located approximately in a plain behind residues with extremely broadened lines (yellow in Fig. 4 ▶). One reason for perturbed chemical shifts without extreme line broadening might be that the chemical shifts between exchanging conformations are very small. The perturbed nuclei include the backbone amide of I18 and its side chain Hδ1 proton (Fig. 2 ▶), which is located in the hydrophobic core of CspB. Therefore, it is unlikely that I18 interacts directly with Y-Box25. We suggest that I18 and most of the residues with perturbed chemical shifts report conformational changes in CspB remote from the ssDNA binding interface.
Several glycine residues of CspB in the loops β3–β4 and β4–β5 show strong perturbations of their 1H and 15N chemical shifts upon binding. The G54A, G54P, and P58A variations in loop β4–β5 lead only to marginal changes in binding, but the G35P mutation in loop β3–β4 reduces the Y-Box25 affinity of CspB by a factor of 7. Therefore, complex formation may induce conformational tension, which can be compensated by readjusting the protein backbone at less constraining glycine positions.
Recently, the solution structure and DNA-binding properties of the cold shock domain of human Y-Box protein YB-1 have been determined by NMR spectroscopy (Kloks et al. 2002). This CSD has 43% sequence identity to CspB and a very similar structure. An NMR titration with the 5-mer ATTGG (Y-box core motif) revealed a KD of about 150 μM and fast chemical exchange between free and complexed protein. The authors identified 21 residues involved in ssDNA binding and the following 15 residues correspond to our findings: 8, 9, 13, 16, 17, 18, 35, 36, 37, 38, 39, 50, 52, 55, and 57 (CspB numbering). This suggests that the binding interface of the prokaryotic cold-shock proteins is conserved in eukaryotic cold-shock domains. It is remarkable that residues at position 19 and 20 (indicated by blue bonds with increased radius in Fig. 4 ▶) of the proposed RNP1 motif (Schröder et al. 1995) show no interaction with ssDNA in the NMR titration experiments of CspA, CspB, and YB-1.
Cerdan et al. performed a similar NMR titration experiment of the high-mobility group domain of Drosophila melanogaster with a bulge DNA, and found as well strong line broadening for protein and DNA nuclei directly located at the binding interface (Cerdan et al. 2001). These protein/DNA complexes are also in fast exchange on the NMR chemical shift time scale and, therefore, show gradual shifts of the resonances during the NMR titration. In contrast, proteins such as the transcriptional coactivator PC4 revealed complexes in the slow exchanging regime, where two separate sets of resonances appear, one for the free protein and one for the protein/DNA complex (Werten et al. 1999).
It has been proposed that proteins have evolved to balance their function and stability (Meiering et al. 1992; Schreiber et al. 1994; Shoichet et al. 1995). For the mutants of this study there is no coupling between these properties. F30 and F38 of CspB are essential for tight ssDNA binding but not for protein stability (Schindler et al. 1998). G54A on the other hand, has a decreased Gibbs free energy of unfolding but the effect on Y-Box25 binding is small. Finally, the substitution of F15 or F17 with alanine reduces both affinity and stability.
Many complexes of proteins with nucleic acids have been investigated to date, and changes upon point mutations have been observed in koff (Hoopes et al. 1998; Katsamba et al. 2001), kon (Dong et al. 1999), or both rates (Neylon et al. 2000). The binding affinities of the examined CspB variants vary due to altered dissociation rate constants whereas the association rates remain nearly constant. This might be related to the “RNA chaperone” function of CspB by a rather fast, but unspecific interaction with stretches of mRNA. Productive interactions with U-rich stretches would result in a tight binding because of a reduced dissociation rate.
Materials and methods
Materials
The ssDNA fragment Y-Box25 was purchased from Interactiva AG (Germany) and was used throughout the entire study. The Y-box core motif is included and underlined in the sequence 5′-ATCCTACTGATTGGCCAAGGTGCTG-3′ (Graumann and Marahiel 1994). Y-Box25 concentrations were calculated from the absorbance at 260 nm. The extinction coefficient at 260 nm (268330 M−1cm−1) was calculated by adding the extinction coefficients of the individual nucleotides (Wallace and Miyada 1987).
