Abstract
L-Aspartate-β-semialdehyde dehydrogenase (ASADH) catalyzes the reductive dephosphorylation of β-aspartyl phosphate to L-aspartate-β-semialdehyde in the aspartate biosynthetic pathway of plants and micro-organisms. The aspartate pathway produces fully one-quarter of the naturally occurring amino acids, but is not found in humans or other eukaryotic organisms, making ASADH an attractive target for the development of new antibacterial, fungicidal, or herbicidal compounds. We have determined the structure of ASADH from Vibrio cholerae in two states; the apoenzyme and a complex with NADP, and a covalently bound active site inhibitor, S-methyl-L-cysteine sulfoxide. Upon binding the inhibitor undergoes an enzyme-catalyzed reductive demethylation leading to a covalently bound cysteine that is observed in the complex structure. The enzyme is a functional homodimer, with extensive intersubunit contacts and a symmetrical 4-amino acid bridge linking the active site residues in adjacent subunits that could serve as a communication channel. The active site is essentially preformed, with minimal differences in active site conformation in the apoenzyme relative to the ternary inhibitor complex. The conformational changes that do occur result primarily from NADP binding, and are localized to the repositioning of two surface loops located on the rim at opposite sides of the NADP cleft.
Keywords: Enzyme mechanism, enzyme structure, X-ray crystallography, L-aspartate-β-semialdehyde dehydrogenase, S-methyl-L-cysteine sulfoxide, Vibrio cholerae
L-Aspartate-β-semialdehyde dehydrogenase (ASADH, EC 1.2.1.11) catalyzes the reductive dephosphorylation of β-aspartyl phosphate (βAP) to L-aspartate-β-semialdehyde (ASA) as the second step in the aspartate biosynthetic pathway (Cohen 1983; Viola 2001). ASA is either further reduced to homoserine, a precursor of threonine, isoleucine, and methionine, or it is condensed with pyruvate to produce lysine after seven enzyme-catalyzed reactions. Lysine is derived from the decarboxylation of diaminopimelic acid, a cross-linking component for bacterial cell walls (Patte 1983). The aspartate pathway is not found in humans or other eukaryotic organisms, making consumption of these amino acids essential to survival. Thus, the preceding enzymes in the pathway are potentially attractive targets for the development of new antimicrobial compounds. Alternatively, manipulation of the flux through the aspartate pathway could result in plants that produce superior yields of these nutritionally important amino acids. Because the product of the ASADH-catalyzed reaction occupies the first metabolic branch point, an understanding of the reaction mechanism for this enzyme is of central interest for either upregulation or inhibition of the entire aspartate pathway.
Studies on Salmonella typhimurium (Galan et al. 1990) and Legionella pneumophila (Harb and Kwaik 1998) demonstrated that perturbations of the asd gene encoding for ASADH can be lethal to a micro-organism. An experimental S. typhimurium vaccine, using a balanced-lethal host-vector system based upon a deletion mutation of the asd gene, has given encouraging results in mice (Kang et al. 2002). There is continued interest in the development of effective microbial ASADH inhibitors (Cox et al. 2001, 2002), and the first purification of a plant ASADH has now been reported (Paris et al. 2002). The structure of Escherichia coli ASADH has been solved, both as the apoenzyme (Hadfield et al. 1999) and as an inhibitor–coenzyme complex (Hadfield et al. 2001). As a continuation of this work, we have expressed and purified ASADH from a number of infectious micro-organisms and have begun their characterization (Moore et al. 2002).
In this article, we focus on ASADH from Vibrio cholerae, the causative agent of cholera, a severe diarrhoeal disease known for its explosive outbreaks, rapid onset, and high morbidity (Wachsmuth et al. 1994; Faruque et al. 1998). Contaminated waters in various regions of the world have allowed pathogenic strains of V. cholerae to thrive, resulting in sporadic outbreaks and seven pandemics in the last two centuries (Wachsmuth et al. 1994). The prevailing model of bacterial cells containing only one chromosome, based primarily on studies of E. coli, is rapidly changing with the discovery of numerous micro-organisms containing multiple chromosomes. One such organism is V. cholerae El Tor, maintaining both a large and a small chromosome (∼3 million and 1 million bp, respectively). Both chromosomes have been sequenced by whole-genome random sequencing (Heidelberg et al. 2000). Chromosome 1 contains most genes involved in DNA replication, pathogenicity, biosynthetic pathways, and cell-wall biosynthesis. However, some genes thought to be essential to normal cellular function (e.g., certain genes encoding ribosomal proteins) are found only on chromosome 2. A significant number of ORFs are duplicated on both chromosomes suggesting a high level of crossover between the two chromosomes. Interestingly, there are two asd genes in chromosome 1 and none in chromosome 2. We have purified both enzymes encoded by theses genes, and each displays significant ASADH activity (Moore et al. 2002). We now report the structure of a V. cholerae ASADH (vcASADH) in two states, the apoenzyme and an NADP–inhibitor complex, and an analysis and comparison of the E. coli and V. cholerae enzyme structures.
