Abstract
Secreted fungal RNases, represented by RNase T1, constitute a family of structurally related proteins that includes ribotoxins such as α-sarcin. The active site residues of RNase T1 are conserved in all fungal RNases, except for Phe 100 that is not present in the ribotoxins, in which Leu 145 occupies the equivalent position. The mutant Leu145Phe of α-sarcin has been recombinantly produced and characterized by spectroscopic methods (circular dichroism, fluorescence spectroscopy, and NMR). These analyses have revealed that the mutant protein retained the overall conformation of the wild-type α-sarcin. According to the analyses performed, Leu 145 was shown to be essential to preserve the electrostatic environment of the active site that is required to maintain the anomalous low pKa value reported for the catalytic His 137 of α-sarcin. Enzymatic characterization of the mutant protein has revealed that Leu 145 is crucial for the specific activity of α-sarcin on ribosomes.
Keywords: Cytotoxic protein, ribonuclease, ribotoxins, RNase T1, RNase U2
Ribotoxins are a group of fungal-secreted ribosome-inactivating proteins that are among the most potent inhibitors of translation known (Lamy et al. 1992; Kao et al. 2001). They are cytotoxic proteins because, in the presence of their target cells, the endocytosed ribotoxin molecules inactivate the ribosome, leading to cell death via apoptosis (Olmo et al. 2001). The inactivation of the ribosome is due to the ribonucleolytic cleavage of a single phosphodiester bond of the larger rRNA (Schindler and Davies 1977; Endo et al. 1983) located within a universally conserved sequence, known as the sarcin/ricin loop (Correll et al. 1998, 1999). This cleavage releases an RNA fragment known as the α-fragment (Schindler and Davies 1977; Endo et al. 1983). Thus, ribotoxins are very specific ribonucleases displaying a common mode of action (Kao et al. 2001; Martínez-Ruiz et al. 2001), which is related to a high degree of sequence identity (Martínez-Ruiz et al. 1999) and an almost identical three-dimensional structure (Yang and Moffat 1996; Pérez-Cañadillas et al. 2000).
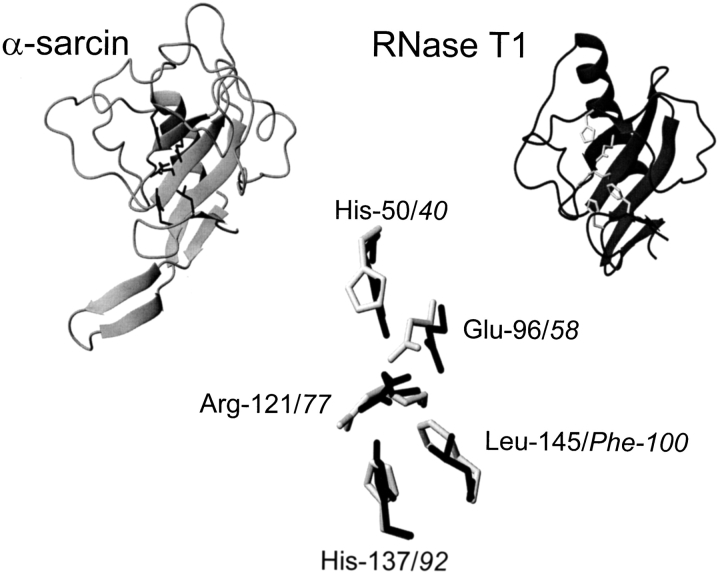
α-Sarcin (Olson and Goerner 1965) is the best characterized member of the group of ribotoxins (Gasset et al. 1994; Martínez-Ruiz et al. 2001) that belongs to the larger family of fungal secreted ribonucleases, usually represented by RNase T1 (Steyaert 1997; Yoshida 2001). These proteins exhibit different enzymatic activities but all of them share a common structural fold (Yang and Moffat 1996; Pérez-Cañadillas et al. 2000; Martínez-Ruiz et al. 2001). Thus, the fold present in α-sarcin shows the same architecture and connectivity of its secondary structure elements as that of RNase T1 (Fig. 1 ▶). This identity extends to the residues considered as catalytic (Fig. 1 ▶), and both α-sarcin and RNase T1 behave as cyclizing ribonucleases (Steyaert 1997; Lacadena et al. 1998), the overall reaction being composed of two steps, cyclization followed by cleavage of the 2′,3′ cyclic product formed. However, RNase T1 is a much less specific ribonuclease than α-sarcin (Steyaert 1997) and has not been reported to be cytotoxic. Nevertheless, the highly specific activity of α-sarcin is only observed at low protein concentrations (in the nanomolar range) (Endo et al. 1983), but micromolar concentrations produce a less specific cleavage of the substrates (Endo et al. 1983), showing only preference for the 3′-side of purines. Thus, although rather inefficiently, α-sarcin can also act as a rather nonspecific RNase when assayed against naked rRNA, homopolyribonucleotides, or dinucleotides (Endo et al. 1983; Lacadena et al. 1998; Kao et al. 2001; Martínez-Ruiz et al. 2001).
Figure 1.
Diagrams corresponding to the structures of α-sarcin and RNase T1 showing catalytic residues (His 50, Glu 96, Arg 121, His 137, in black for α-sarcin; His 40, Glu 58, Arg 77, His 92, in grey for RNase T1), constructed from the atomic coordinates deposited in the Protein Data Bank (PDB, references 1DE3 and 1RNT for α-sarcin and RNase T1, respectively). Both structures have been fitted to the coordinates of the peptide bond atoms of the residues. Leu 145 of α-sarcin and Phe 100 of RNase T1 are also shown. Images were generated by the MOLMOL program (Koradi et al. 1996).
The catalysis by RNase T1 has been largely studied, and it is well established that residues Glu 58 and His 92 serve as the base and acid catalysts, whereas Tyr 38, His 40, and Arg 77 seem to contribute to the stabilization of the transition state of the transesterification reaction (Steyaert 1997). These residues are conserved in all fungal RNases (Martínez-Ruiz et al. 1999; Yoshida 2001), including the ribotoxins. In α-sarcin, they correspond to Glu 96 and His 137, for the base and acid catalysts, and to Tyr 48, His 50, and Arg 121, for the other three. In fact, except for Tyr 48, the role of these α-sarcin residues has also been determined (Pérez-Cañadillas et al. 1998; Lacadena et al. 1999; Masip et al. 2001). Besides these similarities, there are some intriguing differences between α-sarcin and RNase T1 active sites. The most remarkable one is the low intrinsic pKa of the catalytic His 137 of the ribotoxin (pKa 5.8) (Pérez-Cañadillas et al. 1998), in comparison with the much higher value (pKa 7.8) for the corresponding His 92 of RNase T1 (Inagaki et al. 1981). In close proximity to this catalytic His 92, there is another conserved residue (Phe 100), which also contributes to the catalysis exerted by RNase T1 (Doumen et al. 1996). Phe 100 is conserved in all fungal RNases except for the ribotoxins (Martínez-Ruiz et al. 1999; Yoshida 2001) in which the amino acid leucine appears at the equivalent position. In this work, this α-sarcin residue, Leu 145, has been mutated to Phe.