Protein purification
CspB from B. subtilis and CspB variants were overexpressed in E. coli K38 using the bacteriophage T7 RNA polymerase promotor system (Schindelin et al. 1992) and purified as described previously (Schindler et al. 1995) with minor modifications. The respective molar extinction coefficients were determined as 5800 M−1cm−1, 7300 M−1cm −1, and 11,700 M−1cm −1 for wild-type, F15Y, and F30W CspB according to (Gill and von Hippel 1989).
NMR spectroscopy
NMR experiments were performed at 15°C on a Bruker DRX600 spectrometer. The protein concentration was 0.6 mM uniformly 15N enriched CspB dissolved in 20 mM sodium cacodylate/HCl, 3 mM MgCl2, pH 7.0 in 9:1 H2O:D2O. The protein–ssDNA titration experiment was carried out by successive addition of small aliquots of a 14-mM unlabeled Y-Box25 stock solution dissolved in the same buffer as the protein. Complex formation was monitored by recording a 15N-decoupled 1D 1H and a 2D 1H/15N-HSQC spectrum with WATERGATE solvent suppression (Piotto et al. 1992) after each Y-Box25 addition step. For the 2D spectra a combination of sensitivity enhancement and pulsed-field gradient coherence selection was applied with 1024 × 256 complex points (Palmer et al. 1991; Kay et al. 1992; Schleucher et al. 1994). All spectra were processed on a Silicon Graphics O2 workstation using Felix 97 (MSI).
Fluorescence spectroscopy
Fluorescence intensities were measured with a Hitachi F-4010 spectrofluorometer equipped with a thermostated cell holder, which was attached to a circulating water bath. All experiments were carried out at 15°C in 50 mM sodium cacodylate/HCl, 100 mM KCl, pH 7.0. For the determination of the dissociation constant (0.25 μM protein) the fluorescence of Trp8 was measured at 343 nm and excited at 280 nm. An excitation wavelength of 300 nm was used for a titration under stoichiometric conditions (10 μM CspB). The initial volume of the titration experiments was 1.5 mL so that the dilution effect of the successive addition of highly concentrated Y-Box25 solutions was below 4%. Samples were gently stirred during the titration. The tryptophan fluorescence intensity was corrected for inner filter effects, buffer fluorescence and dilution. The changes of the fluorescence intensity with increasing ssDNA concentration reveal hyperbolic binding isotherms, which were analyzed according to the binding equation 1 (Lohman and Bujalowski 1991; Eftink 1997):
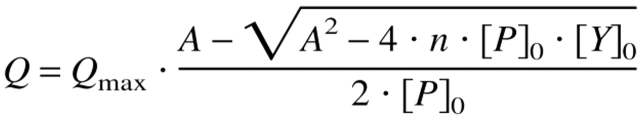 |
1 |
with A = KD + [P]0 + n[Y]0, where Q is the quenching of the intrinsic fluorescence intensity of Trp8 after each addition of Y-Box25; Qmax represents the maximum quenching upon complete saturation of the protein with ssDNA; [P]0 and [Y]0 are the overall CspB and Y-Box25 concentration, respectively; n is the stoichiometry of the protein–ssDNA complex (number of protein molecules bound to one molecule Y-Box25); and KD is the equilibrium dissociation constant of the complex. The binding model (Equation 1) does not take the degeneracy of multiple binding sites within one ssDNA molecule into account, and therefore, the obtained results depict apparent equilibrium constants.
To exclude the influence of random collisions to the fluorescence quenching effect a titration experiment with N-acetyl-tryptophan-amide was performed under the same experimental conditions (data not shown). There was no substantial quenching of the N-acetyl-tryptophan-amide fluorescence indicating that the decrease of the fluorescence intensity of Trp8 results from specific interactions between CspB and Y-Box25.