Results and Discussion
Structural overview of ASADH from V. cholerae
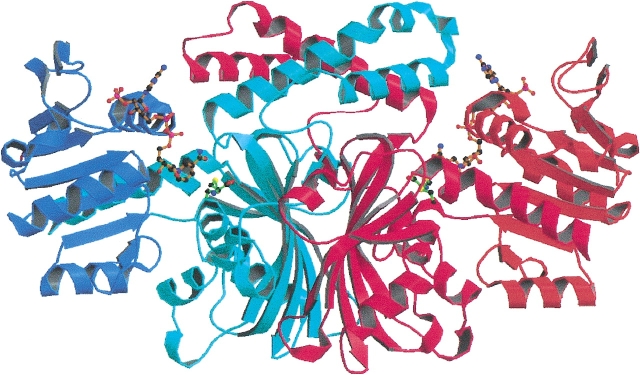
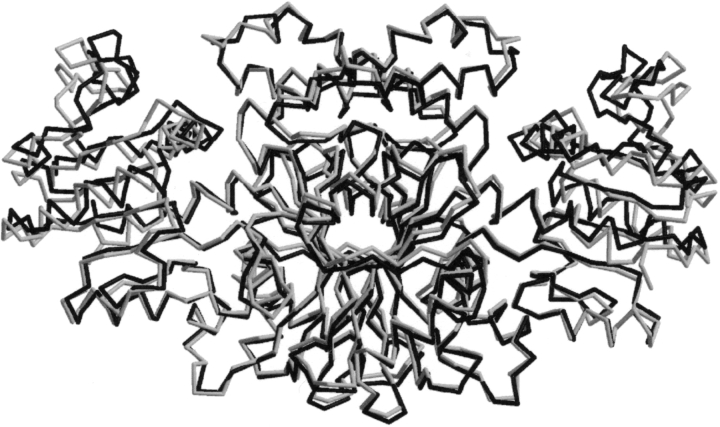
The structure of ASADH from V. cholerae has been solved to 2.8 Å and that of the ternary complex with NADP and an active site-directed inactivator, S-methyl-L-cysteine sulfoxide (SMCS) refined at 1.84 Å. As shown in Figure 1 ▶, vcASADH presents as a functional dimer. The N-terminal domain of each monomer provides the binding site for NADP and the C-terminal region forms the substrate-binding pocket. The V. cholerae enzyme shares 66% sequence identity and 80% similarity with the previously determined E. coli ASADH (Hadfield et al. 1999, 2001). Both proteins display remarkably similar folds (Fig. 2 ▶), with the most notable differences observed in the positioning of two surface loops (Fig. 3 ▶), each of which appears to play a role in binding and catalysis. There is well-defined electron density for the complete dimer in each structure except for the C-terminal Lys370, which was not modeled due to lack of convincing density.
Figure 1.
Ribbon diagram of V. cholerae ASADH dimer with NADP and SMCS bound. The N-terminal domains are shown in darker blue and orange. The central C-terminal domains are shown in red and light blue. Drawings were rendered with Molscript and Raster3D.
Figure 2.
Overlay of the backbone drawings of V. cholerae (light) and E. coli (dark) ASADH ternary complex structures. Drawing was created with XtalView.
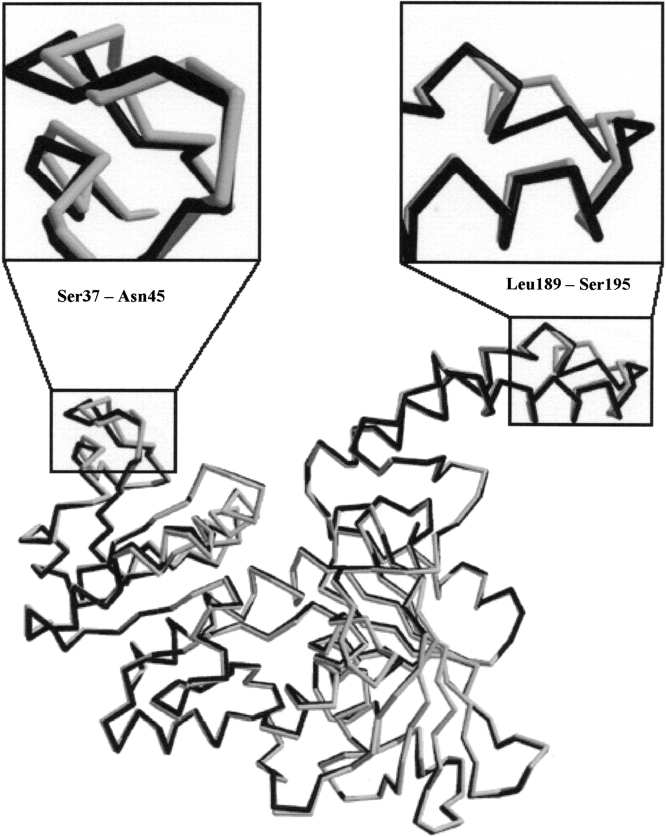
Figure 3.