Results
Purification and structural characterization of L145F
The L145F mutant variant of α-sarcin was purified to homogeneity as determined by its behavior on SDS-PAGE, where a single immunoreactive band was observed after reaction with polyclonal antibodies anti(α-sarcin) in Western-blot analysis, coincident with the single protein band detected after Coomassie Blue staining of the gel. In addition, only one colorless band was detected after electrophoresis on a poly(A)-containing gel (zymogram, performed at both pH 5.0 and 7.0), revealing the absence of contaminating poly(A)-degrading activities (see following). The amino acid composition of the purified protein was also in agreement with the mutation performed. The protein was obtained in a yield of 5–7 mg per liter of induced original bacterial culture.
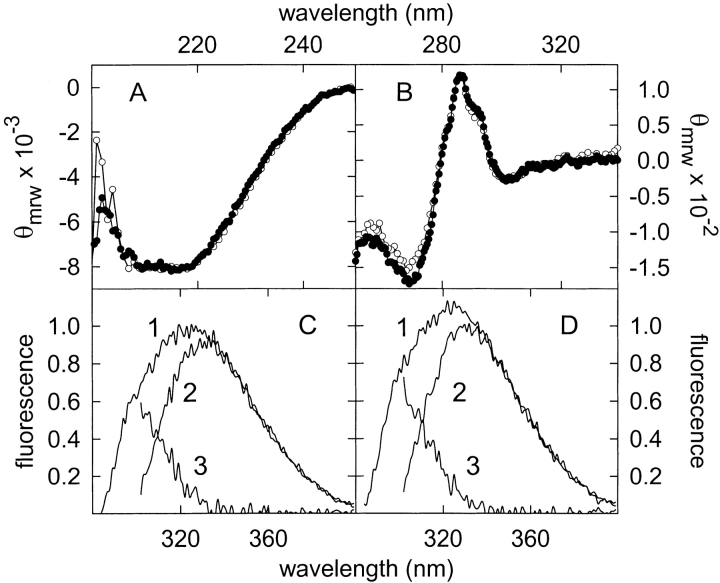
The absorbance as well as the near- and far-UV circular dichroism (CD) spectra of the L145F mutant were indistinguishable from those already reported for wild-type (WT) α-sarcin (Fig. 2A,B ▶) (Martínez del Pozo et al. 1988; Lacadena et al. 1994). The fluorescence emission spectra, using excitation at both 275 and 295 nm, of the L145F mutant were also similar to those of the WT protein, although a slight enhancement of 1.1-fold for both Tyr and Trp contributions were observed (Fig. 2C,D ▶). The presence of Trp 51, Tyr 48, and Tyr 106 in the vicinity of the mutated residue would explain these minor emission changes.
Figure 2.
Circular dichroism spectra in the far-UV (A) and near-UV (B) regions of wild-type (WT) α-sarcin (solid circles) and L145F mutant (open circles). θmrw, mean residue weight ellipticity values in units of degree × cm2 × dmole−1. Fluorescence emission spectra of WT α-sarcin (C) and L145F (D); 1, for excitation at 275 nm; 2 (tryptophan contribution), for excitation at 295 nm and further normalization at wavelengths above 380 nm where the contribution of tyrosine is negligible; 3 (tyrosine contribution), calculated as the difference spectra (spectrum 1 minus spectrum 2). Fluorescence is expressed in arbitrary units considering the intensity at the wavelength of the emission maximum of the WT protein, for excitation at 275 nm, as 1. All the spectra were recorded at 25°C.
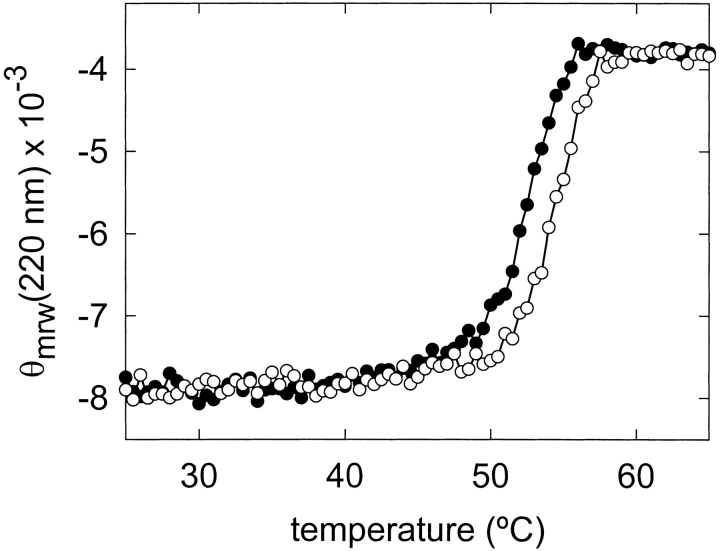
The thermal denaturation profile of the L145F mutant, obtained by CD measurements at 220 nm, revealed that this protein form showed a slightly increased conformational stability in comparison with the WT protein (Fig. 3 ▶). Thus, the Tm value of the L145F mutant was increased by only 2°C (54°C against 52°C for the WT protein at neutral pH; Lacadena et al. 1999; García-Ortega et al. 2001; Masip et al. 2001), which corresponded to an increased stability of only about 3.5 kJ/mole, estimated from the corresponding ΔΔG values (Becktel and Schellman 1987).
Figure 3.
Thermal denaturation profiles of wild-type α-sarcin (solid circles) and L145F (open circles). The measurements were performed by continuously recording the mean residue weight ellipticity at 220 nm, θmrw (220), in units of degree × cm2 × dmole−1.
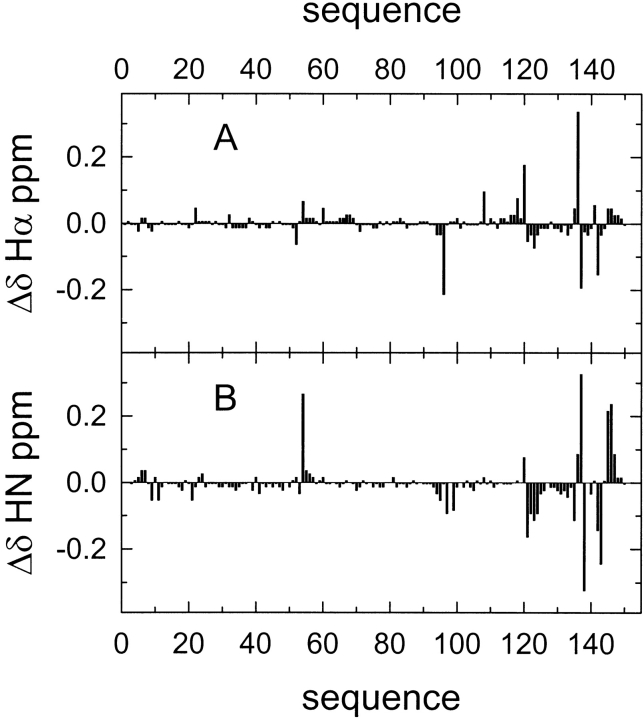
A comparison of the backbone HN and Hα chemical shifts of WT α-sarcin and L145F mutant is shown in Figure 4 ▶. Most of the chemical shift variations were located at space positions close to the mutation performed. Remarkably, the Asn 54 HN chemical shift represented one of the largest variations observed. The results from the Nuclear Magnetic Resonance (NMR) study strongly indicated that the substitution of the native residue Leu 145 by Phe did not induce major conformational changes in the enzyme. Additionally, the Nuclear Overhauser effect (NOE) pattern of the active site residues, His 50, Glu 96, and His 137, was also preserved.
