Abstract
Pressure-induced unfolding of a molten globule (MG) was studied in a residue-specific manner with 1H-15N two-dimensional NMR spectroscopy using a variant of human α-lactalbumin (α-LA), in which all eight cysteines had been replaced with alanines (all-Ala α-LA). The NMR spectrum underwent a series of changes from 30 to 2000 bar at 20°C and from −18°C to 36°C at 2000 bar, showing a highly heterogeneous unfolding pattern according to the secondary structural elements of the native structure. Unfolding began in the loop part of the β-domain, and then extended to the remainder of the β-domain, after which the α-domain began to unfold. Within the α-domain, the pressure stability decreased in the order: D-helix ∼ 310-helix > C-helix ∼ B-helix > A-helix. The D-helix, C-terminal 310-helix and a large part of B- and C-helices did not unfold at 2000 bar, even at 36°C or at −18°C. The results verify that the MG state consists of a mixture of variously unfolded conformers from the mostly folded to the nearly totally unfolded that differ in stability and partial molar volume. Not only heat but also cold denaturation was observed, supporting the view that the MG state is stabilized by hydrophobic interactions.
Keywords: Molten globule; α-lactalbumin; all-Ala α-lactalbumin; high-pressure NMR; partial molar volume; pressure unfolding, heat and cold denaturation
A protein in solution is a dynamic entity, able to adopt a variety of conformation between the native (N) and the fully denatured (U) states. One of the conformations frequently observed in globular proteins under mildly denaturing conditions is a compact denatured state, sometimes called the molten globule (MG), defined as a state with native-like secondary structure, but lacking fixed side-chain packing (Dolgikh et al. 1981; Ohgushi and Wada 1983). Although different views are expressed over the term “molten globule,” we retain this term for the compact denatured state that we deal with in the present report (Kuwajima 1989). The polypeptide chain in a MG is only loosely packed, typically showing a radius of gyration (Rg) of ∼10% larger than the corresponding Rg for the native state (Kataoka et al. 1997; Kamatari et al. 1999). The polypeptide chain must be hydrated, but the state of hydration should be different from that of the fully unfolded state U, and spatial heterogeneity in hydration is also possible. As its chain fold is loose, it must have ample motions leading to fluctuating packing density and volume fluctuations. Thus, the partial molar volume and compressibility are of general concern for the MG state (Chalikian et al. 1995; Kharakoz 1997). In accordance with this, molten globules generally give a poorly resolved and broadened 1H NMR spectra and weak or no signals in 1H-15N two-dimensional HSQC NMR spectra; this is considered to arise from the heterogeneous polypeptide conformations which exchange with life times on the order of ∼mseconds (Baum et al. 1989).
Typical molten globules are found in vitro at low pH (Ohgushi and Wada 1983; Ptitsyn 1991; Ptitsyn and Uversky 1994) and often with high salt concentration. They are also found in the presence of alcohol (Kamatari et al. 1998, 1999). Molten globules have also been found as transient intermediates in protein folding when starting from a highly denatured state such as that produced in the presence of a high concentration of a denaturant (Ptitsyn 1995). Under physiologic conditions, the molten globule is more stable than the unfolded state, probably as a result of residual hydrophobic interactions. This view is supported partly by the observation of cold denaturation of the MG in some proteins (Gursky and Atkinson 1996; Maldonado et al. 1998). Generally, MG-like structures are often considered to be produced in cold denaturation (Ptitsyn 1995), in heat denaturation (Chattopadhyay and Mazumdar 2000), or in pressure denaturation (Zhang et al. 1995; Ruan et al. 1997; Jonas et al. 1998; Lassalle et al. 2000; Kitahara et al. 2002). In living cells, the MG state is likely to be present in equilibrium with the native state, and may be actively involved in various biologic processes such as targeting, transport, and aggregation (Bychkova and Ptitsyn 1993).
In the present study, our purpose is to examine the MG state through volumetric properties using high-pressure NMR. First of all, we would like to know if pressure alone can unfold the MG. This would simply tell us whether the partial molar volume of the MG is larger or smaller than that of the unfolded state, U, and may provide information about the hydration and chain packing. We are also interested in knowing details of cooperativity in the MG → U transition. The nature of interactions that stabilize the MG is also of interest. As a protein system, we choose all-Ala α-LA, namely human α-lactalbumin (α-LA) in which all of its eight cysteine residues forming the four disulfide bridges have been replaced with alanines (Peng et al. 1995). In a previous study carried out at 1 bar, all-Ala α-LA at pH 2 was shown to form a typical molten globule with a hydrodynamic radius of 2.17 nm close to that (1.97 nm) of the native conformer (Redfield et al. 1999). It lacks close packing of the side chains, and has a native-like helical content and a bipartite structure, with the α-helical domain expanded but adopting a native-like fold and the β-domain partially unfolded, similar to the A-state of α-LA (Alexandrescu et al. 1993; Chyan et al. 1993; Wu et al. 1995; Schulman et al. 1997; Troullier et al. 2000). In this previous study, urea-induced unfolding of the MG of all-Ala α-LA was followed with 1H-15N 2D HSQC NMR spectroscopy, demonstrating noncooperative unfolding behavior of this MG (Redfield et al. 1999). Here, we use pressure as a new perturbant and examine the pressure response of the MG state in a residue-specific manner using 1H-15N 2D HSQC NMR spectroscopy.