Overlay of subunit A (dark) and subunit B (light) shows that the major conformational change between the open and closed conformation is the repositioning of surface loops Leu189–Ser195 and Ser37–Asn45 at the rim of the NADP cleft.
Reaction of ASADH with S-methyl-L-cysteine sulfoxide
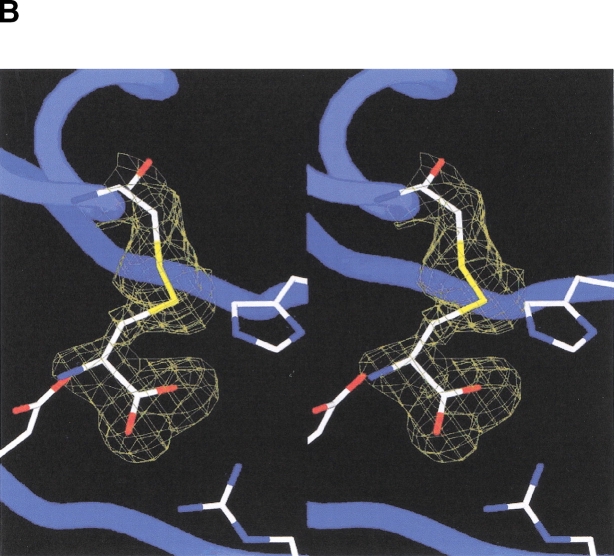
Incubation of vcASADH with NADP and SMCS [CH3—S(=O)—CH2—CH(NH3+)—COO−] prior to crystallization was expected to yield a covalently-inactivated enzyme complex. Indeed, the refined model of the vcASADH complex shows that each active site in the dimer contains NADP and is covalently modified by the inhibitor, displaying continuous density extending from Cys134. (The amino acid sequence numbers used are for V. cholerae ASADH, and are typically one lower than that of E. coli. For example, Cys134 in the V. cholerae sequence corresponds to Cys135 in the E. coli sequence.) However, no density is observed for either the methyl group or the oxygen atom originally attached to the SMCS sulfur atom (Fig. 4B ▶). The electron density of the active site is consistent with that of L-cysteine covalently attached in a disulfide linkage. This observation is supported by electrospray mass spectrometry, which confirms that treatment of vcASADH with S-methyl-L-cysteine sulfoxide results in a modified enzyme with an increased mass of only 120, consistent with a covalently bound cysteine with no oxygen or methyl group attached to sulfur, instead of the expected increase of 151 for SMCS. Inactivation of ASADH by SMCS (Ki = 0.18 mM; Karsten and Viola 1991a) is reversible by addition of excess DTT or β-mercaptoethanol. Incubation of ASADH with either S-methyl-L-cysteine or L-cystine also results in the inactivation of the enzyme. These observations are each consistent with the formation of a disulfide linkage at Cys134.
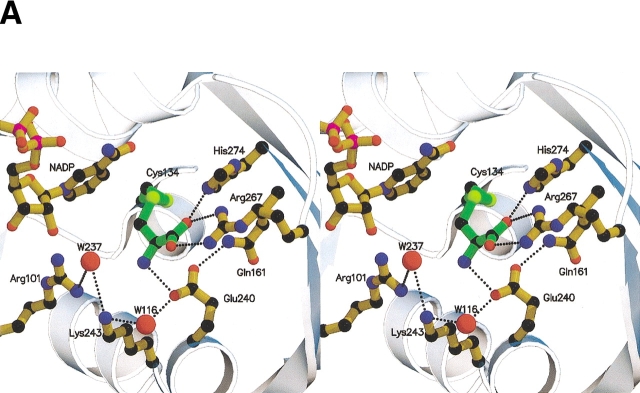
Figure 4.
(A) Stereoview of the active site of V. cholerae ASADH with bound NADP and inhibitor. Pictures rendered with Molscript and Raster3D. (B) Stereoview of the Fo–Fc map at 1.84 Å resolution, using phases calculated from the final refined model omitting the covalent inactivator SMCS and contoured at 2.0 σ. No density is observed for an oxygen or methyl group attached to the inhibitor sulfur atom. Figure created with Spock.
The E. coli ASADH ternary complex showed that only one subunit was modified by SMCS (Hadfield et al. 2001), suggesting that when one active site is bound, the conformation of the second subunit changes making it less accommodating to inhibitor (or substrate) binding. In contrast, the corresponding vcASADH structure clearly shows that SMCS is bound at both active sites. The efficacy of SMCS as an inhibitor is related to the precise orientation of the carboxyl and amino group in the active site, as well as to the susceptibility of the sulfoxide to nucleophilic attack. However, an electrophilic sulfoxide moiety itself is not enough to cause ASADH inhibition, as demonstrated by the inability of dimethyl sulfoxide to inhibit ASADH at concentrations well above that used for SMCS. After SMCS binds to the preformed catalytic pocket the sulfoxide moiety is likely attacked by the active site Cys134. There is precedent for this in the mechanism of methionine sulfoxide reductase, in which an active site thiol attacks the sulfoxide moiety of methionine sulfoxide to form a sulfoxyanion (Lowther et al. 2000). The sulfoxide oxygen of SMCS is likely lost as water, thus leaving a methyl sulfonium species. Nucleophilic attack of the methyl group by the excess DTT that is necessary to keep the enzyme stabilized and in a reducing environment would convert the sulfonium to the uncharged disulfide bond that is observed.