Figure 4.
Comparison of the 1H NMR chemical shifts of wild-type α-sarcin and the L145F mutant. Chemical shift variations (Δδ = δmut − δwt) in the protein backbone resonances for Hα (α-protons) (A) and HN (peptide bond protons) (B), at pH 6.0 and 35°C, are shown along the protein sequence.
Ribonucleolytic activity of L145F
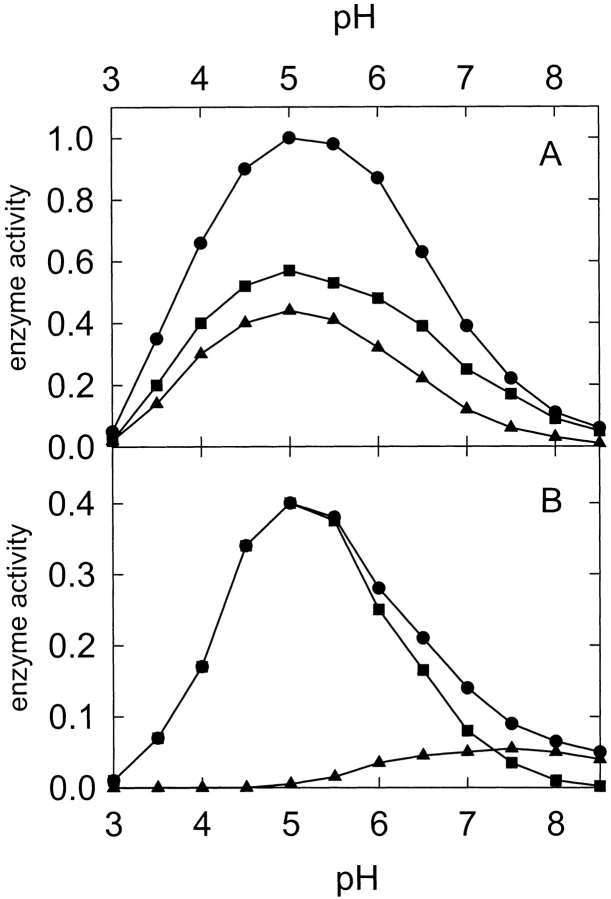
The characteristic activity of ribotoxins is manifested by the release of the α-fragment from rRNA when these proteins act on ribosomes. This specific ribonuclease activity of ribotoxins is also observed when oligonucleotides mimicking the sarcin/ricin loop are used as substrates (Kao et al. 2001). In addition, α-sarcin displays nonspecific ribonuclease activity against poly(A) as substrate, which is routinely used to detect any contaminating RNase present in their preparations (zymogram), as mentioned earlier. Finally, the activity of ribotoxins can be also quantified by measuring the hydrolysis of dinucleotides such as ApA (Lacadena et al. 1998). All of these assays were performed for the L145F mutant, indicating that the ribonuclease machinery of α-sarcin was conserved but also revealing noteworthy differences. Thus, WT α-sarcin and the L145F mutant showed very similar Km values (40 and 44 μM, respectively) but different kcat (2.7 × 10−4 s−1 for the WT protein and 8.9 × 10−5 s−1 for the mutant) when assayed against ApA at pH 5.0. No variation in the KM value has been reported from acid to neutral pH for WT α-sarcin (Lacadena et al. 1999) and only a small variation was observed for the KM value of the L145F mutant (from 44 ± 6 to 37 ± 7 μM). This assay was also used to obtain the pH profile of the ribonuclease activity (Fig. 5 ▶). Both WT and the L145F mutant showed an optimum activity at pH 5.0, but they were different in terms of the Adenosine Monophosphate (AMP) production (Fig. 5 ▶). The reaction of α-sarcin on ApA is composed of two steps: transphosphorylation (adenosine + 2′,3′-cyclic Adenosine Monophosphate (cAMP) are the reaction products) followed by hydrolysis of the cyclic mononucleotide producing 3′-AMP (Lacadena et al. 1998). Although WT α-sarcin displayed a similar profile for both steps, the pH profile for the hydrolysis step catalyzed by the L145F mutant is shifted toward a neutral pH (Fig. 5 ▶).
Figure 5.
pH dependence of the activity of α-sarcin (A) and L145F mutant (B) for the production of adenosine (circles), 2′,3′cAMP (squares) and 3′-AMP (triangles) using ApA as substrate. Activity is expressed in arbitrary units, considering the production of adenosine of the wild-type protein at pH 5.0 as unit.
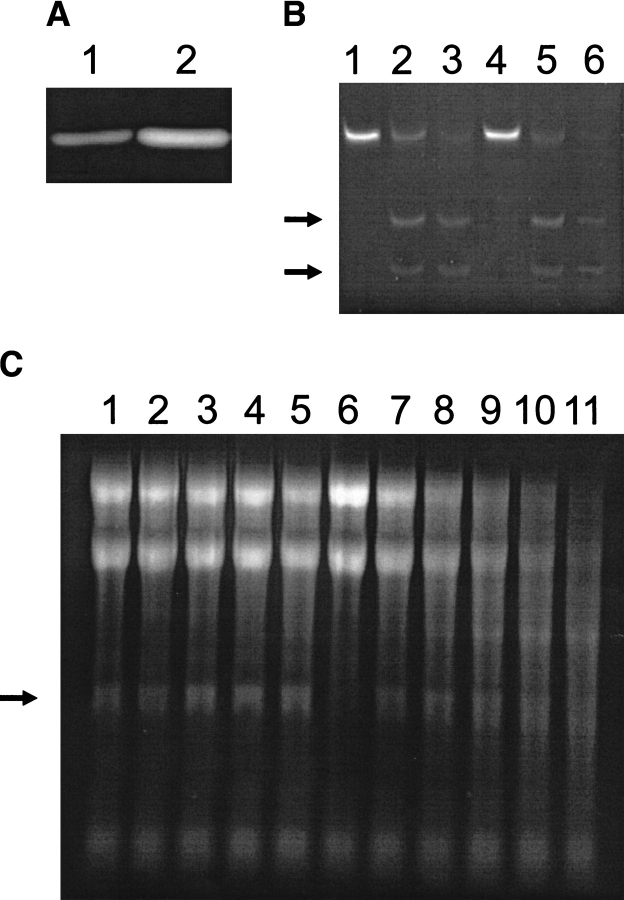
The mutant specifically degraded a synthetic 35mer oligonucleotide mimicking the sarcin/ricin loop (Fig. 6B ▶). Two bands corresponding to the 14mer and 21mer oligonucleotides resulting from the specific hydrolysis of a single phosphodiester bond were observed for both WT protein and L145F variant, although nonspecific cleavage of this synthetic substrate was observed when the assayed protein concentration was increased (Fig. 6B ▶). However, the L145F mutant showed lower specificity for the degradation of rRNA in ribosomes (Fig. 6C ▶). Although the α-fragment was observed when low concentrations of L145F were tested, nonspecific degradation of rRNA was produced with increasing protein concentration. The WT protein also produced nonspecific degradation of rRNA but a 1000-fold higher concentration was required. This fact is in agreement with the observation that the L145F mutant displayed higher activity against poly(A) than did the WT protein (twofold and fivefold at pH 7.0 and 5.0, respectively) (Fig. 6A ▶). These results differ from those obtained for the hydrolysis of ApA (the L145F mutant displayed about one-third the activity against the dinucleotide than did the WT protein).