Heat-induced equilibrium unfolding transitions
Thermal denaturation curves were recorded with a Jasco J600A spectropolarimeter equipped with the Jasco Peltier element PTC-348WI. The temperature was measured with a sensor, which was directly inserted into the cell and the accuracy was verified by using a calibrated precision thermometer (Brand). The heating rate was 1°C per minute and the protein concentration typically 4 μM in 50 mM sodium cacodylate/HCl, 100 mM KCl, pH 7.0. Unfolding was monitored by the decrease of the ellipticity at 222.6 nm. Reversibility was examined by subsequent cooling of the sample to the starting temperature. The temperature transition curves were analyzed according to a two-state mechanism between the folded and unfolded conformation as described previously (Mayr et al. 1993). To derive the Gibbs free energy of unfolding at 15°C a linear extrapolation method was used.
Stopped-flow kinetic experiments
A DX.17MV sequential mixing stopped-flow spectrometer from Applied Photophysics was used for all kinetic experiments. The observation chamber has a path length of 2 mm. To absorb scattered light from the excitation beam a 5-mm cell with acetone was inserted between the observation chamber and the emission photomultiplier. The association kinetics was observed by the change in fluorescence intensity above 320 nm after excitation at 295 nm to account for the inner filter effect of Y-Box25. The zero time point and the dead time of mixing of the stopped-flow instrument were determined by using the procedure suggested by Tonomura et al. (1978).
All experiments were performed in 50 mM sodium cacodylate/HCl, 100 mM KCl, pH 7.0 at 15°C. Association reactions were initiated by a rapid 1:10 mixing of a 16.5 μM Y-Box25 solution (end concentration 1.5 μM) with a solution containing 6.6 μM to 23.1 μM protein (end concentration 6 to 21 μM) to obtain pseudo first-order conditions. Kinetic traces were collected at least eight times at identical conditions and averaged. Each set of stopped-flow data was analyzed according to a single exponential equation.
Acknowledgments
We thank P. Rösch for NMR time at the DRX 600 in Bayreuth, R. Jaenicke for the clone of Csp from Thermotoga maritima, D. Perl and T. Schindler for several CspB variants, and R. Maier, A. Martin, M. Rape, and F.X. Schmid for helpful discussions. This research was supported by grants from the Deutsche Forschungsgemeinschaft (Ba 1821/1–1: Ba 1821/2–1).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Abbreviations
ssDNA, single-stranded DNA
HSQC, 2D 1H/15N heteronuclear single quantum coherence spectroscopy
RNP, ribonucleoprotein
OB-fold, oligonucleotide- and/or oligosaccharide-binding fold
CspA, cold-shock protein A from Escherichia coli
CspB, cold-shock protein B from Bacillus subtilis
CSPs, cold-shock proteins
CSD, cold-shock domain
D2O, deuterium oxide
Y-Box25, 25-mer ssDNA fragment containing the Y-Box motif ATTGG
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.0219703.
References
- Berg, O.G., Winter, R.B., and von Hippel, P.H. 1981. Diffusion-driven mechanisms of protein translocation on nucleic acids. 1. Models and theory. Biochemistry 20 6929–6948. [DOI] [PubMed] [Google Scholar]
- Bernasconi, C.F. 1976. Relaxation kinetics. Academic Press, New York.