NADP binding
The binding and orientation of NADP to ASADH is characterized by a number of interactions. There is hydrogen bonding of the Gln350 amide nitrogen to the amide carbonyl of NADP, stabilized by Ser164, which hydrogen bonds to both Gln350 via its side-chain hydroxyl and the NADP amide group via its main-chain carbonyl group. The hydrogen bonding between the Asn133 amide nitrogen and Glu240 is present in both the NADP-bound and unbound state, therefore not forming as a consequence of NADP binding as previously proposed. The side-chain hydroxyls of Ser37 and Thr36 form new hydrogen bonds with the 2′-phosphate of NADP, while the Ser35 hydroxyl forms a water-mediated hydrogen bond with phosphate. The main-chain amide nitrogens of Val12 and Met11 bind to pyrophosphate moiety of NADP, as does the main-chain nitrogen of Ala168 via a water molecule. The side-chain carbonyl group of Gln72 interacts with the 3′ hydroxyl of the adenine ribose. Another notable difference between the binding of NADP in E. coli and V. cholerae is that in the vcASADH Lys243 is at least 7 Å away from either ribose hydroxyl group and does not form hydrogen bonds with them. It is more likely that this lysine plays a direct role in stabilizing the phosphate of βAP.
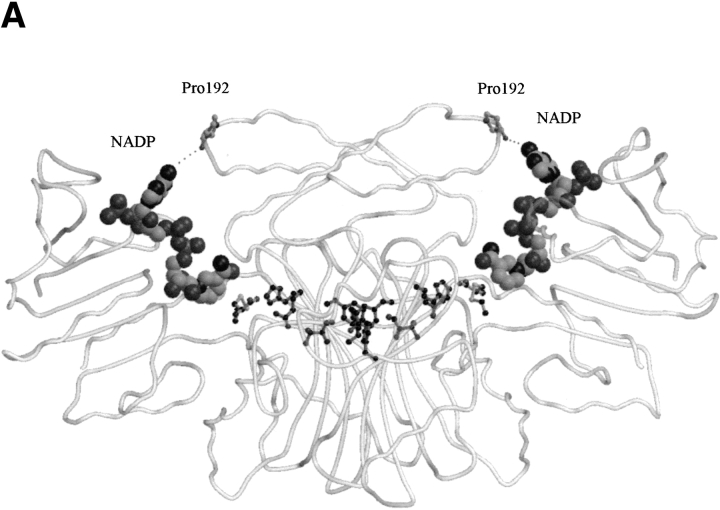
Examination of the E. coli ASADH structure resulted in the identification of an internal loop from Asp230 to Glu240, located near the active site, which is disordered in the apoenzyme. The driving force for ordering this loop was proposed to be the formation of new hydrogen bonds during NADP binding. While the loop does cross the active site, it is already ordered in the V. cholerae apoenzyme, and retains essentially the same conformation in the ternary complex. The active site appears to be preformed to accommodate the substrate, and it is only when NADP binds that the cleft closes around it. Previously identified hydrogen bonds between Ser98 and Arg240 (E. coli numbering) that formed upon complex formation are not seen in the new structures. In fact, the corresponding arginine is replaced by Lys239 in vcASADH. When NADP is enveloped by vcASADH there is a conformational change, primarily in the repositioning of a surface loop (Leu189–Ser195) with respect to NADP. This is best observed as the main-chain Pro192 carbonyl of one subunit moves roughly 3 Å closer to the NADP adenine ring in the opposing subunit, forming a new hydrogen bond with the amino group (Fig. 5A ▶). On the opposite side of the cleft, Arg9 is repositioned to form a new hydrogen bond with the 2′-phosphate of NADP while also forming a π-charge interaction with the adenine ring of NADP. The distance between Cα of Arg9 and the main-chain carbonyl of Pro192 is particularly diagnostic of this domain closure. In both the E. coli and V. cholerae apoenzymes that distance is approximately 13.5 Å. In the E. coli ternary complex this proline carbonyl to arginine Cα distance shortens to 10.7 Å in both subunits, each containing NADP, but only one of which is SMCS modified. Interestingly, the V. cholerae ternary complex, having NADP and SMCS bound in both active sites, displays an open conformation in one subunit and a closed conformation in the other. In the closed subunit, the Pro192 carbonyl to Arg9 Cαdistance is 10.3 Å, indicative of domain closure, while this distance is 13.1 Å in the open subunit, causing asymmetry between the two monomers. This can be observed by the differing positions of the critical loops (Leu189–Ser195, Ser37–Asn45) in the closed versus open conformations (Fig. 3 ▶).
Figure 5.