Figure 6.
Ribonuclease activity assays of wild-type (WT) α-sarcin and its L145F mutant. (A) Zymogram assay of the ribonucleolytic activity against poly(A), at pH 7.0, of 100 ng of WT α-sarcin (lane 1) and the L145F (lane 2) mutant. When the zymogram was performed at pH 5.0, qualitatively identical results were obtained. (B) Activity assay on the 35mer oligonucleotide mimicking the sarcin/ricin loop (SRL-RNA). This substrate (lanes 1 and 4) was incubated in the presence of 65 ng of WT protein (lane 2) or L145F (lane 5) and 650 ng of these two proteins (lanes 3 and 6, respectively). The reaction mixture was analyzed by denaturing 19% polyacrylamide gels and stained with ethidium bromide. The two arrows indicate the 21mer and 14mer oligonucleotides resulting from the specific hydrolysis of a single phosphodiester bond of the sequence. (C) Ribosome inactivating activity assay. The highly specific activity of α-sarcin is shown by the release of the 400 nt α-fragment (arrow) from the 28S rRNA of eukaryotic ribosomes. Cell-free reticulocyte lysates (lane 6) were incubated in the presence of different amounts of WT protein (lanes 1–5; 100, 250, 500, 750, and 1000 ng) or L145F (lanes 7–11; 100, 250, 500, 750, and 1000 ng). The reaction mixture was analyzed in agarose gels and stained with ethidium bromide.
However, it is difficult to compare in quantitative terms both assays because they were performed under different conditions (in solution for ApA and as a zymogram for the polynucleotide); in addition, it would be also necessary to consider the polymeric character of the poly(A) substrate.
Cytotoxic activity of L145F
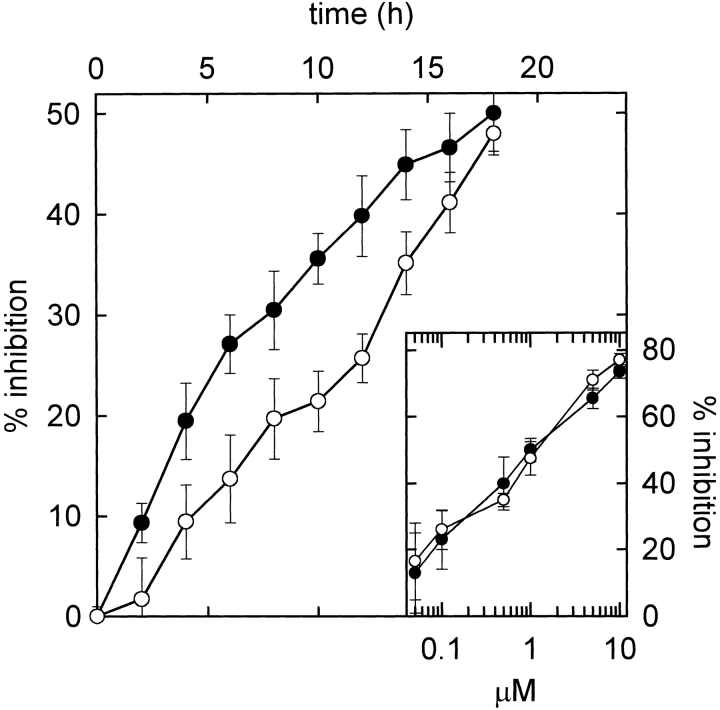
The L145F mutant retained the cytotoxic activity but a significant difference was observed in terms of the kinetics of the protein biosynthesis inhibition promoted by the two protein forms. This assay was performed on a human rhabdomyosarcoma cell line (Olmo et al. 2001), one of the tumor cell lines for which α-sarcin is cytotoxic (Turnay et al. 1993). The determination of the IC50 (concentration of toxin required for 50% protein biosynthesis inhibition) value is usually performed after an incubation of the cytotoxin and the cells for 18 h (Olmo et al. 2001). Under these conditions, the IC50 values of the WT protein and the L145F mutant were coincident (Fig. 7 ▶). However, at shorter incubation times, the L145F mutant displayed lower cytotoxicity than the WT protein (Fig. 7 ▶).
Figure 7.
Protein biosynthesis inhibition promoted by wild-type (WT) α-sarcin (solid circles) and the L145F mutant (open circles) at different incubation times. The protein concentration assayed in both cases was 1 μM. (Inset) Dependence of the protein biosynthesis inhibition on the concentration of WT (solid circles) and L145 (open circles) forms after 18 h incubation time. The data are expressed as percentages of control samples in the absence of toxin and are the average ± S.D. of three independent experiments with two different samples.
NMR titration of catalytic residues
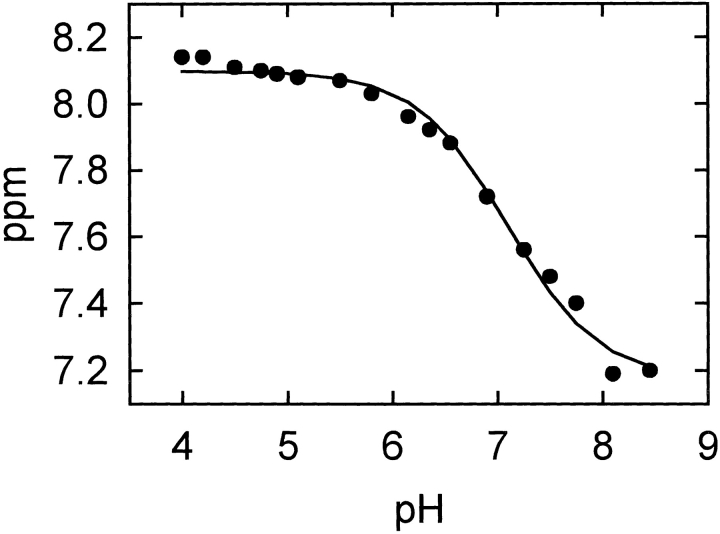
Finally, the pKa values of the three catalytic residues of α-sarcin were determined by NMR titration of the L145F mutant (Table 1). Only small differences were detected for His 50 and Glu 96; but a large shift, from 5.8 in the WT protein to 7.1 in the mutant, was observed for His 137 (Fig. 8 ▶).