- Billeter, M., Qian, Y.Q., Otting, G., Müller, M., Gehring, W., and Wüthrich, K. 1993. Determination of the nuclear magnetic resonance solution structure of an Antennapedia homeodomain–DNA complex. J. Mol. Biol. 234 1084–1093. [DOI] [PubMed] [Google Scholar]
- Brandi, A., Pietroni, P., Gualerzi, C.O., and Pon, C.L. 1996. Post-transcriptional regulation of CspA expression in Escherichia coli. Mol. Microbiol. 19 231–240. [DOI] [PubMed] [Google Scholar]
- Brandi, A., Spurio, R., Gualerzi, C.O., and Pon, C.L. 1999. Massive presence of the Escherichia coli “major cold-shock protein” CspA under non-stress conditions. EMBO J. 18 1653–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerdan, R., Payet, D., Yang, J.C., Travers, A.A., and Neuhaus, D. 2001. HMG-D complexed to a bulge DNA: An NMR model. Protein Sci. 10 504–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didier, D.K., Schiffenbauer, J., Woulfe, S.L., Zacheis, M., and Schwartz, B.D. 1988. Characterization of the cDNA encoding a protein binding to the major histocompatibility complex class II Y box. Proc. Natl. Acad. Sci. 85 7322–7326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, F., Spott, S., Zimmermann, O., Kisters-Woike, B., Muller-Hill, B., and Barker, A. 1999. Dimerisation mutants of Lac repressor. I. A monomeric mutant, L251A, that binds Lac operator DNA as a dimer. J. Mol. Biol. 290 653–666. [DOI] [PubMed] [Google Scholar]
- Draper, D.E. and Reynaldo, L.P. 1999. RNA binding strategies of ribosomal proteins. Nucleic Acid Res. 27 381–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eftink, M.R. 1997. Fluorescence methods for studying equilibrium macromolecule–ligand interactions. Methods Enzymol. 278 221–257. [DOI] [PubMed] [Google Scholar]
- Feng, W., Tejero, R., Zimmerman, D.E., Inouye, M., and Montelione, G.T. 1998. Solution NMR structure and backbone dynamics of the major cold-shock protein (CspA) from Escherichia coli: Evidence for conformational dynamics in the single-stranded RNA-binding site. Biochemistry 37 10881–10896. [DOI] [PubMed] [Google Scholar]
- Foster, M.P., Wuttke, D.S., Radhakrishnan, I., Case, D.A., Gottesfeld, J.M., and Wright, P.E. 1997. Domain packing and dynamics in the DNA complex of the N-terminal zinc fingers of TFIIIA. Nat. Struct. Biol. 4 605–608. [DOI] [PubMed] [Google Scholar]
- Gill, S.C. and von Hippel, P.H. 1989. Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 182 319–326. [DOI] [PubMed] [Google Scholar]
- Goldenberg, D., Azar, I., and Oppenheim, A.B. 1996. Differential mRNA stability of the cspA gene in the cold-shock response of Escherichia coli. Mol. Microbiol. 19 241–248. [DOI] [PubMed] [Google Scholar]
- Gomis-Rüth, F.X., Sola, M., Acebo, P., Parraga, A., Guasch, A., Eritja, R., Gonzalez, A., Espinosa, M., del Solar, G., and Coll, M. 1998. The structure of plasmid-encoded transcriptional repressor CopG unliganded and bound to its operator. EMBO J. 17 1704–1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graumann, P. and Marahiel, M.A. 1994. The major cold shock protein of Bacillus subtilis CspB binds with high affinity to the ATTGG- and CCAAT sequences in single stranded oligonucleotides. FEBS Lett. 338 157–160. [DOI] [PubMed] [Google Scholar]
- Graumann, P., Wendrich, T.M., Weber, M.H., Schröder, K., and Marahiel, M.A. 1997. A family of cold shock proteins in Bacillus subtilis is essential for cellular growth and for efficient protein synthesis at optimal and low temperatures. Mol. Microbiol. 25 741–756. [DOI] [PubMed] [Google Scholar]
- Graumann, P.L. and Marahiel, M.A. 1998. A superfamily of proteins that contain the cold-shock domain. Trends Biochem. Sci. 23 286–290. [DOI] [PubMed] [Google Scholar]