The special feature of using pressure as a perturbant, in comparison with temperature or chemical perturbation, is understood straightforwardly in the following. The Gibbs energy determining the thermodynamic equilibrium among different conformers of a protein in solution is driven by pressure according to the following relation,
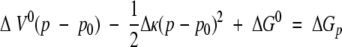 |
1 |
where ΔG° and ΔV° are differences in the Gibbs energy and partial volume at 1 bar, respectively, and Δκ denotes the difference in compressibility. Pressure simply changes the conformational equilibrium by acting on volumetric properties, while denaturants such as urea and GdnHCl directly perturb the interaction energy and entropy embedded in ΔG° and change the conformation (Wu and Wang 1999; AbouAiad et al. 1997). That is, pressure drives the equilibrium to increase the population of the lower volume conformer relative to the higher volume conformer (Weber and Drickamer 1983; Inoue et al. 2000). If conformers with different partial molar volumes coexist but are hidden in the system, applying pressure may disclose their presence.
Results
Pressure-induced changes in15N-1H HSQC 2D NMR spectra
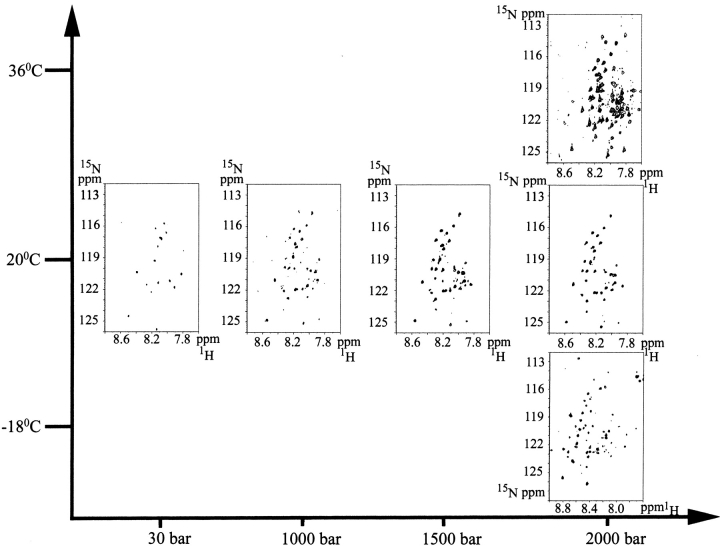
Figure 1 ▶ shows fully reversible 15N-1H HSQC spectral changes of all-Ala α-LA (pH 2) under various conditions of temperature and pressure. All-Ala α-LA has 124 residues, of which 121 backbone amide groups are potentially observable as cross-peaks in the HSQC spectrum. We note that at 20°C at 30 bar, only 16 cross-peaks are observed in the narrow region of 7.8 ppm–8.5 ppm for 1H. Under similar conditions (pH 2, 1 bar), 37 cross-peaks were previously observed (Redfield et al. 1999). The difference between the two experiments is attributable to the lower signal-to-noise ratio due to a small sample volume (∼20 μL) in the active part of the high pressure cell; the 16 peaks observed here are the most intense peaks observed in the previous study (Redfield et al. 1999). The loss of intensity of many cross-peaks (105 peaks remain undetected) has been considered typical for a molten globule due to the restriction of mobility of its disordered but compact polypeptide chain (Baum et al. 1989; Schulman et al. 1997). The region of the polypeptide chain giving rise to the 16 cross-peaks is less restricted in its mobility, due to local unfolding, giving observable signals even though the majority of the chain is in the compact molten globule state.
Figure 1.
15N-1H HSQC-spectra of all-Ala α-lactalbumin (pH 2) as a function of pressure and temperature (20°C, 36°C, or −18°C).
In 8 M urea, cross-peaks from all residues of all-Ala α-LA are observable in the HSQC spectrum, and have been assigned to specific amino acid residues using a three-dimensional 15N-edited NOESY-HSQC experiment (Redfield et al. 1999). The 16 cross-peaks observed at 20°C at 30 bar were assigned to specific amino acid residues by comparison with the spectrum collected in the absence of urea in the previous study (Redfield et al. 1999). These are localized to the N-terminus and the loop part of the β-domain (Fig. 2 ▶), indicating that these regions are locally unfolded; this is consistent with the earlier finding that the β-domain is more unstructured than the α-domain in the α-LA MG (Schulman et al. 1997; Redfield et al. 1999).