Table 1.
Apparent pKa values of relevant catalytic residues in WT α-sarcin, L145F mutant, and RNase T1
| Residue | WT | L145F | RNase T1a |
| His 50 | 7.7 ± 0.2 | 7.9 ± 0.2 | 7.9 ± 0.01 |
| Glu 96 | 5.2 ± 0.1 | 5.0 ± 0.1 | 4.3 ± 0.05 |
| His 137 | 5.8 ± 0.1 | 7.1 ± 0.1 | 7.8 ± 0.01 |
a The equivalent residues to H50, E96, and H137 of α-sarcin in RNase T1 are H40, E58, and H92, respectively. Values from Inagaki et al. (1981).
(WT) Wild type.
Figure 8.
Chemical shift titration curve of His 137 Hɛ1 proton of α-sarcin L145F mutant in the pH range of 4.0–8.5. The pKa value of 7.1 was determined by a nonlinear least-squares fit (solid line) of the experimental pH titration curve to the Henderson-Hasselbach derived equation.
Discussion
RNase T1 and α-sarcin display a similar acid-base enzymatic mechanism (Steyaert 1997; Lacadena et al. 1998), with a very similar geometry of the active site residues when the three-dimensional structures of the two proteins are compared (Fig. 1 ▶). The pKa values for the ionization of the two histidines and the glutamic acid involved in the catalysis have been determined for both proteins (Table 1). These pKa values are very similar in both cases except for the low value of α-sarcin His 137. It is well known that the pKa values of ionizable residues are highly dependent on their local environments. Specifically, it has been recently described that the ionization of histidines in some proteins and in peptide model compounds is dependent on the ionic strength of the medium and the nature of the counterions used (Kao et al. 2000; Lee et al. 2002). This dependence is especially dramatic for histidine residues with significantly depressed pKa values (Lee et al. 2002), as in the case of α-sarcin His 137 (Table 1). In RNase T1, the imidazole ring of the equivalent residue (His 92) is stacked with the aromatic ring of Phe 100 (Doumen et al. 1996). In α-sarcin, this stacking interaction is not possible due to the presence of Leu 145 instead of Phe. As a consequence, the protonated imidazole ring of His 137 would not be as stable as that of His 92 in RNase T1 (Pérez-Cañadillas et al. 1998, 2000). Therefore, substitution of this Leu residue by Phe in the L145F mutant could allow establishment of a π-cation interaction between both residues that would contribute to the stabilization of the mentioned protonated imidazole ring. This new interaction alone could explain the increased His 137 pKa value observed in the L145F mutant (Table 1), a value that is much closer to that reported for RNase T1 His 92 (Inagaki et al. 1981).
Indeed, α-sarcin His 137 is deeply buried (solvent accessibility ∼7%) in a special environment because loop 5 (residues Thr 138 to Glu 144) folds toward the active center (Pérez-Cañadillas et al. 2000). At least two hydrogen bonds involving His 137 (Asn 8 δ21–His 137 O and Gly 143 O–His 137 Hδ1) contribute to maintain the orientation of loop 5 shielding this residue from the solvent (Pérez-Cañadillas et al. 2000). In RNase T1 the corresponding loop (residues Thr 93 to Asn 99) folds in the opposite direction and the catalytic His 92 is more exposed (solvent accessibility ∼22%) (Inagaki et al. 1981; Steyaert 1997). It is noteworthy that the residues within the α-sarcin loop 5 are among those showing highest NMR chemical shift variations on the mutation performed (Fig. 4 ▶), indicating that they have suffered local conformational rearrangements in the L145F mutant. These changes could lead to a more open conformation of the α-sarcin active site, and the greater solvent accessibility of His 137 in the L145F mutant could explain its increased pKa value.
It is quite generally accepted that ribotoxins would have evolved from extracellular microbial RNases by means of natural genetic engineering (Kao and Davies 1995, 1999, 2000). Then, what would have been the selective pressure on the ribotoxin family to introduce a Leu residue instead of the Phe conserved within the other nontoxic microbial RNases? This question is especially relevant considering that the L145F mutant is a slightly more stable protein (Fig. 3 ▶). One of the most obvious explanations would be to consider that this low pKa would be related to the pH profile of the enzymatic activity of α-sarcin. Actually, this profile is rather narrow and centered around pH 5.0, when the substrate used is the dinucleotide ApA (Pérez-Cañadillas et al. 1998; Lacadena et al. 1999). When dinucleotides are also used as substrates, RNase T1 has a more extended profile with maximum activity within the 4.0 to 7.0 pH range (Inagaki et al. 1981). However, WT α-sarcin and its L145F mutant displayed very similar pH profiles for the cleavage of ApA, although the WT protein was about 2.5- to 3.0-fold more active (Fig. 5 ▶). This result indicated that the acid-base mechanism of α-sarcin is not strongly affected by the individual pKa of the catalytic His 137.
Within this same idea, the L145F mutant retained the characteristic high specificity in cleaving a 35-mer oligonucleotide mimicking the rRNA sarcin/ricin loop (Fig. 6B ▶). However, it was more active against a nonspecific homopolymeric substrate such as poly(A) (Fig. 6A ▶) and, although it was still able to release the α-fragment when assayed against intact ribosomes, it lacked the high stringency of the WT protein (Fig. 6C ▶). This different behavior can be explained in molecular terms as follows. First, it has been recently shown how the NH2-terminal β-hairpin of α-sarcin (Leu 7 to Arg 22) is required for the recognition of the SRL in the ribosomes, but not for the hydrolysis of the 35-mer oligonucleotide (García-Ortega et al. 2002). Second, analysis of the crystal structure of restrictocin, a ribotoxin homologous to α-sarcin, bound to different analogs that mimic the SRL (Yang et al. 2001), reveal how this protein contacts the identity SRL element (a bulged G, G 15 in the 35-mer oligonucleotide) responsible for the specific recognition of the cleavage site, through Lys residues (Lys 111, 112, and 114 in α-sarcin) in a loop equivalent to loop 3 in α-sarcin. Thus, the L145F mutant would retain the specificity of α-sarcin because its NH2-terminal β-hairpin and the Lys residues of loop 3 remain practically unchanged (Fig. 4 ▶).