(A) The active sites in the dimer are interconnected through a π-stacking interactions and hydrogen bonds. One active site is in the open conformation and the other is closed as shown by the different Pro192 to NADP distances. NADP is shown as a spacefilling model. (B) Diagram of residues involved in the pathway that is proposed to allow communication between the active sites, with the bound inhibitor SMCS shown in lighter shading.
Active site analysis of conserved binding residues
The well-established assignment of Cys134 as the active site nucleophile is confirmed by the vcASADH complex structure. In this complex with SMCS the inactivator carboxyl group is engaged in a bidentate interaction with the terminal nitrogens of Arg267 and Nɛ of His274 (Fig. 4 ▶). The inhibitor amino group interacts directly with Glu240. The side-chain amide nitrogen of Asn133 and Water116 also interact with Glu240 to stabilize the inhibitor-bound active site. The active site hydrogen-bonding network is extended through Water237, which bridges between Arg102 and Lys243, occupying the position that we propose phosphate occupies during catalysis. It is likely that active site differences that have been noted between the V. cholerae ternary complex and the E. coli complex are not the result of a difference in mechanism between the two enzymes, but rather a more precise modeling of the higher resolution data that is available for the V. cholerae complex. All of these active site groups identified thus far are highly conserved among the currently known ASADH enzymes.
Catalysis
The proposed role of His274 as the acid/base catalyst that facilitates deprotonation of Cys134 is well supported by this structure. The Nɛ atom is 3.1 Å from the sulfur atom of Cys134 in both subunits, perfectly placed for proton abstraction (Fig. 4 ▶). Attack of the thiolate at the carbonyl of βAP (or Sγ of SMCS) yields a tetrahedral intermediate, stabilized by hydrogen bonding to active site water molecules. Collapse of the initial tetrahedral intermediate and the concomitant expulsion of phosphate, stabilized by Arg102 and Lys243, gives a relatively stable thioester intermediate. Hydride transfer from bound NADPH leads to the formation of a second tetrahedral intermediate, the collapse of which produces ASA and the free thiol of Cys134. Gln161 is not likely to play a role in positioning His274 (Ouyang and Viola 1995) because it is too far away in both V. cholerae structures. In the current model, Gln161 has no interaction with the substrate amino group, mainly due to differences in the way that the inhibitor amino group was fit into the electron density for this enzyme relative to E. coli ASADH (Hadfield et al. 2001). Mutations of Gln161 to Asn and His, conducted with E. coli ASADH before structural data were available (Ouyang and Viola 1995), show a 13-fold and threefold reduction in kcat for the Asn and His mutants, respectively. The Km values for both variants were unaffected. We postulate that Gln161 plays a more supplemental role in catalysis by forming a hydrogen bond between its amide nitrogen and the strategically located Glu240 carboxyl group that hydrogen bonds to the SMCS amino group.
Communication between the active sites
The current mechanism proposed for ASADH favors a model of alternating site reactivity orchestrated by communication between the active sites. It has been proposed (Biellmann et al. 1980), based upon affinity labeling studies with covalent inactivators, that the mechanism for ASADH was an “alternating” or “half-of-sites” reactivity in which the binding order is NADPH followed by the substrate βAP and the order of release is ASA before NADP. Kinetic experiments further demonstrated that E. coli ASADH has a preferred, but not obligatory, binding order with NADP binding before the substrate ASA (Karsten and Viola 1991b).
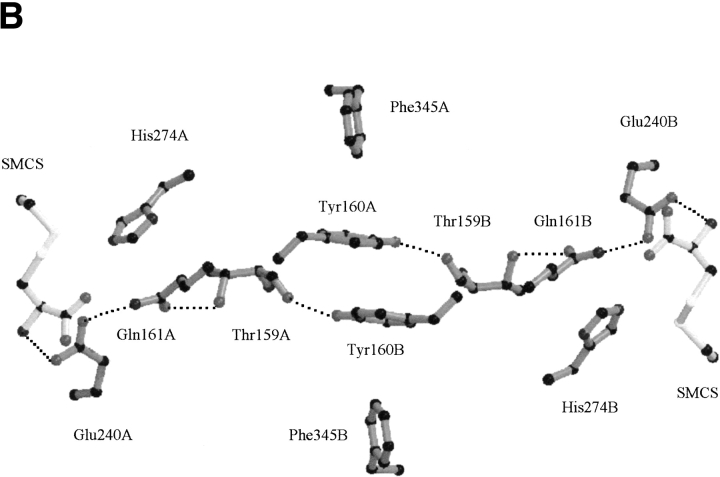
There are a number of electrostatic interactions between the subunits that serve to maintain the conformational integrity of the dimer. The Glu241 adjacent to active site Glu240 is engaged in a bidentate interaction with Arg330 of the opposite subunit. In a similar manner, Asp261 is engaged with Arg332 of the opposing subunit. From this ensemble of interactions, we have identified a plausible route by which one active site could relay information to the other. There are π-stacking interactions between Tyr160 of opposite subunits in the center of the dimer. Both Tyr160 aromatic rings are slightly offset relative to each other, each flanked by Phe345 of the same subunit in a perpendicular π-stacking interaction (Fig. 5B ▶). The hydroxyl group of Tyr160 from one subunit is hydrogen bonded to the main-chain Thr159 carbonyl of the opposing subunit, which in turn, is hydrogen bonded through its side-chain hydroxyl group to the amide carbonyl oxygen of active site residue Gln161. This results in a symmetrical 4-amino acid bridge linking the active site glutamines in adjacent subunits (Fig. 5B ▶). Catalytic and binding events between the active sites are likely synchronized by perturbations along this route. Thus, interaction between the subunits is maintained not only by the previously observed (Hadfield et al. 2001) hydrophobic core at the center of the enzyme, but also by a conduit of hydrogen bonding interactions spanning the entire length of the dimer interface as well as significant electrostatic interactions between the subunits.