- Grzesiek, S., Stahl, S.J., Wingfield, P.T., and Bax, A. 1996. The CD4 determinant for downregulation by HIV-1 Nef directly binds to Nef. Mapping of the Nef binding surface by NMR. Biochemistry 35 10256–10261. [DOI] [PubMed] [Google Scholar]
- Hebraud, M. and Potier, P. 1999. Cold shock response and low temperature adaptation in psychrotrophic bacteria. J. Mol. Microbiol. Biotechnol. 1 211–219. [PubMed] [Google Scholar]
- Henkin, T.M. 1996. Control of transcription termination in prokaryotes. Annu. Rev. Genet. 30 35–57. [DOI] [PubMed] [Google Scholar]
- Hingorani, M.M. and O’Donnell, M. 2000. Sliding clamps: A (tail)ored fit. Curr. Biol. 10 25–29. [DOI] [PubMed] [Google Scholar]
- Hoopes, B.C., LeBlanc, J.F., and Hawley, D.K. 1998. Contributions of the TATA box sequence to rate-limiting steps in transcription initiation by RNA polymerase II. J. Mol. Biol. 277 1015–1031. [DOI] [PubMed] [Google Scholar]
- Horvath, M.P., Schweiker, V.L., Bevilacqua, J.M., Ruggles, J.A., and Schultz, S.C. 1998. Crystal structure of the Oxytricha nova telomere end binding protein complexed with single strand DNA. Cell 95 963–974. [DOI] [PubMed] [Google Scholar]
- Iwahara, J., Wojciak, J.M., and Clubb, R.T. 2001. Improved NMR spectra of a protein–DNA complex through rational mutagenesis and the application of a sensitivity optimized isotope-filtered NOESY experiment. J. Biomol. NMR. 19 231–241. [DOI] [PubMed] [Google Scholar]
- Jiang, W., Jones, P., and Inouye, M. 1993. Chloramphenicol induces the transcription of the major cold shock gene of Escherichia coli, cspA. J. Bacteriol. 175 5824–5828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, W., Hou, Y., and Inouye, M. 1997. CspA, the major cold-shock protein of Escherichia coli, is an RNA chaperone. J. Biol. Chem. 272 196–202. [DOI] [PubMed] [Google Scholar]
- Jones, P.G. and Inouye, M. 1994. The cold-shock response—A hot topic. Mol. Microbiol. 11 811–818. [DOI] [PubMed] [Google Scholar]
- Jones, P.G., VanBogelen, R.A., and Neidhardt, F.C. 1987. Induction of proteins in response to low temperature in Escherichia coli. J. Bacteriol. 169 2092–2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, P.G., Krah, R., Tafuri, S.R., and Wolffe, A.P. 1992. DNA gyrase, CS7.4, and the cold shock response in Escherichia coli. J. Bacteriol. 174 5798–5802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsamba, P.S., Myszka, D.G., and Laird-Offringa, I.A. 2001. Two functionally distinct steps mediate high affinity binding of U1A protein to U1 hairpin II RNA. J. Biol. Chem. 276 21476–21481. [DOI] [PubMed] [Google Scholar]
- Kay, L., Keifer, P., and Saarinen, T. 1992. Pure absorption gradient enhanced heteronuclear single quantum correlation spectroscopy with improved senitivity. J. Am. Chem. Soc. 114 10663–10665. [Google Scholar]
- Kloks, C.P., Spronk, C.A., Lasonder, E., Hoffmann, A., Vuister, G.W., Grzesiek, S., and Hilbers, C.W. 2002. The solution structure and DNA-binding properties of the cold-shock domain of the human Y-box protein YB-1. J. Mol. Biol. 316 317–326. [DOI] [PubMed] [Google Scholar]
- Koradi, R., Billeter, M., and Wüthrich, K. 1996. MOLMOL: A program for display and analysis of macromolecular structures. J. Mol. Graph. 14 51–55. [DOI] [PubMed] [Google Scholar]
- Lohman, T.M. and Bujalowski, W. 1991. Thermodynamic methods for model-independent determination of equilibrium binding isotherms for protein–DNA interactions: Spectroscopic approaches to monitor binding. Methods Enzymol. 208 258–290. [DOI] [PubMed] [Google Scholar]