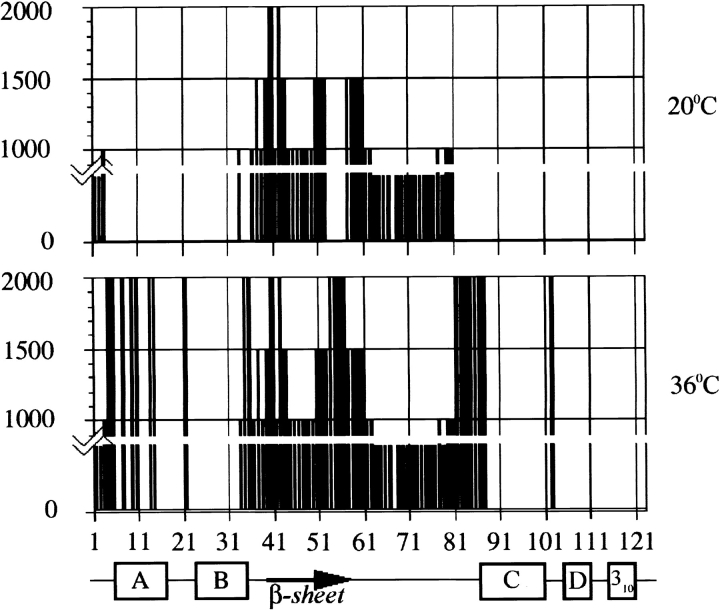
Figure 2.
Histograms showing the stepwise appearance of the 15N-1H HSQC cross-peaks of all-Ala α-lactalbumin (pH 2) as a function of pressure and temperature. The height of the bar indicates the pressure at which the cross-peak first appears. Cross-peaks are visible from residues 1, 2, 63–66, 68–76, 78 (16 peaks) at 30 bar, 20°C; from residues 1–3, 33, 36, 38, 41, 44–49, 61–66, 68–80 (32 peaks) at 1000 bar, 20°C; from residues 1–3, 33, 36–39, 41, 43–52, 57–66, 68–80 (42 peaks) at 1500 bar, 20°C; from residues 1–3, 33, 36–52, 57–66, 68–80 (44 peaks) at 2000 bar, 20°C; from residues 1–5, 7, 9–10, 13–14, 21, 33–66, 68–87, 102 (66 peaks) at 2000 bar, 36°C.
The number of observable cross-peaks increased as pressure was increased to 2000 bar at 20°C (Fig. 1 ▶), showing increased unfolding of the chain. At 1000 bar, 32 cross-peaks were detected, out of which 26 peaks were assigned to residues in the β-domain (Fig. 2 ▶). The remaining six peaks belong to the α-domain, including the three N-terminal residues. Thr 33 is located at the end of the B-helix and Tyr 36 and Thr 38 in the loop linking the B-helix to the β-domain. By increasing the pressure to 1500 bar, 42 cross-peaks became observable; the 10 additional peaks are assigned to residues in the β-domain (Fig. 2 ▶). A further increase of pressure to 2000 bar led to the appearance of only two more signals assignable to residues in the β-domain (Fig. 2 ▶). Many of these additional peaks correspond to the weaker peaks observed at 1 bar and 20°C in a previous study (Redfield et al. 1999). In summary, at 2000 bar signals from 90% (36 residues) of the β-domain were observed, whereas only 8.4% (residues) of the signals were detected from the α-domain, showing that the unfolding was still not complete.
When we increased the temperature to 36°C at 2000 bar, 66 peaks became visible in the HSQC spectrum (Fig. 2 ▶). The increase in the number of peaks with increasing temperature can arise from thermal unfolding of the MG, but may also result from a sharpening of peaks due to decreased solvent viscosity or from an increase in the rate of interconversion within the family of conformers defining the MG (Kim et al. 1999). Cross-peaks from all the residues of the β-domain were observed at 36°C. Twenty-five cross-peaks were observed from the α-domain, corresponding to 30% unfolding of the α-domain. The majority (15 peaks) of the α-domain signals arises from loop regions, particularly those linking the α-domain to the β-domain. Ten peaks originate from helical regions; six from the A-helix (Lys 5, Glu 7, Ser 9, Gln 10, Lys 13, and Asp 14), two from the end of the B-helix (Thr 33, Ser 34), and two from the beginning of the C-helix (Thr 86, Asp 87), immediately adjacent to the loops that connect the α-domain to the β-domain. No cross-peaks were observed from the D- and C-terminal 310 helices or for the central parts of the B- and C-helices at 36°C at 2000 bar, indicating that these helices constitute the most stable part of the protein.