Materials and methods
Crystallization of the apoenzyme
Recombinant V. cholerae El Tor N16961 (TIGR locus VC2036, Swiss-Pro Q9KQG2) ASADH was purified as described (Moore et al. 2002), concentrated by ultrafiltration (Millipore) to 12 mg/mL, and dialyzed against 10 mM HEPES, 1 mM EDTA, 1 mM DTT at pH 7.0. Initial crystallization conditions were tested by hanging-drop vapor-diffusion using the PEG/Ion™ Screen (Hampton Research). After further optimization, crystals of the apoenzyme were grown at 20°C in 1:1 mixtures of enzyme and precipitant solution (18% PEG 3350, 0.2 M sodium acetate and 0.1 M Tris at pH 8.5). The best crystals, often reaching 0.4 mm in the largest dimension, were produced in about 7 d, and were indexed in the P4332 space group with unit cell dimensions a = b = c = 141.8 Å and one monomer in the asymmetric unit. Harvesting solutions for freezing the crystals were 18% PEG 3350, 0.2 M sodium acetate, and 20% glycerol in 0.1 M sodium citrate, pH 5.6.
Crystallization of the ternary complex
The ASADH inhibitor S-methyl-L-cysteinesulfoxide (SMCS) was purchased from Karlan Research Products, and was found to be at least 99% pure by 1H and 13C NMR analysis. DTNB titration of the SMCS showed no (<0.1%) free thiol in the sample. Crystals of the enzyme in the presence of 5 mM NADP and 5 mM SMCS were grown at 20°C from 1:1 mixtures of enzyme solution (12 mg/mL) and precipitant (22% PEG 3350, 0.2 M sodium acetate and 0.1 M sodium citrate, pH 5.6). Tetragonal crystals belonging to the P41212 space group with unit cell dimensions a = b = 107.1 Å and c = 153.1 Å and one dimer in the asymmetric unit formed after 10 d. Prior to freezing, crystals were soaked without DTT for 1 h in 22% PEG 3350, 0.2 M sodium acetate, 5 mM SMCS, 5 mM NADP, 20% glycerol and 0.1 M sodium citrate, pH 5.6.
Data collection and processing
A data set was collected from a single frozen crystal of apoenzyme on a Quantum-IV imaging plate (detector distance 200 mm; 0.75° oscillation per image) at Argonne National Laboratory (APS) BioCARS beamline (14-D). For the ternary ASADH/NADP/SMCS complex, a complete data set up to 1.8 Å was collected at SSRL Stanford synchrotron beamline 5A (detector distance 150 mm; 1° oscillation per image). The images were processed and scaled using the program HKL2000 (Otwinowski and Minor 1997). The data collection statistics for each of these data sets are summarized in Table 1.
Table 1.
Data collection statistics
| Data set | Native enzyme | NADP/SMCS complex |
| Wavelength (Å) | 1.0 | 1.0 |
| Temperature (K) | 90 | 90 |
| Resolution range (Å) | 50–2.77 | 50–1.84 |
| No. of observations | ||
| Measured | 785,297 | 1,811,452 |
| Unique | 24,543 | 149,813 |
| Completeness | ||
| Overall | 99.1 | 99.5 |
| Last shell | 99.5 | 95.4 |
| Rsymm (%)a | 5.4 | 8.4 |
| Average 〈I/σ〉 | 33.5 | 19.1 |
aRsymm = 100 (∑hkl∑i|Ihkl,i − 〈Ihkl〉|∑hkl,i〈Ihkl〉) where Ihkl,i is the intensity of an individual measurement of the reflection with Miller indices h, k, and l and 〈Ihkl〉 is the mean intensity of that reflection.