However, these results also show that the L145F mutant is a less specific but slightly more active ribonuclease (Fig. 6A,C ▶). The earlier mentioned different accessibility of the active site might account for this lack of absolute specificity observed in the mutant. In fact, a specific role for the Asn 8-His 137 hydrogen bond in determining the accessibility of His 137 in the native active site was proposed earlier. This bond is also present in the restrictocin crystal structure (Yang and Moffat 1996), and it has been reported that mutating the corresponding Asn residue (Asn 7 in restrictocin) to Ala results in the production of a more active enzyme form at the expense of reducing substrate specificity (Kao and Davies 2000). This result was also interpreted by these authors as being due to the elimination of the existing hydrogen bond between Asn 7 and the catalytic His 136 in the restrictocin mutant. Furthermore, another conserved residue within the family of the extracellular microbial ribonucleases is Asn 54 of α-sarcin. The equivalent residue in RNase T1 (Asn 44) has been proven to be directly involved in the recognition of the substrate guanine moiety (Kumar and Walz 2001). One of the residues showing the largest NMR chemical shift variation in the L145F mutant of α-sarcin is Asn 54 (Fig. 4 ▶), which would be in perfect agreement with the idea of a conformational rearrangement of the L145F mutant active site leading to a less specific enzyme.
Finally, it seems clear that substitution of α-sarcin Leu 145 by Phe results in the modification of the complex relationships existing between the residues in the active site and loop 5, producing a more efficient ribonuclease but a less specific ribotoxin. In fact, the mutant still recognizes the ribosome, but it does not identify the target bond as efficiently as the WT α-sarcin, resulting in the cleavage of many other rRNA phosphodiester bonds (Fig. 6C ▶). If such is the case, one should expect the L145F mutant to be less cytotoxic than the WT protein when assayed against intact cells, despite its higher nonspecific activity on ribosomes. This is easily explained just by considering that the substrate concentration for the WT protein would be only one bond per ribosome, whereas for the L145F mutant that concentration would be much higher due to its lower specificity. Cleavage of only that single phosphodiester bond located in the SRL is enough to inactivate the ribosome. Thus, the number of ribosomes that could be inactivated by a protein molecule in a short period of time would be higher for the WT protein than for the L145F mutant. This was in fact the result obtained (Fig. 7 ▶). The WT protein displayed a higher cytotoxic activity for protein biosynthesis inhibition at short periods of time, but both proteins, WT and L145F, displayed similar cytotoxicity for longer treatments against the target cells.
In summary, α-sarcin would be a highly engineered ribonuclease. The evolution would give loops for the interaction with membranes, an NH2-terminal hairpin for recognition of the ribosome, and the substitution of Phe by Leu to give specificity at the resulting molecule. Additionally, the change of Phe by Leu from RNase T1 to ribotoxins modifies the pKa of the catalytic His 137, revealing a major role of that residue in preserving the unique accessibility and electrostatic properties of the α-sarcin active site.
Materials and methods
DNA manipulations
All materials and reagents were molecular biology grade. Cloning procedures, including oligonucleotide-site directed mutagenesis, and bacteria manipulations were carried out according to standard methods, as described previously (Lacadena et al. 1994, 1999; Martínez-Ruiz et al. 2001). The mutagenic primer used was 5′-CAGGGCGAATTTAAGCTCTGC-3′. The base that changes the α-sarcin Leu 145 codon to Phe is underlined.
Protein production and purification
Escherichia coli BL21(DE3) cells cotransformed with a thioredoxin-producing (pT-Trx) and the α-sarcin mutant plasmids were used to produce and purify the mutant, as described for the WT protein (Lacadena et al. 1994; García-Ortega et al. 2000). Fungal WT α-sarcin was produced and purified according to methods previously reported (Olson and Goerner 1965; Martínez-Ruiz et al. 2001). Polyacrylamide gel electrophoresis of proteins, protein hydrolysis, and amino acid analysis were also performed according to standard procedures (Lacadena et al. 1994).
Spectroscopic characterization
Absorbance measurements were carried out on a Uvikon 930 spectrophotometer at 100 nm/min scanning speed, at room temperature, and in 1-cm optical-path cells. CD spectra were obtained on a Jasco 715 spectropolarimeter at 0.2 nm/sec scanning speed; 0.1- and 1.0-cm optical-path cells were used in the far and near UV, respectively. Mean residue weight ellipticities were expressed in units of degree × cm2 × dmole−1. Thermal denaturation profiles were obtained by measuring the temperature dependence of the ellipticity at 220 nm in the 25°–85°C range; the temperature was continuously changed at a rate of 0.5°C/min. Fluorescence emission spectra were obtained on an SLM Aminco 8000 spectrofluorimeter at 25°C and in 0.2-cm optical-path cells. All these determinations were performed as described previously (Martínez del Pozo et al. 1988; Lacadena et al. 1994, 1999; Masip et al. 2001).
NMR experiments
1H NMR spectra of the L145F mutant was assigned using standard sequential assignment procedures (Wüthrich 1986) and comparing with the previous assignment of the WT α-sarcin (Campos-Olivas et al. 1996). NMR experiments were performed on a Bruker Avance 600 spectrometer. Data were collected with a 1.5 mM sample at 35°C and pH 6.0. 1H homonuclear total correlated spectroscopy (TOCSY) (Bax and Davies 1985) with a mixing time of 60 msec, and Nuclear Overhauser enhancement spectroscopy (NOESY) spectra (Kumar et al. 1980) with a 50-msec mixing time, were recorded by standard methods. The effect of pH titration on the chemical shift of the imidazole protons was determined by analyzing a set of 1D H-NMR spectra in D2O recorded at 17 pH values ranging from 4.0 to 8.5. Ambiguities were solved with the help of NOESY and TOCSY spectra recorded at different pH values and by comparison to the titration curves of the WT protein (Pérez-Cañadillas et al. 1998) and other single mutants. The pKa value of His 137 was then determined by a nonlinear least-squares fit of the experimental pH titration curve of the Hɛ1 proton to the Henderson-Hasselbach derived equation as described previously (Pérez-Cañadillas et al. 1998).