Additional cross-peaks are also observed when the temperature is lowered from 20°C to −18°C at 2000 bar (Fig. 1 ▶). At 1 bar, the number of peaks observed is found to decrease as the temperature is lowered from 20°C to 10°C (unpublished). The increase in the number of observed peaks at −18°C may be an indication of cold denaturation of the MG at high pressure. Unfortunately, site-specific assignments of these signals could not be made, because of the large low field shifts of all the cross-peaks in both 1H and 15N. However, the spectra in Figure 1 ▶ indicate possible structural differences between the heat- and cold-denaturated states.
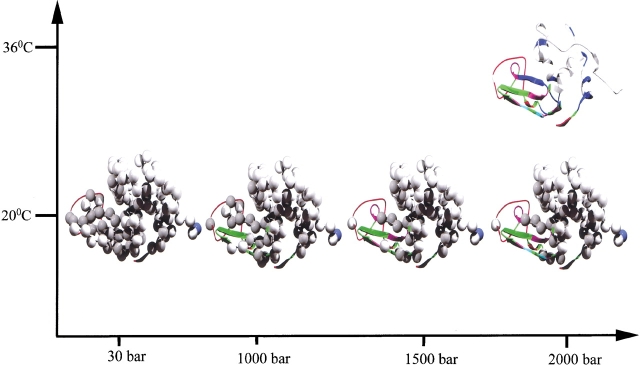
The unfolding events observed for the MG are shown schematically in a graphic illustration on the native structure of α-LA in Figure 3 ▶, in which progressive unfolding is shown by the ribbon representation of parts of the structure. It must be emphasized that the phenomenon is reversible with pressure and temperature and that all measurements were done in equilibrium defined by Equations 1 or 2.
Figure 3.
Schematic illustration of the gradual unfolding of the molten globule of all-Ala α-lactalbumin (pH 2) with increasing pressure, drawn on the native structure of α-lactalbumin. Ribbons show residues whose signals are visible in the 15N-1H HSQC spectra. The colors indicate the pressure at which the signal begins to appear (red: 30 bar; green: 1000 bar; violet: 1500 bar; cyan: 2000 bar; blue: 2000 bar at −18°C or 36°C). Balls represent residues invisible in the 15N-1H HSQC-spectra.
Pressure-induced changes in1H and 15N chemical shifts
Pressure-induced chemical shift changes of amide 1H and 15N chemical shifts can give information concerning the exposure of the amide group to the solvent water (Akasaka et al. 2001; Akasaka and Yamada 2001). The shifts were followed for those 1H/15N cross-peaks that appear throughout all the pressures (30–2000 bar) at 20°C (Fig. 4 ▶). The shifts were almost linear with pressure, with a slope varying from 1 * 10−4 to 5.5 * 10−4 ppm/bar for 15N and from 2 * 10−5 to 8 * 10−5 ppm/bar for 1H.
Figure 4.
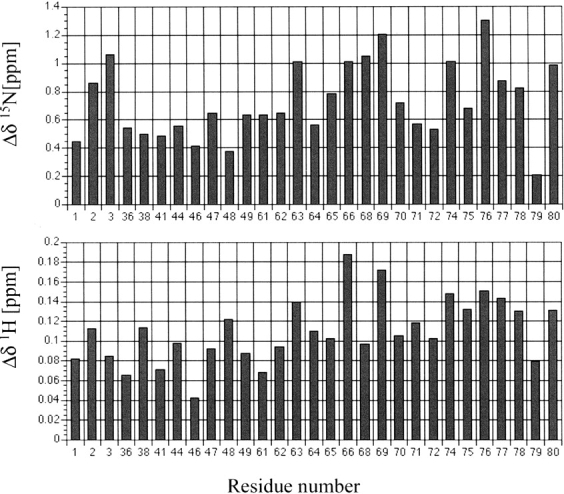
Histograms showing the pressure-induced 1H and 15N chemical shifts Δδ (2000–30 bar) for individual amide groups of all-Ala α-lactalbumin (pH 2).
Both 1H and 15N pressure shifts are all positive (low-field shifts), showing the typical character of a disordered polypeptide chain (Kamatari et al. 2001). The average shifts at 2000 bar were calculated to be 0.11 ppm and 0.73 ppm for 1H and 15N, respectively. The 1H shift values are nearly twice as large as typical values for internally hydrogen-bonded amides in folded proteins (average 0.059 ppm/2 kbar in BPTI and 0.051 ppm/2 kbar in hen lysozyme), but are close to those in a random coiled peptide (average 0.108 ppm/2 kbar in melittin in water) (Akasaka et al. 2001) and to those that are not internally hydrogen bonded in folded proteins (average 0.098 ppm/2 kbar in BPTI and 0.101 ppm/2 kbar in hen lysozyme) (Li et al. 1998; Akasaka et al. 1999; Kamatari et al. 2001). The larger low-field shifts for 1H can be explained by favorable hydrogen bonding interactions of the polypeptide amides with the solvent water molecules (Li et al. 1998; Akasaka et al. 1999). The amides observed at all pressures in the present experiment correspond to the 3 N-terminal residues and residues of the loop region of the β-domain. Thus, the results indicate that these amide groups are exposed to the solvent and hydrogen-bonded with water molecules within the average molten globule structure. This is consistent with the bipartite structure of the all-Ala α-LA MG in which the majority of β-domain residues are not involved in secondary structure.