Structure solution and refinement
The structure of the NADP/SMCS ternary complex was solved by molecular replacement using a subunit of E. coli apo-ASADH (PDB-1brm) as the search model. The V. cholerae apoenzyme structure was subsequently solved by molecular replacement using the V. cholerae ternary ASADH complex as the search model. The CNS package was used for the rotation and translation searches (Brunger et al. 1998). Optimal solutions for the rotation matrix corresponding to two subunits of ASADH per asymmetric unit were obtained for the ternary complex. The first translation search was conducted subsequent to rotation of the search model coordinates according to one of the two rotation matrices. The translation function clearly identified a solution that was well above the next highest peak. The second translation search, performed after noncrystallographic symmetry had been applied to the coordinates of the first subunit, yielded a solution corresponding to the position of the second subunit. After the relative translation between the first and second subunit had been determined, the model containing both subunits was subjected to rigid body refinement considering the two subunits as independent rigid structures. The rigid body refinement led to an R factor of 46% at a resolution between 20.0 and 3.0 Å. Simulated annealing refinement at 2.0 Å was applied to the coordinates file obtained from the rigid body refinement that improved the R factor to 29%. The nonconserved amino acids in the search model were then replaced based on the sequence of the V. cholerae enzyme. Model building was applied using XtalView (McRee 1999). Several iterations of refinement and model building improved the ternary complex structure. The cofactor NADP was then fit to the corresponding difference electron density calculated by using coefficients Fo–Fc.
The final refinement statistics for the structures are listed in Table 2. The refined coordinates have been deposited with the PDB and assigned the entry code 1MC4 for apoenzyme and 1MB4 for the NADP/SMCS complex.
Table 2.
Refinement statistics
| Native enzyme | NADP/SMCS complex | |
| Refinement | ||
| Resolution range (Å) | 40–2.77 | 50–1.84 |
| Ra | 21.5 | 19.2 |
| Rfreeb | 28.1 | 22.9 |
| No. of protein atoms | 2828 | 5656 |
| No. of nonprotein atoms | — | 206 |
| No. of water molecules | 44 | 457 |
| Stereochemistry | ||
| msd for bond lengths (Å) | 0.01 | 0.007 |
| rmsd for bond angles (deg) | 1.6 | 1.4 |
| Residues in the Ramachandran plot | ||
| Most favored region (%) | 85.2 | 90.7 |
| Additional allowed regions (%) | 13.9 | 8.7 |
| Generously allowed regions (%) | 0.9 | 0.6 |
aR = 100(∑h|Fobs− F|/∑h|Fobs|).
bRfree = R for a randomly selected 5% of the data excluded from the refinement.
Mass spectrometry
Mass spectrometry on the enzyme samples were conducted at the Ohio State University CCIC facility. Samples were prepared in a solution containing 50% acetonitrile/50% water, 0.1% formic acid at a concentration of ∼1 pmole/μL and infused into the nanospray source at a rate of 1 μL/min. Optimal ESI conditions were: capillary voltage 3000 V, source temperature 110°C and a cone voltage of 60 V. All ions were scanned over m/z 500 to 3000 with a l -sec integration time, and data was acquired in continuum mode until acceptable averaged data was obtained (10–15 min). ESI data was deconvoluted using MaxEnt I software that was provided by Micromass.
Acknowledgments
This work was supported by a grant (MCB0196107) from the National Science Foundation. The authors thank Dr. Tim Mueser for helpful discussions on the analysis and refinement of the enzyme structures. They acknowledge Andrew Collins, for help with the graphics, and Johanne Le Coq, for NMR and DTNB titration characterization. Kari B. Greenchurch from the OSU mass spectrometry facility is thanked for obtaining the mass spectrum and for helpful discussions. Use of the Argonne National Laboratory at the Advanced Photon Source was supported by U.S. Department of Energy, Office of Energy Research, under Contract No. W-31–109-ENG-38. Portions of this research were carried out at the Stanford Synchrotron Radiation Laboratory, a national user facility operated by Stanford University on behalf of the U.S. Department of Energy, Office of Basic Energy Sciences. We thank the staff members at BioCARS and SSRL for their assistance with data collection.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Abbreviations
ASADH, L-aspartate-β-semialdehyde dehydrogenase
βAP, β-aspartyl phosphate
ASA, L-aspartate-β-semialdehyde
SMCS, S-methyl-L-cysteine sulfoxide
NADP, β-nicotinamide adenine dinucleotide phosphate
DTT, dithiothreitol
vc ASADH, Vibrio cholerae ASADH
ORF, open reading frame
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.0230803.
References
- Biellmann, J.F., Eid, P., and Hirth, C. 1980. Aspartate-β-semialdehyde dehydrogenase from Escherichia coli. Affinity labeling with the substrate analogue L-2-amino-4-oxo-5-chloropentanoic acid: An example of half-site reactivity. Eur. J. Biochem. 104 59–64. [DOI] [PubMed] [Google Scholar]
- Brunger, A.T., Adams, P.D., Clore, G.M., DeLano, W.L., Gros, P., Grosse-Kunstleve, R.W., Jiang, J.S., Kuszewski, J., Nilges, M., Pannu, N.S., Read, R.J., Rice, L.M., Simonson, T., and Warren, G.L. 1998. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D54 905–921. [DOI] [PubMed] [Google Scholar]
- Cohen, G.N. 1983. The common pathway to lysine, methionine, and threonine. In Amino acids: Biosynthesis and genetic regulation (eds. K.M. Herrmann and R.L. Somerville), pp. 147–171. Addison-Wesley, Reading, MA.