Ribonucleolytic activity
The specific ribonucleolytic activity of α-sarcin on ribosomes was followed by detecting the release of the 400-nt α-fragment from a cell-free reticulocyte lysate (Kao et al. 2001). The production of the 400-nt α-fragment was visualized by ethidium bromide staining after electrophoresis on 2.4% agarose gels. The activity of the purified proteins against poly(A) (zymogram) was also assayed as previously described (Kao et al. 2001). Enzymatic hydrolysis of ApA was performed as described before (Lacadena et al. 1998) by analyzing the reaction products (ApA, adenosine, 3′-AMP, and 2′,3′-cAMP) by High Performance Liquid Chromotography (HPLC). All assays were performed with controls to test potential nonspecific degradation of the substrates, which did not occur under the conditions used. The specific cleavage of a synthetic 35-mer RNA by α-sarcin was also studied. The synthesis of this SRL RNA was carried out as described (Kao et al. 2001), by using synthetic and urea-PAGE purified DNA templates, (T7-promoter) 5′-TTCTAATACGACTCACTATAG-3′ and (template) 3′-AA GATTATGCTGAGTGATATCCCTTAGGACGAGTCATGCTC TCCTTGGCGTCCAA-5′, and the AmpliScribe T7 Transcription Kit. The resulting product was purified by electrophoresis on 8% (w/v)-polyacrylamide gel, containing 7 M urea in 45 mM Tris-borate buffer at pH 8.3, containing 1 mM Ethylenediamine tetracetic acid (EDTA) (Sambrook et al. 1989). The assay was performed with 2 μM SRL RNA and incubation for 20 min at 37°C in 10 mM Tris-HCl buffer at pH 7.4 (Kao et al. 2001). The reaction products were detected by ethidium bromide staining after electrophoretic separation on a denaturing 19%(w/v)-polyacrylamide gel. The specific action of α-sarcin produced both 21-mer and 14-mer fragments.
Cytotoxicity assay
This assay was performed essentially as described (Turnay et al. 1993; Olmo et al. 2001) by using human rhabdomyosarcoma cells. Protein synthesis was analyzed by measuring the incorporation of L-[4,5-3H]leucine (166 Ci/mmole). The radioactivity was measured on a Beckman LS 3801 liquid scintillation counter. The results are expressed as percentage of radioactivity incorporation in control samples. A plot of these percentage values versus toxic protein concentration in the cytotoxicity assay allows the calculation of the IC50 values. The reported values correspond to the average of triplicate experiments.
Acknowledgments
This work was supported by grants BMC2000–0551 from the Ministerio de Ciencia y Tecnología (Spain) and PB98–0677 and PM98–0083 from the Ministerio de Educación y Cultura (MEC) (Spain). M.M., L.G.-O., and F.G.-M. are recipients of fellowships from the MEC (Spain) and the Comunidad Autónoma de Madrid (CAM, Spain), respectively.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.0225903.
References
- Bax, A. and Davies, D.G. 1985. MLEV-17 based two-dimensional homonuclear magnetisation transfer spectroscopy. J. Magn. Reson. 65 355–360. [Google Scholar]
- Becktel, W.J. and Schellman, J.A. 1987. Protein stability curves. Biopolymers 26 1859–1877. [DOI] [PubMed] [Google Scholar]
- Campos-Olivas, R., Bruix, M., Santoro, J., Martínez del Pozo, A., Lacadena, J., Gavilanes, J.G., and Rico, M. 1996. 1H and 15N nuclear magnetic resonance assignment and secondary structure of the cytotoxic ribonuclease α-sarcin. Protein Sci. 5 969–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correll, C.C., Munishkin, A., Chan, Y.L., Ren, Z., Wool, I.G., and Steitz, T.A. 1998. Crystal structure of the ribosomal RNA domain essential for binding elongation factors. Proc. Natl. Acad. Sci. 9513436–13441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correll, C.C., Wool, I.G., and Munishkin, A. 1999. The two faces of the Escherichia coli 23 S rRNA sarcin/ricin domain: The structure at 1.11 Δ resolution. J. Mol. Biol. 292 275–287. [DOI] [PubMed] [Google Scholar]
- Doumen, J., Gonciarz, M., Zegers, I., Loris, R., Wyns, L., and Steyaert, J. 1996. A catalytic function for the structurally conserved residue Phe 100 of ribonuclease T1. Protein Sci. 5 1523–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo, Y., Hubert, P.W., and Wool, I.G. 1983. The ribonuclease activity of the cytotoxin α-sarcin: The characteristics of the enzymatic activity of α-sarcin with ribosomes and ribonucleic acids as substrates. J. Biol. Chem. 258 2662–2667. [PubMed] [Google Scholar]
- García-Ortega, L., Lacadena, J., Lacadena, V., Masip, M., de Antonio, C., Martínez-Ruiz, A., and Martínez del Pozo, A. 2000. The solubility of the ribotoxin α-sarcin, produced as a recombinant protein in Escherichia coli, is significantly increased in the presence of thioredoxin. Lett. Appl. Microbiol. 30 298–302. [DOI] [PubMed] [Google Scholar]
- García-Ortega, L., Lacadena, J., Mancheño, J.M., Oñaderra, M., Kao, R., Davies, J., Olmo, N., Martínez del Pozo, A., and Gavilanes, J.G. 2001. Involvement of the amino-terminal β-hairpin of the Aspergillus ribotoxins on the interaction with membranes and non-specific ribonuclease activity. Protein Sci. 10 1658–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Ortega, L., Masip, M., Mancheño, J.M., Oñaderra, M., Lizarbe, M.A., García-Mayoral, M.F., Bruiz, M., Martínez del Pozo, A., and Gavilanes, J.G. 2002. Deletion of the NH2-terminal β-hairpin of the ribotoxin α-sarcin produces a non-toxic but active ribonuclease. J. Biol. Chem. 277 18632–18639. [DOI] [PubMed] [Google Scholar]
- Gasset, M., Mancheño, J.M., Lacadena, J., Turnay, J., Olmo, N., Lizarbe, M.A., Martínez del Pozo, A., Oñaderra, M., and Gavilanes, J.G. 1994. α-Sarcin, a ribosome-inactivating protein that translocates across the membrane of phospholipid vesicles. Current Topics in Peptide and Protein Research 1 99–104. [Google Scholar]
- Inagaki, F., Kawano, Y., Shimada, I., Takahashi, K., and Miyazawa, T. 1981. Nuclear magnetic resonance study on the microenvironments of histidine residues of ribonuclease T1 and carboxymethylated ribonuclease T1. J. Biochem. (Tokyo) 89 1185–1195. [PubMed] [Google Scholar]
- Kao, R. and Davies, J. 1995. Fungal ribotoxins: A family of naturally engineered toxins? Biochem. Cell Biol. 73 1151–1159. [DOI] [PubMed] [Google Scholar]
- ———. 1999. Molecular dissection of mitogillin reveals that the fungal ribotoxins are a family of natural genetically engineered ribonucleases. J. Biol. Chem. 274 12576–12582. [DOI] [PubMed] [Google Scholar]