Discussion
Although volumetric properties of MG states have been studied in some proteins (Nolting et al. 1993; Nolting and Sligar 1993; Kharakoz and Bychkova 1997), their relation to those of a more fully denatured state, U, is obscure. A molecular dynamics study of metmyoglobin (Floriano et al. 1998) suggested that at very high pressure (1.2 GPa) the MG transforms into a fully unfolded state U, inferring that the partial molar volume decreases in the transition MG → U. On the other hand, the volumetric equivalence of the molten globule and the unfolded states has been reported (Kobashigawa et al. 1999). In addition, a controversy exists over the cooperativity of the transition MG → U; although there are reports showing that the process MG → U is cooperative (Ptitsyn and Uversky 1994), noncooperative unfolding of the MG has also been discussed in the literature (Schulman and Kim 1996; Shimizu et al. 1993). In a previous study, urea-induced unfolding of the MG of all-Ala α-LA (Redfield et al. 1999) and wild-type α-LA (Schulman et al. 1997), noncooperative unfolding behavior of the MG was reported.
Our results show that the unfolding of the molten globule conformation of all-Ala α-LA occurs quite heterogeneously over the molecule. The noncooperative unfolding is not only limited to different domains, but also within domains themselves. Different helices unfold under different conditions. The resulting order of pressure stability, partly assisted by temperature, of the helices of all-Ala-α-LA are: D-helix ∼ 310-helix > C-helix ∼ B-helix > A-helix. Furthermore, differential unfolding was observed within a specific α-helix with residues at the beginning and end of the helix unfolding more readily. Overall, a highly noncooperative unfolding picture of the molten-globule state of all-Ala α-LA is presented as a result of pressure perturbation. This picture of heterogeneous unfolding qualitatively agrees with that from urea-induced unfolding of the MG against urea (Redfield et al. 1999), but differs in the details, which is not surprising because of the different mechanisms of destabilization in the two experiments. The results confirm the view that the structure of the MG is heterogeneous not only in the stability as probed by the interaction with urea, but heterogeneous also in partial molar volume, and consequently in atom packing and hydration. This view has been generally accepted, but so far without much evidence.
In general, pressure drives the equilibrium to increase the population of the lower volume conformer relative to the higher one (Weber and Drickamer 1983; Inoue et al. 2000). The experiment showed that pressure turned the MG (equation 1) into conformers with increasing disorder and hydration. This gives straightforward evidence that the partial molar volume of the MG state is significantly larger than that of the U state. It is also important to note that the conformational changes were brought about reversibly with pressure under equilibrium conditions. This means from equation 1 that the MG state at 30 bar coexists with other conformers with partial unfolding at various degrees shown in Figure 2 ▶. The results verify that the MG state consists of a mixture of variously unfolded conformers from the mostly folded to the nearly totally unfolded that differ in stability and partial molar volume. The populations of the latter conformers are small compared to the main MG conformers found at 30 bar, but their fractions become significant at higher pressure because of their smaller partial molar volumes.
The temperature dependence of the conformational stability of a protein, is given by a delicate balance between enthalpy and entropy terms, resulting in an expression for the Gibbs free energy as
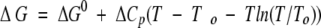 |
2 |
with the slope of the stability curve determined by ΔCp. Although the heat capacity change is a function of different interactions, the positive contribution leading to both heat and cold denaturation results from the hydrophobic effect (Privalov 1990; Blokzijil and Engberts 1993). More extensive denaturation of the MG state appeared to be attained when the temperature of the solution was increased or decreased from 20°C at 2000 bar. This indicates that ΔCp is positive, providing additional support to the notion that hydrophobic interactions play a crucial role in stabilizing the MG structure of all-Ala α-lactalbumin. By utilizing 19F-NMR, Bai et al. (2000) suggested that the stability of the molten globule state of α-LA was largely explained by hydrophobic and van der Waals interactions. Stabilization of a MG by hydrophobic interactions is well known for cytochrome c (Kamiyama et al. 1999). All these results support the general view that hydrophobic interactions are important for the stability of a molten globule.
Materials and methods
Sample preparation
15N-labeled all-Ala α-LA was expressed and purified as described previously (Peng et al. 1995; Schulman et al. 1995). The NMR sample contained 1 mM protein at pH 2 in a solution containing 20 mM maleic acid in a mixture of 90% H2O/10% D2O. As internal chemical shift standards, 1.25 mM 2,2-dimethyl-2-silapentane-5-sulfonate (DSS) and 0.625 mM dioxan were added.