- Lopez, M.M. and Makhatadze, G.I. 2000. Major cold shock proteins, CspA from Escherichia coli and CspB from Bacillus subtilis, interact differently with single-stranded DNA templates. Biochim. Biophys. Acta 1479 196–202. [DOI] [PubMed] [Google Scholar]
- Lopez, M.M., Yutani, K., and Makhatadze, G.I. 1999. Interactions of the major cold shock protein of Bacillus subtilis CspB with single-stranded DNA templates of different base composition. J. Biol. Chem. 274 33601–33608. [DOI] [PubMed] [Google Scholar]
- ———. 2001. Interactions of the cold shock protein CspB from Bacillus subtilis with single-stranded DNA. Importance of the T base content and the position within the template. J. Biol. Chem. 276 15511–15518. [DOI] [PubMed] [Google Scholar]
- Mayr, L.M., Landt, O., Hahn, U., and Schmid, F.X. 1993. Stability and folding kinetics of ribonuclease T1 are strongly altered by the replacement of cis-proline 39 with alanine. J. Mol. Biol. 231 897–912. [DOI] [PubMed] [Google Scholar]
- Meiering, E.M., Serrano, L., and Fersht, A.R. 1992. Effect of active site residues in barnase on activity and stability. J. Mol. Biol. 225 585–589. [DOI] [PubMed] [Google Scholar]
- Murzin, A.G. 1993. OB (oligonucleotide/oligosaccharide binding)-fold: Common structural and functional solution for non-homologous sequences. EMBO J. 12 861–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa, A., Nakashima, T., Taniguchi, M., Hosaka, H., Kimura, M., and Tanaka, I. 1999. The three-dimensional structure of the RNA-binding domain of ribosomal protein L2; A protein at the peptidyl transferase center of the ribosome. EMBO J. 18 1459–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newkirk, K., Feng, W., Jiang, W., Tejero, R., Emerson, S.D., Inouye, M., and Montelione, G.T. 1994. Solution NMR structure of the major cold shock protein (CspA) from Escherichia coli: Identification of a binding epitope for DNA. Proc. Natl. Acad. Sci. 91 5114–5118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neylon, C., Brown, S.E., Kralicek, A.V., Miles, C.S., Love, C.A., and Dixon, N.E. 2000. Interaction of the Escherichia coli replication terminator protein (Tus) with DNA: A model derived from DNA-binding studies of mutant proteins by surface plasmon resonance. Biochemistry 39 11989–11999. [DOI] [PubMed] [Google Scholar]
- Palmer III, A.G., Cavanagh, J., Wright, P.E., and Rance, M. 1991. Sensitivity improvement in proton-detected 2-dimensional heteronuclear correlation NMR-spectroscopy. J. Magn. Reson. 93 151–170. [Google Scholar]
- Pelletier, M., Miller, M.M., and Read, L.K. 2000. RNA-binding properties of the mitochondrial Y-box protein RBP16. Nucleic Acids Res. 28 1266–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phadtare, S. and Inouye, M. 1999. Sequence-selective interactions with RNA by CspB, CspC and CspE, members of the CspA family of Escherichia coli. Mol. Microbiol. 33 1004–1014. [DOI] [PubMed] [Google Scholar]
- Phadtare, S., Alsina, J., and Inouye, M. 1999. Cold-shock response and cold-shock proteins. Curr. Opin. Microbiol. 2 175–180. [DOI] [PubMed] [Google Scholar]
- Phillips, R.J., Hickleton, D.C., Boehmer, P.E., and Emmerson, P.T. 1997. The RecB protein of Escherichia coli translocates along single-stranded DNA in the 3′ to 5′ direction: A proposed ratchet mechanism. Mol. Gen. Genet. 254 319–329. [DOI] [PubMed] [Google Scholar]
- Piotto, M., Saudek, V., and Sklenar, V. 1992. Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J. Biomol. NMR 2 661–665. [DOI] [PubMed] [Google Scholar]
- Richardson, J.P. 1993. Transcription termination. Crit. Rev. Biochem. Mol. Biol. 28 1–30. [DOI] [PubMed] [Google Scholar]
- Sandström, J. 1982. Dynamic NMR spectroscopy. Academic Press, New York.