- Cox, R.J., Hadfield, A.T., and Mayo-Martin, M.B. 2001. Difluoromethylene analogues of aspartyl phosphate: The first synthetic inhibitors of aspartate semi-aldehyde dehydrogenase. Chem. Commun. 18 1710–1711. [DOI] [PubMed] [Google Scholar]
- Cox, R.J., Gibson, J.S., and Martin, M.B. 2002. Aspartyl phosphonates and phosphoramidates: The first synthetic inhibitors of bacterial aspartate-semialdehyde dehydrogenase. Chem. BioChem. 3 874–886. [DOI] [PubMed] [Google Scholar]
- Faruque, S.M., Albert, M.J., and Mekalanos, J.J. 1998. Epidemiology, genetics, and ecology of toxigenic Vibrio cholerae. Microbiol. Mol. Biol. Rev. 62 1301–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galan, J.E., Nakayama, K., and Curtiss, R. 1990. Cloning and characterization of the asd gene of Salmonella typhimurium: Use in stable maintenance of recombinant plasmids in Salmonella vaccine strains. Gene 94 29–35. [DOI] [PubMed] [Google Scholar]
- Hadfield, A.T., Kryger, G., Ouyang, J., Petsko, G.A., Ringe, D., and Viola, R.E. 1999. Structure of aspartate-β-semialdehyde dehydrogenase from Escherichia coli, a key enzyme in the aspartate family of amino acid biosynthesis. J. Mol. Biol. 289 991–1002. [DOI] [PubMed] [Google Scholar]
- Hadfield, A.T., Shammas, C., Kryger, G., Ringe, D., Petsko, G.A., Ouyang, J., and Viola, R.E. 2001. Active site analysis of the potential antimicrobial target aspartate semialdehyde dehydrogenase. Biochemistry 40 14475– 14483. [DOI] [PubMed] [Google Scholar]
- Harb, O.S. and Kwaik, Y.A. 1998. Identification of the aspartate-β-semialdehyde dehydrogenase gene of Legionella pneumolphila and characterization of a null mutant. Infect. Immun. 66 1898–1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidelberg, J.F., Eisen, J.A., Nelson, W.C., Clayton, R.A., Gwinn, M.L., Dodson, R.J., Haft, D.H., Peterson, J.D., Umayam, L., and Gill S.R. 2000. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406 477–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, H.Y., Srinivasan, J., and Curtiss, R. 2002. Immune responses to recombinant pneumococcal PspA antigen delivered by live attenuated Salmonella enterica Serovar typhimurium vaccine. Infect. Immun. 70 1739–1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karsten, W.E. and Viola, R.E. 1991a. Chemical and kinetic mechanism of aspartate-β-semialdehyde dehydrogenase from Escherichia coli. Biochim. Biophys. Acta 1077 209–219. [DOI] [PubMed] [Google Scholar]
- ———. 1991b. Kinetic studies of L-aspartase from Escherichia coli: pH dependent activity changes. Arch. Biochem. Biophys. 287 60–67. [DOI] [PubMed] [Google Scholar]
- Lowther, W.T., Brot, N., Weissbach, H., Honek, J.F., and Matthews, B.W. 2000. Thiol-disulfide exchange is involved in the catalytic mechanism of peptide methionine sulfoxide reductase. Proc. Natl. Acad. Sci. 97 6463–6468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McRee, D.E. 1999. XtalView/Xfit—A versatile program for manipulating atomic coordinates and electron density. J. Struct. Biol. 125 156–165. [DOI] [PubMed] [Google Scholar]
- Moore, R.A., Bocik, W.E., and Viola, R.E. 2002. Expression and purification of L-aspartate-β-semialdehyde dehydrogenase from infectious microorganisms. Protein Expr. Purif. 25 189–194. [DOI] [PubMed] [Google Scholar]
- Otwinowski, Z. and Minor, W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276 307–326. [DOI] [PubMed] [Google Scholar]
- Ouyang, J. and Viola, R.E. 1995. Use of structural comparisons to select mutagenic targets in aspartate-β-semialdehyde dehydrogenase. Biochemistry 34 6394–6399. [DOI] [PubMed] [Google Scholar]
- Paris, S., Wessel, P.M., and Dumas, R. 2002. Overproduction, purification, and characterization of recombinant aspartate semialdehyde dehydrogenase from Arabidopsis thaliana. Protein Expr. Purif. 24 99–104. [DOI] [PubMed] [Google Scholar]
- Patte, J.C. 1983. Diaminopimelate and lysine. In Amino acids: Biosynthesis and genetic regulation (eds. K.M. Herrmann and R.L. Somerville), pp. 213–228. Addison-Wesley, Reading, MA.
- Viola, R.E. 2001. The central enzymes of the aspartate family of amino acid biosynthesis. Acc. Chem. Res. 34 339–349. [DOI] [PubMed] [Google Scholar]
- Wachsmuth, K., Olsvik, O., Evins, G.M., and Popovic, T. 1994. In Vibrio cholerae and cholera: Molecular to global perspective (eds. I.K. Wachsmuth, P.A. Blake, and O. Olsvik), pp. 357–370. ASM Press, Washington, D.C.