High-pressure NMR apparatus
The cell (made of synthetic quartz) has an outer diameter of about 3 mm and an inner diameter of about 1 mm, and can hold a pressure up to 2 kbar. The cell was set in the 5 mm 1H-detected triple-resonance probe on an NMR spectrometer operating at 750 MHz for 1H (DMX-750, Bruker), and was connected through a Cu-Be separator to a kerosene line whose pressure was varied by a remote pressure pump. The pressure in the cell was kept at a desired value between 30 and 2000 bar during signal accumulation. More details about the principles of the high-resolution high-pressure NMR technique can be found elsewhere (Akasaka and Yamada 2001).
NMR measurements
Two-dimensional 1H-15N HSQC (Kay et al. 1992) spectra were measured at pressures between 30–2000 bar and temperatures from −18°C up to 36°C on a Bruker DMX-750 spectrometer operating at a 15N frequency of 76.02 MHz and a 1H frequency of 750.13 MHz. In the 15N dimension 256 increments covering 1441 Hz and in the 1H dimension 2048 points covering 8170 Hz were collected and zero-filled to obtain 512 and 4096 real data points. A total of 200 scans were collected per t1 increment. The data were transformed with a Gaussian window function in the 1H dimension and a sine-bell window function in the 15N dimension. Pressure-induced 1H and 15N chemical shifts between 30 and 2000 bar, Δδ (2000–30 bar), were evaluated for individual amide groups. The experimental error due to the digital resolution and line broadening is expected to be within 10%.
Acknowledgments
This research was supported by a Grant-in-Aid for Scientific Research from the Ministry of Education, Science, Sports and Culture of Japan (to K.A.) and by a fellowship from the Japan Society for the Promotion of Science (JSPS) (to M.W.L). C.R. acknowledges a Research Fellowship from the Biotechnology and Biological Sciences Research Council (U.K.). We thank P.S. Kim and M.A. Milhollen for providing the 15N-labelled all-Ala α- lactalbumin.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Abbreviations
α-LA, α-lactalbumin
all-Ala α-lactalbumin, α-LA with all eight cysteines mutated to alanine
MG, molten globule
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.0221303.
References
- AbouAiad, T., Becker, U., Biedenkap, R., Brengelmann, R., Elsebrock, R., Hinz, H.-J., and Stockhausen, M. 1997. Dielectric relaxation of aqueous solutions of ribonuclease A in the absence and presence of urea. Berichte Bunsen Gesellschaft Phys. Chem. Chem. Phys. 101 1921–1927. [Google Scholar]
- Akasaka, K. and Yamada, H. 2001. Methods in enzymology 338: Nuclear magnetic resonance of biological macromolecules part A. Academic Press, New York.
- Akasaka, K., Li, H., Yamada, H., Li, R., Thorensen, T., and Woodward, C.K. 1999. Pressure response of protein backbone structure. Pressure-induced amide 15N chemical shifts in BPTI. Protein Sci. 81946–1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akasaka, K., Li, H., Dubovskii, P., Kalbitzer, H.R., and Yamada, H. 2001. Structure, dynamics and function of biological macromolecules. IOS Press, Amsterdam, The Netherlands.
- Alexandrescu, A.T., Evans, P.A., Pitkeathly, M., Baum, J., and Dobson, C.M. 1993. Structure and dynamics of the acid-denatured molten globule state of α-lactalbumin: A two-dimensional NMR study. Biochemistry 32 1707–1718. [DOI] [PubMed] [Google Scholar]
- Bai, P., Luo, L., and Peng, Z.-Y. 2000. Side-chain accessibility and dynamics in the molten globule state of α-lactalbumin: A 19F-NMR study. Biochemistry 39 372–380. [DOI] [PubMed] [Google Scholar]
- Baum, J., Dobson, C.M., Evans, P.A., and Hanley, C. 1989. Characterization of a partly folded protein by NMR methods: Studies on the molten globule state of guinea pig α-lactalbumin. Biochemistry 28 7–13. [DOI] [PubMed] [Google Scholar]
- Blokzijil, W. and Engberts, J.B.F.N. 1993. Hydrophobe Effekte—Ansichten und Tatsachen. Angew. Chem. 105 1610–1648. [Google Scholar]
- Bychkova, V.E. and Ptitsyn, O.B. 1993. The molten globule in vitro and in vivo. Chemtracts Biochem. Mol. Biol. 4 133–163. [Google Scholar]
- Chalikian, T.V., Gindikin, V.S., and Breslauer, K.J. 1995. Volumetric characterizations of the native, molten globule and unfolded states of cytochrome c at acidic pH. J. Mol. Biol. 250 291–306. [DOI] [PubMed] [Google Scholar]