- Schindelin, H., Herrler, M., Willimsky, G., Marahiel, M.A., and Heinemann, U. 1992. Overproduction, crystallization, and preliminary X-ray diffraction studies of the major cold shock protein from Bacillus subtilis, CspB. Proteins 14 120–124. [DOI] [PubMed] [Google Scholar]
- Schindelin, H., Marahiel, M.A., and Heinemann, U. 1993. Universal nucleic acid-binding domain revealed by crystal structure of the B. subtilis major cold-shock protein. Nature 364 164–168. [DOI] [PubMed] [Google Scholar]
- Schindelin, H., Jiang, W., Inouye, M., and Heinemann, U. 1994. Crystal structure of CspA, the major cold shock protein of Escherichia coli. Proc. Natl. Acad. Sci. 91 5119–5123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler, T., Herrler, M., Marahiel, M.A., and Schmid, F.X. 1995. Extremely rapid folding in the absence of intermediates: The cold-shock protein from Bacillus subtilis. Nat. Struct. Biol. 2 663–673. [DOI] [PubMed] [Google Scholar]
- Schindler, T., Perl, D., Graumann, P., Sieber, V., Marahiel, M.A., and Schmid, F.X. 1998. Surface-exposed phenylalanines in the RNP1/RNP2 motif stabilize the cold-shock protein CspB from Bacillus subtilis. Proteins 30 401–406. [DOI] [PubMed] [Google Scholar]
- Schindler, T., Graumann, P.L., Perl, D., Ma, S., Schmid, F.X., and Marahiel, M.A. 1999. The family of cold shock proteins of Bacillus subtilis. Stability and dynamics in vitro and in vivo. J. Biol. Chem. 274 3407–3413. [DOI] [PubMed] [Google Scholar]
- Schleucher, J., Schwendinger, M., Sattler, M., Schmidt, P., Schedletzky, O., Glaser, S.J., Sørensen, O.W., and Griesinger, C. 1994. A general enhancement scheme in heteronuclear multidimensional NMR employing pulsed-field gradients. J. Biomol. NMR 4 301–306. [DOI] [PubMed] [Google Scholar]
- Schnuchel, A., Wiltschek, R., Czisch, M., Herrler, M., Willimsky, G., Graumann, P., Marahiel, M.A., and Holak, T.A. 1993. Structure in solution of the major cold-shock protein from Bacillus subtilis. Nature 364 169–171. [DOI] [PubMed] [Google Scholar]
- Schreiber, C., Buckle, A.M., and Fersht, A.R. 1994. Stability and function: Two constraints in the evolution of barstar and other proteins. Structure 2 945–951. [DOI] [PubMed] [Google Scholar]
- Schröder, K., Graumann, P., Schnuchel, A., Holak, T.A., and Marahiel, M.A. 1995. Mutational analysis of the putative nucleic acid-binding surface of the cold-shock domain, CspB, revealed an essential role of aromatic and basic residues in binding of single-stranded DNA containing the Y-box motif. Mol. Microbiol. 16 699–708. [DOI] [PubMed] [Google Scholar]
- Sette, M., van Tilborg, P., Spurio, R., Kaptein, R., Paci, M., Gualerzi, C.O., and Boelens, R. 1997. The structure of the translational initiation factor IF1 from E. coli contains an oligomer-binding motif. EMBO J. 16 1436–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoichet, B.K., Baase, W.A., Kuroki, R., and Matthews, B.W. 1995. A relationship between protein stability and protein function. Proc. Natl. Acad. Sci. 92 452–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonomura, B., Nakatani, H., Ohnishi, M., Yamaguchi-Ito, J., and Hiromi, K. 1978. Test reaction for a stopped-flow apparatus. Anal. Biochem. 84 370–383. [DOI] [PubMed] [Google Scholar]
- Wallace, R.B. and Miyada, C.G. 1987. Oligonucleotide probes for the screening of recombinant DNA libraries. Methods Enzymol. 152 432–442. [DOI] [PubMed] [Google Scholar]
- Werten, S., Wechselberger, R., Boelens, R., van der Vliet, P.C., and Kaptein, R. 1999. Identification of the single-stranded DNA binding surface of the transcriptional coactivator PC4 by NMR. J. Biol. Chem. 274 3693–3699. [DOI] [PubMed] [Google Scholar]
- Yamanaka, K., Fang, L., and Inouye, M. 1998. The CspA family in Escherichia coli: Multiple gene duplication for stress adaptation. Mol. Microbiol. 27 247–255. [DOI] [PubMed] [Google Scholar]