- ———. 2000. Single amino acid substitution affecting the specificity of the fungal ribotoxin mitogillin. FEBS Lett. 466 87–90. [DOI] [PubMed] [Google Scholar]
- Kao, Y.-H., Fitch, C.A., Bhattacharya, S., Sarkisian, C.J., Lecomte, J.T.J., and García-Moreno, E.B. 2000. Salt effects on ionization equilibria of histidines in myoglobin. Biophys. J. 79 1637–1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao, R., Martínez-Ruiz, A., Martínez del Pozo, A., Crameri, R., and Davies, J. 2001. Mitogillin and related fungal ribotoxins. Methods Enzymol. 341 324–335. [DOI] [PubMed] [Google Scholar]
- Koradi, R., Billeter, M., and Wütrich, K. 1996. MOLMOL: A program for display and analysis of macromolecular structures. J. Mol. Graph. 14 51–55. [DOI] [PubMed] [Google Scholar]
- Kumar, A., Ernst, R.R., and Wüthrich, K. 1980. A two-dimensional nuclear Overhauser enhancement (2D NOE) experiment for the elucidation of complete proton-proton cross relaxation networks in biological macromolecules. Biochem. Biophys. Res. Commun. 95 1–6. [DOI] [PubMed] [Google Scholar]
- Kumar, K. and Walz, Jr., F.G. 2001. Probing functional perfection in substructures of ribonuclease T1: Double combinatorial random mutagenesis involving Asn43, Asn44, and Glu46 in the guanine binding loop. Biochemistry 40 3748–3757. [DOI] [PubMed] [Google Scholar]
- Lacadena, J., Martínez del Pozo, A., Barbero, J.L., Mancheño, J.M., Gasset, M., Oñaderra, M., López-Otín, C., Ortega, S., García, J.L., and Gavilanes, J.G. 1994. Overproduction and purification of biologically active native fungal α-sarcin in Escherichia coli. Gene 142 147–151. [DOI] [PubMed] [Google Scholar]
- Lacadena, J., Martínez del Pozo, A., Lacadena, V., Martínez-Ruiz, A., Mancheño, J.M., Oñaderra, M., and Gavilanes, J.G. 1998. The cytotoxin α-sarcin behaves as a cyclizing ribonuclease. FEBS Lett. 424 46–48. [DOI] [PubMed] [Google Scholar]
- Lacadena, J., Martínez del Pozo, A., Martínez-Ruiz, A., Pérez-Cañadillas, J.M., Bruix, M., Mancheño, J.M., Oñaderra, M., and Gavilanes, J.G. 1999. Role of histidine-50, glutamic acid-96 and histidine-137 in the ribonucleolytic mechanism of the ribotoxin α-sarcin. Proteins 37 474–484. [DOI] [PubMed] [Google Scholar]
- Lamy, B., Davies, J., and Schindler, D. 1992. The Aspergillus ribonucleolytic toxins (ribotoxins). In Genetically engineered toxins (ed. A.E. Frankel), pp. 237–257. Marcel Dekker, New York, NY. [PubMed]
- Lee, K.K., Fitch, C.A., Lecomte, J.T.J., and García-Moreno, E.B. 2002. Electrostatic effects in highly charged proteins: Salt sensitivity of pKa values of histidines in staphylococcal nuclease. Biochemistry 41 5656–5657. [DOI] [PubMed] [Google Scholar]
- Martínez del Pozo, A., Gasset, M., Oñaderra, M., and Gavilanes, J.G. 1988. Conformational study of the antitumor protein α-sarcin. Biochim. Biophys. Acta 953 280–288. [DOI] [PubMed] [Google Scholar]
- Martínez-Ruiz, A., Kao, R., Davies, J., and Martínez del Pozo, A. 1999. Ribotoxins are a more widespread group of proteins within the filamentous fungi than previously believed. Toxicon 37 1549–1563. [DOI] [PubMed] [Google Scholar]
- Martínez-Ruiz, A., García-Ortega, L., Kao, R., Lacadena, J., Oñaderra, M., Mancheño, J.M., Davies, J., Martínez del Pozo, A., and Gavilanes, J.G. 2001. RNase U2 and α-sarcin: A study of relationships. Methods Enzymol. 341 335–351. [DOI] [PubMed] [Google Scholar]
- Masip, M., Lacadena, J., Mancheño, J.M., Oñaderra, M., Martínez-Ruiz, A., Martínez del Pozo, A, and Gavilanes, J.G. 2001. Arginine 121 is a crucial residue for the specific cytotoxic activity of the ribotoxin α-sarcin. Eur. J. Biochem. 268 1–8. [DOI] [PubMed] [Google Scholar]
- Olmo, N., Turnay, J., González de Buitrago, G., López de Silanes, I., Gavilanes, J.G., and Lizarbe, M.A. 2001. Cytotoxic mechanism of the ribotoxin α-sarcin. Induction of cell death via apoptosis. Eur. J. Biochem. 268 2113–2123. [DOI] [PubMed] [Google Scholar]
- Olson, B.H. and Goerner, G.L. 1965. α-Sarcin, a new antitumor agent. I. Isolation, purification, chemical composition, and the identity of a new amino acid. Appl. Microbiol. 13 314–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Cañadillas, J.M., Campos-Olivas, R., Lacadena, J., Martínez del Pozo, A., Gavilanes, J.G., Santoro, J., Rico, M., and Bruix, M. 1998. Characterization of pKa values and titration shifts in the cytotoxic ribonuclease α-sarcin by NMR. Relationship between electrostatic interactions, structure and catalytic function. Biochemistry 37 15865–15876. [DOI] [PubMed] [Google Scholar]
- Pérez-Cañadillas, J.M., Santoro, J., Campos-Olivas, R., Lacadena, J., Martínez del Pozo, A., Gavilanes, J.G., Rico, M., and Bruix, M. 2000. The highly refined solution structure of the cytotoxic ribonuclease α-sarcin reveals the structural requirements for substrate recognition and ribonucleolytic activity. J. Mol. Biol. 299 1061–1073. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. 1989. Molecular cloning: A laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Schindler, D.G. and Davies, J.E. 1977. Specific cleavage of ribosomal RNA caused by α-sarcin. Nucleic Acids Res. 4 1097–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steyaert, J. 1997. A decade of protein engineering on ribonuclease T1. Atomic dissection of the enzyme-substrate interaction. Eur. J. Biochem. 247 1–11. [DOI] [PubMed] [Google Scholar]
- Turnay, J, Olmo, N., Jiménez, A., Lizarbe, M.A., and Gavilanes, J.G. 1993. Kinetic study of the cytotoxic effect of α-sarcin, a ribosome inactivating protein from Aspergillus giganteus, on tumour cell lines: Protein biosynthesis inhibition and cell binding. Mol. Cell. Biochem. 122 39–47. [DOI] [PubMed] [Google Scholar]
- Wüthrich, K. 1986. NMR of proteins and nucleic acids. Wiley, New York.
- Yang, X.J. and Moffat, K. 1996. Insights into specificity of cleavage and mechanism of cell entry from the crystal structure of the highly specific Aspergillus ribotoxin, restrictocin. Structure 4 837–852. [DOI] [PubMed] [Google Scholar]
- Yang, X., Gérczei, T., Glover, L.T., and Correll, C.C. 2001. Crystal structures of restrictocin-inhibitor complexes with implications for RNA recognition and base flipping. Nat. Struct. Biol. 8 968–973. [DOI] [PubMed] [Google Scholar]
- Yoshida, H. 2001. The ribonuclease T1 family. Methods Enzymol. 341 28–41. [DOI] [PubMed] [Google Scholar]