- Chattopadhyay, K. and Mazumdar, S. 2000. Structural and conformational stability of horseradish peroxidase: Effect of temperature and pH. Biochemistry 39 263–270. [DOI] [PubMed] [Google Scholar]
- Chyan, C.-L., Wormland, C., Dobson, C.M., Evans, P.A., and Baum, J. 1993. Structure and stability of the molten globule state of guinea-pig α-lactalbumin: A hydrogen exchange study. Biochemistry 32 5681–5691. [DOI] [PubMed] [Google Scholar]
- Dolgikh, D.A., Gilmanshin, R.I., Brazhnikov, V.E., Bychkova, V.E., Semisotnov, G.V., Venyaminov, S.Y., and Ptitsyn, O.B. 1981. α-lactalbumin: Compact state with fluctuating tertiary structure? FEBS Lett. 136 311–315. [DOI] [PubMed] [Google Scholar]
- Floriano, W.B., Nascimento, M.A.C., Domont, G.B., and Goddard III, W.A. 1998. Effects of pressure on the structure of metmyoglobin: Molecular dynamics predictions for pressure unfolding through a molten globule intermediate. Protein Sci. 7 2301–2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gursky, O. and Atkinson, D. 1996. High- and low-temperature unfolding of human high-density apolipoprotein A-2. Protein Sci. 5 1874–1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue, K., Yamada, H., Akasaka, K., Herrmann, C., Kremer, W., Maurer, T., Doeker, R., and Kalbitzer, H.R. 2000. Pressure-induced local unfolding of the Ras-binding domain of RalGEF. Nat. Struct. Biol. 7 547–550. [DOI] [PubMed] [Google Scholar]
- Jonas, J., Ballard, L., and Nash, D. 1998. High-resolution, high-pressure NMR studies of proteins. Biophys. J. 75 445–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamatari, Y.O., Konno, T., Kataoka, M., and Akasaka, K. 1998. The methanol-induced transition and the expanded helical conformation in hen lysozyme. Protein Sci. 7 681–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamatari, Y.O., Ohji, S., Konno, T., Seki, Y., Soda, K., Kataoka, M., and Akasaka, K. 1999. The compact and expanded denatured conformations of apomyoglobin in the methanol–water solvent. Protein Sci. 8 873–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamatari, Y.O., Yamada, H., Akasaka,, K., Jones, J.A., Dobson, C.M., and Smith, L.J. 2001. The response of native and denatured hen lysozyme to high pressure studied by 15N/1H NMR spectroscopy. Eur. J. Biochem. 268 1782–1793. [PubMed] [Google Scholar]
- Kamiyama, T., Sadahide, Y., Nogusa, Y., and Gekko, K. 1999. Polyol-induced molten globule of cytochrome c: An evidence for stabilization by hydrophobic interaction. Biochim. Biophys. Acta 1434 44–57. [DOI] [PubMed] [Google Scholar]
- Kataoka, M., Kuwajima, K., Tokunaga, F., and Goto, Y. 1997. Structural characterization of the molten globule of α-lactalbumin by solution x-ray scattering. Protein Sci. 6 442–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay, L.E., Keifer, P., and Saarinen, T.J. 1992. Pure absorption gradient enhanced heteronuclear single quantum correlation spectroscopy with improved sensitivity. J. Am. Chem. Soc. 114 10663–10665. [Google Scholar]
- Kharakoz, D.P. 1997. Partial volumes and compressibilities of extended polypeptide chains in aqueous solution: Additivity scheme and implication of protein unfolding at normal and high pressure. Biochemistry 36 10276–10285. [DOI] [PubMed] [Google Scholar]
- Kharakoz, D.P. and Bychkova, V.E. 1997. Molten globule of human α-lactalbumin: Hydration, density, and compressibility of the interior. Biochemistry 36 1882–1890. [DOI] [PubMed] [Google Scholar]
- Kim, S., Bracken, C., and Baum, J. 1999. Characterization of millisecond time-scale dynamics in the molten globule state of α-lactalbumin by NMR. J. Mol. Biol. 294 551–560. [DOI] [PubMed] [Google Scholar]
- Kobashigawa, Y., Sakurai, M., and Nitta, K. 1999. Effect of hydrostatic pressure on unfolding of α-lactalbumin: Volumetric equivalence of the molten globule and unfolded state. Protein Sci. 8 2765–2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwajima, K., Mitani, M., and Sugai, S. 1989. Characterization of the critical state in protein folding. J. Mol. Biol. 206 547–561. [DOI] [PubMed] [Google Scholar]
- Kitahara, R., Yamahada, H., Akasaka, K., and Wright, P.E. 2002. High pressure NMR reveals that apomyoglobin is an equilibrium mixture from the native to the unfolded. J. Mol. Biol. 320 311–319. [DOI] [PubMed] [Google Scholar]
- Lassalle, M.W., Yamada, H., and Akasaka, K. 2000. The pressure-temperature free energy-landscape of Staphylococcal nuclease monitored by 1H NMR. J. Mol. Biol. 298 293–302. [DOI] [PubMed] [Google Scholar]
- Li, H., Yamada, H., and Akasaka, K. 1998. Effect of pressure on individual hydrogen bonds in proteins. Basic pancreatic trypsin inhibitor. Biochemistry 37 1167–1173. [DOI] [PubMed] [Google Scholar]
- Maldonado, S., Jimenez, M.A., Langdon, G.M., and Sancho, J. 1998. Cooperative stabilization of a molten globule apoflavodoxin fragment. Biochemistry 37 10589–10596. [DOI] [PubMed] [Google Scholar]
- Nolting, B. and Sligar, G.S. 1993. Adiabatic compressibility of molten globule. Biochemistry 32 12319–12323. [DOI] [PubMed] [Google Scholar]
- Nolting, B., Jiang, M., and Sligar, S.G.J. 1993. The acidic molten globule state of α-lactalbumin probed by sound velocity. J. Am. Chem. Soc. 115 9879–9882. [Google Scholar]
- Ohgushi, M. and Wada, A. 1983. “Molten-globule state”: A compact form of globular proteins with mobile side-chains. FEBS Lett. 164 21–24. [DOI] [PubMed] [Google Scholar]
- Peng, Z.-Y., Wu, L.C., and Kim, P.S. 1995. Local structure preferences in the α-lactalbumin molten globule. Biochemistry 34 3248–3252. [DOI] [PubMed] [Google Scholar]
- Privalov, P.L. 1990. Cold denaturation of proteins. Crit. Rev. Biochem. Mol. Biol. 25 281–303. [DOI] [PubMed] [Google Scholar]
- Ptitsyn, O.B. 1991. How does protein synthesis give rise to the 3D-structure? FEBS Lett. 285 176–181. [DOI] [PubMed] [Google Scholar]
- ———. 1995. Structures of folding intermediates. Curr. Opin. Struct. Biol. 5 74–78. [DOI] [PubMed] [Google Scholar]
- Ptitsyn, O.B. and Uversky, V.N. 1994. The molten globule is a third thermodynamical state of protein molecules. FEBS Lett. 341 15–18. [DOI] [PubMed] [Google Scholar]
- Redfield, C., Schulman, B.A., Milhollen, M.A., Kim, P.S., and Dobson, C.M. 1999. α-Lactalbumin forms a compact molten globule in the absence of disulfide bonds. Nat. Struct. Biol. 6 948–952. [DOI] [PubMed] [Google Scholar]
- Ruan, K., Lange, R., Bec, N., and Balny, C. 1997. A stable denatured state of trypsin induced by high hydrostatic pressure. Biochem. Biophys. Res. Commun. 239 150–154. [DOI] [PubMed] [Google Scholar]
- Schulman, B.A. and Kim, P.S. 1996. Proline scanning mutagenesis of a molten globule reveals non-cooperative formation of a proteins overall topology. Nat. Struct. Biol. 3 682–687. [DOI] [PubMed] [Google Scholar]
- Schulman, B.A., Redfield, C., Peng, Z.-Y., Dobson, C.M, and Kim, P.S. 1995. Different subdomains are most protected from hydrogen exchange in the molten globule and native states of human α-lactalbumin. J. Mol. Biol. 253 651–657. [DOI] [PubMed] [Google Scholar]
- Schulman, B.A., Kim, P.S., Dobson, C.M., and Redfield, C. 1997. A residue-specific NMR view of the non-cooperative unfolding of a molten globule. Nat. Struct. Biol. 4 630–634. [DOI] [PubMed] [Google Scholar]
- Shimizu, A., Ikeguchi, M., and Sugai, S. 1993. Unfolding of the molten globule state of α-lactalbumin studied by 1H NMR. Biochemistry 32 13198–13203. [DOI] [PubMed] [Google Scholar]
- Troullier, A., Reinstaedler, D., Dupont, Y., Naumann, D., and Forge, V. 2000. Transient non-native secondary structures during the refolding of α-lactalbumin detected by infrared spectroscopy. Nat. Struct. Biol. 7 78–85. [DOI] [PubMed] [Google Scholar]
- Weber, G. and Drickamer, H.G. 1983. The effect of high pressure upon proteins and other biomolecules. Q. Rev. Biophys. 16 89–112. [DOI] [PubMed] [Google Scholar]
- Wu, J.-W. and Wang, Z.-X. 1999. New evidence for the denaturant binding model. Protein Sci. 8 2090–2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, L.C., Peng, Z.Y., and Kim, P.S. 1995. Bipartite structure of the α-lactalbumin molten globule. Nat. Struct. Biol. 4 281–286. [DOI] [PubMed] [Google Scholar]
- Zhang, J., Peng, X., Jonas, A., and Jonas, J. 1995. NMR study of the cold, heat, and pressure unfolding of ribonuclease A. Biochemistry 34 8631–8641. [DOI] [PubMed] [Google Scholar]